Abstract
Lung cancer is the most common cause of cancer-associated mortality. MicroRNAs (miRNAs), as oncogenes or tumor suppressor genes, serve crucial roles not only in tumorigenesis, but also in tumor invasion and metastasis. Although miRNA-let-7a (let-7a) has been reported to suppress cell growth in multiple cancer types, the biological mechanisms of let-7a in lung adenocarcinoma are yet to be fully elucidated. In the present study, the molecular roles of let-7a in lung adenocarcinoma were investigated by detecting its expression in lung adenocarcinoma tissues and exploring its roles in the regulation of lung cancer cell proliferation. Let-7a expression was identified to be downregulated in lung adenocarcinoma tissues compared with normal tissues. Overexpression of let-7a effectively suppressed cancer cell proliferation, migration and invasion in H1299 and A549 cells. Let-7a also induced cell apoptosis and cell cycle arrest. Furthermore, let-7a significantly inhibited cell growth by directly regulating cyclin D1 signals. This novel regulatory mechanism of let-7a in lung adenocarcinoma provides possible avenues for future targeted therapies of lung cancer.
Keywords: let-7a, cyclin D1, non-small cell lung cancer, cell proliferation, metastasis
Introduction
The increasing rate of cancer morbidity has become a serious public health issue worldwide (1). It has been reported that there were 14.1 million newly diagnosed cancer cases, and 8.2 million cases of cancer-associated mortality in 2012 (2). Lung cancer is among the most common malignant tumors and the leading cause of cancer-associated cases of mortality worldwide (3). In lung cancer, non-small cell lung cancer (NSCLC) accounts for ~80% of all clinical cases, with a poor prognosis and a 5-year survival rate as low as 15% (4). Treatment of lung cancer usually includes a combination of surgery, radiation therapy and chemotherapy. Although considerable advances have been made in lung cancer research, there is still an urgent need to improve understanding of the molecular pathogenesis of lung cancer, particularly to identify novel therapeutic targets (5).
MicroRNAs (miRNAs) are a class of highly conserved non-coding RNAs of ~20–22 nucleotides in length (6). The regulatory roles of miRNAs in gene expression are associated with their pairing to the 3′untranslated region (3′UTR) of their target mRNAs (7–9). Following processing by the consecutive actions of Drosha and Dicer ribonucleases, miRNAs are subsequently loaded onto RNA-induced silencing complexes to interact with their target mRNAs and induce post-transcriptional silencing (2,9,10). Numerous studies have indicated that miRNAs perform their functions by regulating various biological processes, including cell proliferation, differentiation, angiogenesis and apoptosis (2,11–13). MiRNAs also serve as oncogenes or tumor suppressor genes. Let-7a, as a tumor-suppressive miRNA, has been identified to be deregulated in multiple types of malignant cells. For instance, overexpression of let-7a was demonstrated to suppress the proliferation, migration and invasion of gastric cancer cells by downregulating the expression of pyruvate kinase muscle isozyme M2 (11,14).
As a dysplastic disease, cancer formation is largely attributed to the deregulation of cell proliferation, which is strictly controlled by checkpoints of the cell cycle. G1/S and G2/M are the classic cell cycle checkpoints (11,15). As a member of the cyclin family, cyclin D1 is an important regulator of cell proliferation. Cyclin D1 reaches a peak level at the G1 stage, indicating that it is involved in the checkpoint of G1/S. Therefore, overexpression of cyclin D1 may lead to transition through the G1/S checkpoint and promotion of cell proliferation, which may eventually lead to the formation of cancer (16).
Although it has been observed that certain miRNAs are unconventionally expressed in lung cancer and associated with poor outcomes, the role of let-7a in lung cancer has not yet been fully elucidated (17). The present study aimed to investigate the effects of let-7a on cell proliferation, apoptosis, migration and invasion in A549 and H1299 cells.
Materials and methods
Lung adenocarcinoma tissues
The present study was conducted between August 1, 2014 and July 31, 2015 at the Inpatient Department of Medical Oncology, Yantai Shan Hospital, The Teaching Hospital of Binzhou Medical University (Yantai, China). The experiments were performed according to the relevant guidelines of the Code of Ethics of the World Medical Association for experiments involving humans and the Medical Ethics Committee of Binzhou Medical University (18).
A total of 20 patients (10 males and 10 females), who were pathologically diagnosed with lung adenocarcinoma for the first time and had not yet received chemotherapy, were included in the present study. Fresh lung adenocarcinoma and control tissues were obtained from the patients who underwent surgery. Written informed consent was obtained from all patients prior to the collection of lung tissue samples. The control and lung adenocarcinoma tissues were collected from the same patients, and control samples were normal adjacent control tissues.
Quantitative polymerase chain reaction (qPCR)
Lung adenocarcinoma cells were incubated with let-7a-mimics or let-7a-inhibitor and harvested 48 h after miRNA treatment. Small RNA was isolated using RNAiso for Small RNA reagent (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed as previously described (14). The primers used to amplify let-7a (Shanghai GenePharm Co., Ltd., Shanghai, China) were as follows: Forward, 5′-ACACTCCAGCTGGGTGAGGTAGTAGGTTGT-3′ and reverse, 5′-AACATGTACAGTCCATGGATG-3′. Human 5S rRNA served as the positive control. The primers used to amplify 5S rRNA (GenePharm Co., Ltd.) were as follows: Forward, 5′-GCCATACCACCCTGAACG-3′ and reverse, 5′-AACATGTACAGTCCATGGATG-3′. Total RNA was isolated using TRIzol reagent (Takara Biotechnology Co., Ltd.). The primers used for amplifying cyclin D1 (GenePharm Co., Ltd.) were as follows: Forward, 5′-CTGGCCATGAACTACCTGGA-3′ and reverse, 5′-GTCACACTTGATCACTCTGG-3′. qPCR was performed on an RG3000 system (Qiagen GmbH, Hilden, Germany) under the following reaction conditions: Initial denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec, annealing at 60°C for 20 sec and extension at 72°C for 30 sec. GAPDH cDNA served as the positive control (18). The primers used for amplifying GAPDH (Shanghai GenePharm, Co., Ltd.) were as follows: Forward, 5′-GTCTTCACCACCATGGAGAAGG-3′ and reverse, 5′-GCCTGCTTCACCACCTTCTTGA-3′.
Construction of cyclin D1-3′UTR GFP plasmid
The sequence of cyclin D1-3′ UTR was obtained from GenBank. The primers were designed using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) and then synthesized by Shanghai GenePharma Co., Ltd. The primers used to amplify cyclin D1-3′ UTR were as follows: forward, 5′-TGCTCTAGATGAATTCTTATCCCCTGCCC-3′ and reverse, 5′-CGCGGATCCAAGAGAAGAGGGACACAGCC-3′. The amplification template was human genomic DNA. Then, cyclin D1-3′ UTR was inserted into the pcDNA3.1-GFP-neo (+) expression vector.
Cell culture and transfection
Lung adenocarcinoma cell lines (A549 and H1299) were obtained from the Shanghai Institute of Cell Biology (Shanghai, China). HBE 135E6E7 cells were obtained from the American Type Culture Collection (human bronchus epithelial; ATCC® CRL-2741; Manassas, VA, USA). The cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. Cells (2×105) were seeded into 6-well plates. Let-7a mimics, let-7a inhibitors, negative controls and si-cyclin D1 were synthesized by Shanghai GenePharma Co., Ltd. The target sequence of si-cyclin D1 was as follows: Sense, 5′-CAAACAGAUCAUCCGCAA-3′ and antisense, 5′-UUUGCGGAUGAUCUGUU-3′. Transfection was performed in triplicate at ~60% confluence using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol (19).
MTT assay
Cell proliferation assays were conducted using a modified colorimetric MTT assay as previously described (20). All procedures were repeated three times. Cell colony formation rate was assessed using a plate colony formation assay. The plate was gently washed and stained with crystal violet. Then, the number and size of colonies was analyzed.
Apoptosis assays
An apoptosis assay was performed 48 h after the oligonucleotides were transfected into lung adenocarcinoma cells. The assay was performed using Annexin V-FITC/PI (BD Pharmingen; BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's protocol (19).
Cell cycle analysis by flow cytometry
A cell cycle assay was performed 48 h after transfection. The cells were collected using a Cell Cycle Detection kit (Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China) according to the manufacturer's protocols and counted by flow cytometry (21).
Western blot analysis
Western blot analysis was performed as previously described (22). The antibodies used were as follows: Rabbit antibodies against GAPDH (1:800; cat. no. AP0063), Rb (1:800; cat. no. BS1311), p-Rb (1:800; cat. no. BS4164), Bcl-2 (1:800; cat. no. BS1511), Bax (1:800; cat. no. BS2538; all from Bioworld Technology, Inc., St. Louis Park, MN, USA) and caspase-9 (1:1,000; cat. no. 9502), caspase-8 (1:1,000; cat. no. 9748) and caspase-3 (1:1,000; cat. no. 9662; all from Cell Signaling Technology, Inc., Danvers, MA, USA). GAPDH was used as an internal reference.
Cell migration and invasion assays
An invasion assay was performed using a modified two-chamber plate with a pore size of 8.0 µm (23) as previously described (14). The Transwell migration assay was conducted according to the same protocol as the invasion assay, with the exception that the cell suspension was added into the upper chamber directly, without Matrigel.
Statistical analysis
SPSS 22.0 software (IBM Corp., Armonk, NY, USA) was used to analyze the significance of all results. One-way analysis of variance (ANOVA) was used to analyze the differences among three or more groups. A post-hoc test of ANOVA was conducted by performing a Tukey's test. Correlations were calculated with Spearman's rank correlation coefficiant. Group means were compared using an unpaired, two-sided, Student's t-test. Wilcoxen signed-rank test was used to compare the expression of let-7a and cyclin D1 in para-carcinoma and carcinoma tissues. Array data of cyclin D1 were downloaded from data link(s): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13213. Overall survival was determined using Kaplan-Meier survival analysis by a log-rank test. P<0.05 was considered to indicate a statistically significant difference (11,23). All experiments were performed in triplicate, and all data are expressed as the mean ± standard deviation.
Results
Let-7a is downregulated in human lung adenocarcinoma tissues
Although let-7a has previously been investigated in other types of cancer (24), the molecular mechanism of let-7a in lung adenocarcinoma remains unclear. In the present study, we first collected the clinicopathological features of patients with lung adenocarcinoma (Table I). Lung cancer tumor-node-metastasis (TNM) staging was based on the 8th edition of the Union for International Cancer Control (UICC) (25). The number of nuclear fission images or proliferation index, necrosis range, invasion status and other parameters were used to determine the lung cancer grade according to the tumor structure and cell atypia in H&E stained sections (26–29). Then, let-7a expression was analyzed in lung adenocarcinoma tissues to investigate the roles of let-7a in lung cancer. The current results indicated that let-7a levels were significantly lower in lung adenocarcinoma tissues (n=20) compared with para-carcinoma tissues (2 cm from the para-carcinoma tissues, n=20, Fig. 1A; P<0.001). The current results indicated that let-7a is involved in the progression of lung adenocarcinoma.
Table I.
The clinicopathological features of the patients with lung cancer.
| Sample no. | Sex | Age (years) | Tumor stage | TNM stage | Tumor grade |
|---|---|---|---|---|---|
| 1 | Female | 61 | IIA | T2aN1M0 | G1 |
| 2 | Female | 36 | IA | T1aN0M0 | G1 |
| 3 | Female | 67 | IA | T1aN0M0 | G1 |
| 4 | Female | 64 | IB | T2aN0M0 | G1 |
| 5 | Female | 48 | IA | T1aN0M0 | G1 |
| 6 | Female | 61 | IA | T1aN0M0 | G1 |
| 7 | Female | 71 | IA | T1aN0M0 | G1 |
| 8 | Female | 42 | IB | T2aN0M0 | G1 |
| 9 | Female | 61 | IA | T1aN0M0 | G1 |
| 10 | Female | 44 | IA | T1aN0M0 | G1 |
| 11 | Male | 56 | IA | T1aN0M0 | G1 |
| 12 | Male | 69 | IA | T1aN0M0 | G1 |
| 13 | Male | 59 | IA | T1aN0M0 | G1 |
| 14 | Male | 71 | IA | T1aN0M0 | G1 |
| 15 | Male | 68 | IA | T1aN0M0 | G1 |
| 16 | Male | 61 | IB | T2aN0M0 | G1 |
| 17 | Male | 53 | IA | T1aN0M0 | G1 |
| 18 | Male | 64 | IIA | T1bN0M0 | G1 |
| 19 | Male | 64 | IA | T1aN0M0 | G1 |
| 20 | Male | 75 | IA | T1aN0M0 | G1 |
TNM, tumor-node-metastasis.
Figure 1.
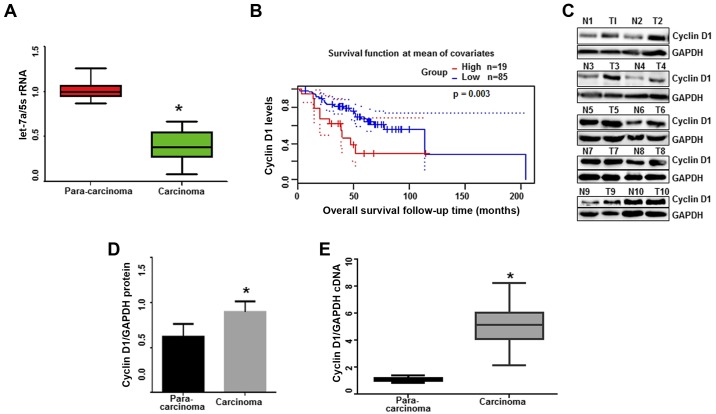
Let-7a is markedly downregulated in human lung adenocarcinoma tissues. (A) Let-7a expression in lung adenocarcinoma tissues (n=20) and para-carcinoma tissues (n=20) was revealed by qRT-PCR. *P<0.001, vs. para-carcinoma. (B) Overall survival was determined through Kaplan-Meier survival analysis. The low survival rate of cancer patients was related to high cyclin D1 expression. Kaplan-Meier survival analysis was performed to determine the significance of cyclin D1 expression in the outcome of lung cancer patients from the GSE13213 dataset. (C) We detected the expression of cyclin D1 protein in lung adenocarcinoma, and found that the expression of cyclin D1 was considerably higher in lung adenocarcinoma tissues (n=10) than that in para-carcinoma tissues. (D) The mean protein expression levels of cyclin D1 in tumor and normal para-carcinoma tissues. *P<0.001. (E) The mRNA levels of cyclin D1 in para-carcinoma and carcinoma tissue samples obtained from the 20 patients with lung cancer, *P<0.0001.
Kaplan-Meier survival analysis was performed to determine the significance of cycin D1 expression in affecting the outcome of lung cancer patients from the GSE13213 dataset. The poor survival time of patients was not significantly correlated with age (P=0.360) or sex (P=0.278). Notably, patients with high cyclin D1 levels exhibited short survival time (P=0.003; Fig. 1B) through a log-rank test analysis, suggesting that higher cyclin D1 levels increase the risk of lung adenocarcinoma. Then, the expression of cyclin D1 was evaluated in lung adenocarcinoma tissues. The results indicated that the expression of cyclin D1 was considerably higher in lung adenocarcinoma tissues (n=10) compared with para-carcinoma tissues (2 cm from the para-carcinoma tissues, n=10, Fig. 1C). Subsequently, we assessed the protein and mRNA expression levels of cyclin D1 in para-carcinoma and carcinoma tissue samples, and our results revealed that cyclin D1 levels were significantly higher in lung adenocarcinoma tissues (n=20) than those in para-carcinoma tissues (Fig. 1D and E; P<0.0001). These results indicated that lower levels of let-7a and higher levels of cyclin D1 may be associated with the development of lung adenocarcinoma.
Let-7a is downregulated in lung adenocarcinoma cell lines
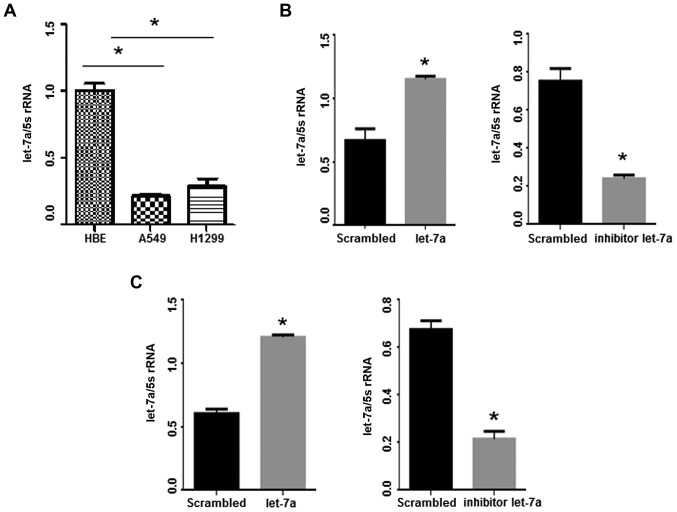
Let-7a levels were detected in lung adenocarcinoma cell lines (A549 and H1299) and control human bronchus epithelial (HBE) cell line by qPCR. Notably, let-7a levels were reduced in the two lung adenocarcinoma cell lines compared with the HBE cells (Fig. 2A; P<0.001). Let-7a-mimic oligo treatment led to significant upregulation of let-7a compared with the control group (P<0.05). Conversely, the expression of let-7a was significantly reduced in lung adenocarcinoma cell lines following let-7a-inhibitor transfection (P<0.05; Fig. 2B and C).
Figure 2.
Let-7a is markedly downregulated in A549 and H1299 cells. (A) Let-7a expression in A549, H1299 and control HBE cells was revealed by qRT-PCR, *P<0.001. (B and C) The levels of let-7a were detected after let-7a-mimic and let-7a-inhibitor transfection in A549 and H1299 cells. *P<0.05, vs. the scrambled control group.
Let-7a directly targets cyclin D1
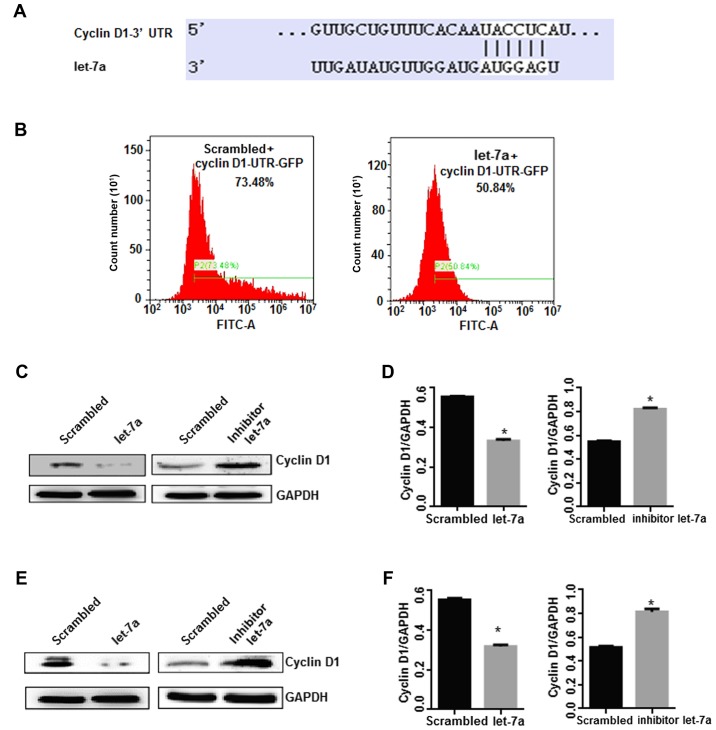
To elucidate the molecular mechanisms of let-7a in the suppression of the growth of lung adenocarcinoma cells, the putative target gene of let-7a was identified. It was indicated that cyclin D1 was a target gene of let-7a with high probability using TargetScan software online (Fig. 3A; http://www.targetscan.org/vert_71/). Then, a cyclin D1-3′ UTR GFP plasmid was constructed to transfect cells with let-7a. The results indicated that GFP fluorescence intensity and positive rate decreased significantly in let-7a-treated cells compared with the control treatment (P<0.05; Fig. 3B), indicating that cyclin D1-3′ UTR was directly targeted by let-7a. Next, the effects of let-7a on cyclin D1 expression in lung cancer cell lines were investigated by western blot analysis. Notably, the expression of cyclin D1 was significantly reduced in lung adenocarcinoma cell lines following overexpression of let-7a (P<0.05). Conversely, the let-7a inhibitor resulted in the significant upregulation of cyclin D1 compared with the control group (P<0.05; Fig. 3C-F). Cyclin D1 is a positive regulator of cell cycle progression (11), and these results indicated that let-7a suppresses lung cancer cell growth by regulating the pathway of cyclin D1.
Figure 3.
Let-7a directly regulates cyclin D1 expression. (A) The 3′ UTR of the cyclin D1 gene was targeted by let-7a using TargetScan software. (B) Cyclin D1−3′UTR GFP plasmid was constructed to transfect cells with let-7a. The results revealed that GFP fluorescence intensity and the positive rate were decreased in A549 cells. P<0.05 vs. the scrambled control. (C and D) Both upregulation and downregulation experiments of let-7a were performed to validate that cyclin D1 was targeted by let-7a in A549 cells. *P<0.01 vs. the scrambled control. (E and F) The effects of let-7a-mimics and let-7a-inhibitor on the expression of cyclin D1 were analyzed in H1299 cells. *P<0.01 vs. the scrambled control. GAPDH was used as an internal loading control.
Let-7a inhibits cellular proliferation and colony formation
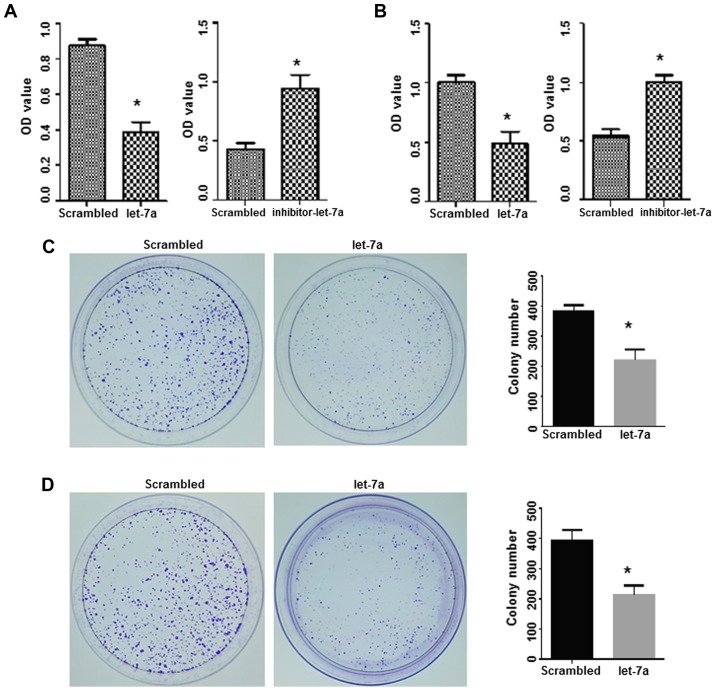
To further assess the potential inhibitory roles of let-7a in lung adenocarcinoma, the effects of let-7a on cell proliferation and colony formation were studied. An MTT assay indicated that the proliferation of lung adenocarcinoma cells in the let-7a-treated groups was significantly decreased compared with the scrambled oligo-treated cells (P<0.05). In contrast, the let-7a-inhibitor caused a significant increase in cell proliferation compared with the scrambled oligo treatment (P<0.05; Fig. 4A and B).
Figure 4.
Upregulation of let-7a suppresses cell proliferation and colony formation. (A and B) The MTT assay assessed the viability of A549 and H1299 cells that were transfected with let-7a-mimics and let-7a-inhibitor, respectively. *P<0.05 vs. the scrambled control. (C) The effects of let-7a on the colony formation of A549 cells. *P<0.01 vs. the scrambled control. (D) The effects of let-7a on the colony formation of H1299 cells. *P<0.01 vs. scrambled control.
A colony formation assay was performed to evaluate the long-term effect of let-7a on cell proliferation. Compared with the scrambled control treatment, a smaller size and a decreased number of clones were observed in the let-7a-treated cells, suggesting that the upregulation of let-7a significantly suppressed the colony formation ability of A549 and H1299 cells (Fig. 4C and D).
Let-7a regulates cell apoptosis and apoptosis-associated proteins
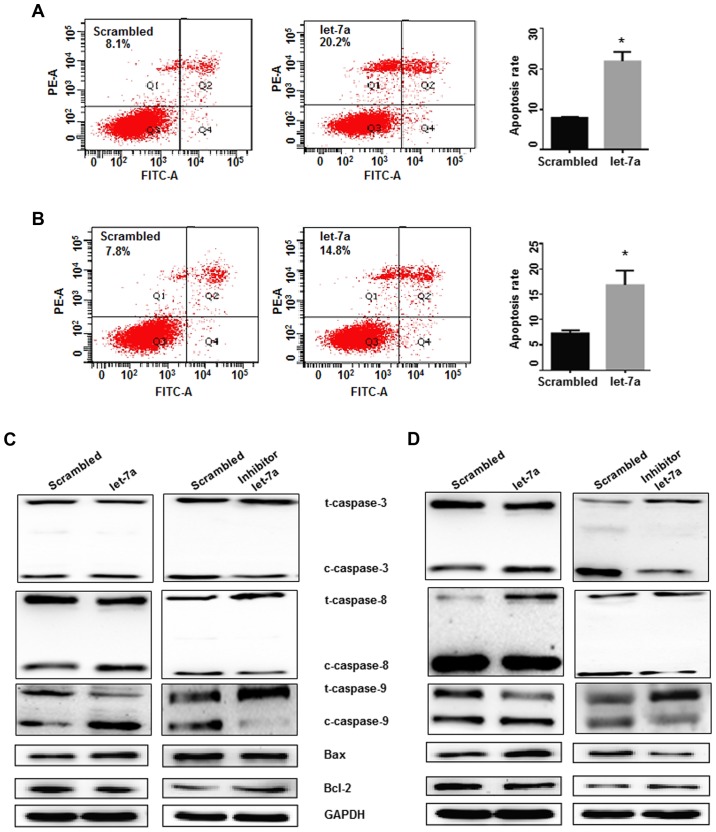
Cell apoptosis was detected following transfection of lung adenocarcinoma cells with let-7a mimics and controls. Cell apoptosis was significantly increased in let-7a-treated A549 and H1299 cells, indicating that let-7a overexpression induced lung adenocarcinoma cell apoptosis (P<0.05; Fig. 5A and B). To further characterize the possible mechanisms of let-7a in inducing cell apoptosis, the apoptotic genes, Bcl-2 (as an anti-apoptotic protein) and Bax (as a pro-apoptotic protein), were studied. It was determined that let-7a treatment significantly downregulated Bcl-2 expression and upregulated the expression of Bax and cleaved caspase-3, −8 and −9, compared with the control treatment in A549 and H1299 cells. In contrast, the opposite trends for these proteins were observed in cells transfected with the let-7a-inhibitor (Fig. 5C and D). These findings demonstrated that the Bcl-2/Bax pathways and the activation of caspase-3, −8 and −9 were constitutively downregulated by let-7a overexpression, and that the Bcl-2 family proteins are involved in the apoptosis induced by let-7a.
Figure 5.
The effects of let-7a on cell apoptosis and gene expression. (A) Cell apoptosis in A549 cells was determined by flow cytometry. *P<0.05. (B) The effects of let-7a on apoptosis were analyzed in H1299 cells. *P<0.05. (C) Let-7a significantly downregulated Bcl-2 expression and upregulated the expression of Bax, cleaved-caspase-3, −8 and −9, compared with the scrambled control treatment in A549 cells. The let-7a-inhibitor increased the Bcl-2 levels and reduced Bax, and decreased cleaved-caspase-3, −8 and −9. (D) The expression of Bcl-2, Bax and cleaved-caspase-3, −8 and −9 was detected in the H1299 cell line. GAPDH was used as an internal control.
Let-7a functions as an inhibitor of cell cycle progression
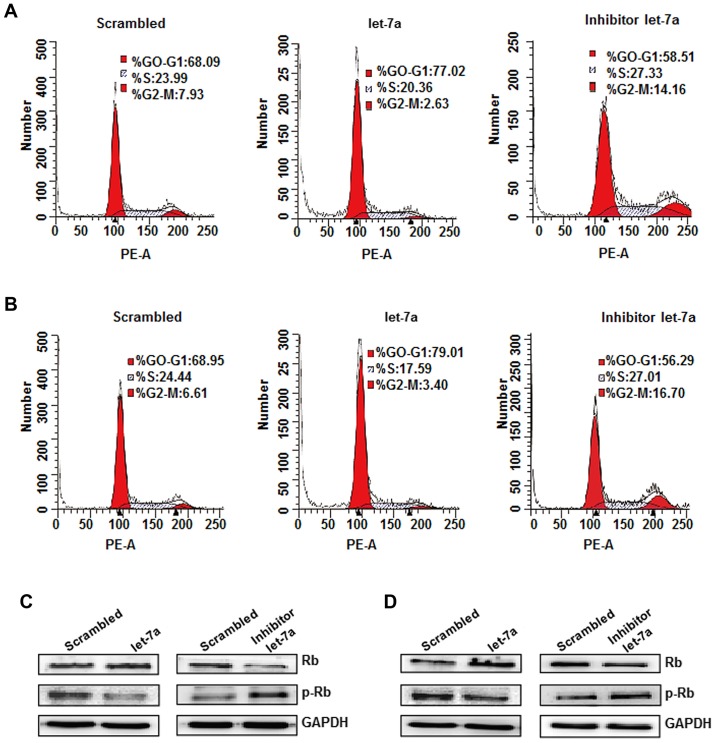
The effects of let-7a on cell cycle regulation were evaluated by flow cytometric analysis (30). The results indicated that the distribution of the cell cycle in A549 and H1299 cells was notably affected by let-7a overexpression compared with the scrambled group. The cell cycle profile indicated that 77.02% of A549 cells and 79.01% of H1299 cells were arrested at the G0/G1 phase at 48 h after let-7a treatment, compared with 68.09% of NC-transduced A549 cells and 68.95% of NC-transduced H1299 cells. However, the let-7a inhibitor-transduced cells exhibited a smaller percentage of arrest at the G0/G1 phase and increased cells inhibited in the G2/M phase compared with let-7a treatment (Fig. 6A and B). These experiments demonstrated that let-7a exerts an inhibitory effect on cell cycle progression. To investigate the potential molecular mechanism of let-7a in cell cycle arrest, the expression of cell cycle molecules, namely cyclin D1, Rb and p-Rb, was detected. Let-7a treatment significantly decreased the expression of cyclin D1 and p-Rb, but increased the Rb protein expression compared with the controls. The opposite trends were observed in let-7a inhibitor-transduced cells (Fig. 6C and D). Therefore, let-7a induces G0/G1 arrest in A549 and H1299 cells, which may be mediated by downregulation of cyclin D1 and upregulation of Rb.
Figure 6.
Let-7a inhibits cell cycle progression. (A) Representative cell cycle histograms show the percentage of A549 cells in each phase of the cell cycle after miR-negative control (scrambled), let-7a-mimics or let-7a-inhibitor transfection. (B) Let-7a resulted in cell cycle arrest at the G0/G1 in H1299 cells, compared to the control. (C) The expression of Rb and p-Rb proteins were analyzed by western blotting in A549 cells. (D) The expression of Rb and p-Rb proteins in H1299 cells. GAPDH was used as an internal control.
Let-7a inhibits cell migration and invasion
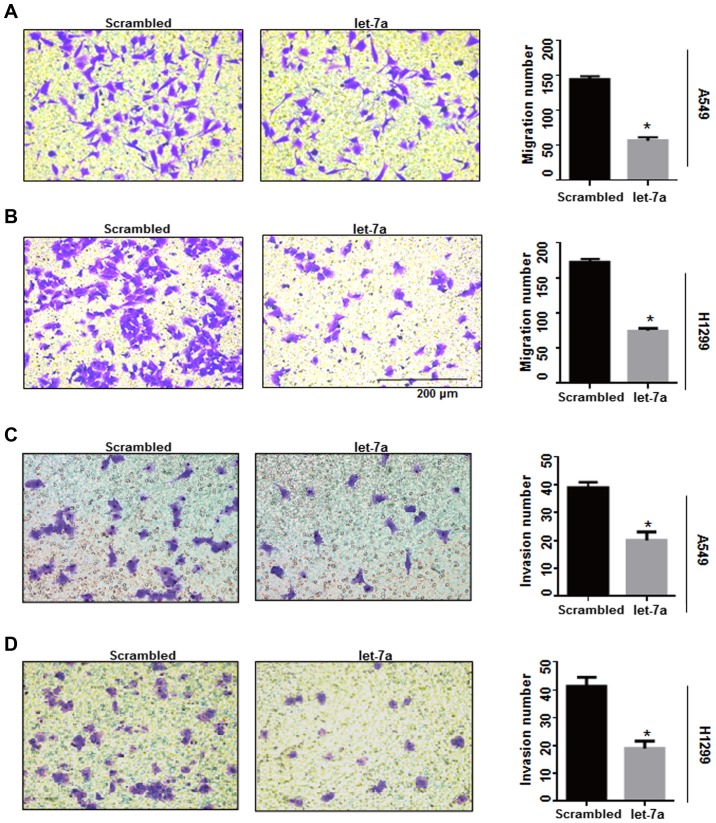
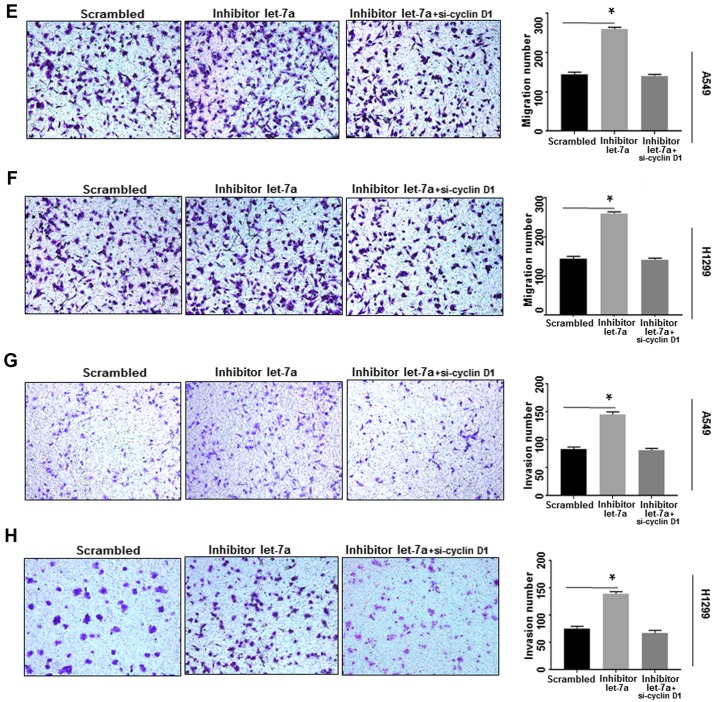
Cyclin D1-associated factors may affect the migration and invasion potential of breast cancer cells (31). In the present study, Transwell experiments were performed to detect whether let-7a inhibits lung cancer cell migration and invasion by influencing cyclin D1-associated factors. The number of migrated cells in let-7a-treated A549 and H1299 cells was significantly reduced compared with the controls (P<0.0001; Fig. 7A and B), while the let-7a inhibitor increased the number of migrated cells. There was no significant increase in the rate of migration in A549 and H1299 cells co-transfected with let-7a-inhibitor and si-cyclin D1 (P<0.001; Fig. 7E and F). Furthermore, an invasion assay demonstrated that invasive ability was significantly decreased in let-7a-transfected A549 and H1299 cells compared with the control groups (P<0.0001; Fig. 7C and D), and the effects of let-7a on invasion could be reversed by knockdown of cyclin D1 using siRNA (P<0.001; Fig. 7G and H). These results indicated that let-7a inhibits cell migration and invasion by targeting cyclin D1.
Figure 7.
Let-7a suppresses cancer cell migration. (A) A migration assay was performed to assess the roles of let-7a in A549 cells. The number of A549 cells was reduced in let-7a-treated cells compared to the scrambled group. *P<0.0001 vs. scrambled control. (B) Upregulation of let-7a significantly decreased the number of H1299 migrated cells compared to the scrambled group. *P<0.0001. (C) In A549 cells, let-7a mimics significantly reduced the number of A549 invaded cells to the Transwell chamber compared to the scrambled group as determined by Matrigel invasion assay. *P<0.01. (D) The same result was confirmed in H1299 cells. *P<0.01. Let-7a suppresses cancer cell migration. (E and F) Let-7a inhibitor improved cell migration ability, which was reversed by cyclin D1-siRNA in A549 and H1299 cell lines. *P<0.01. (G and H) Cell invasion ability was improved by let-7a-inhibitor, which was reversed by cyclin D1-siRNA in A549 and H1299 cell lines. *P<0.01.
Discussion
As small non-coding RNAs, miRNAs are able to influence numerous biological processes through post-transcriptional regulation of gene expression (18). Over the past two decades, numerous studies have indicated miRNA changes are associated with cancer biology, including cancer formation, development and metastasis (18,32). Let-7 miRNAs are underexpressed in the blood of patients with NSCLC (33) and expressed at lower levels in lung cancer (34). Similarly, the current results indicated that the expression of let-7a in lung adenocarcinoma tissues is reduced, suggesting that loss of let-7a may be a key factor in lung cancer development.
Gain-of-function and loss-of-function experiments were performed to observe the effects of let-7a on the biological characteristics of A549 and H1299 cells. It was determined that let-7a treatment inhibits proliferation and induces apoptosis of lung adenocarcinoma cells, via a decrease of cyclin D1. It was previously demonstrated that cyclin D1/cyclin-dependent kinase 4 may interact with filamin A and impact the migration and invasion potential of breast cancer cells (31). In the present study, it was determined that migration and invasion abilities were reduced following let-7a upregulation in A549 and H1299 cells.
Aberrant expression of cyclin D1 is observed frequently in a variety of tumor types, and cyclin D1 is regarded as a prognostic marker in multiple cancer types (35,36). Recently, researchers have aimed to explore the roles of miRNAs in regulating cyclin D1. Lower expression of miR-138 increases cyclin D1 expression, which serves a key function in the pathogenesis of OLP mucosal disease (37). MiRNA-520a-3p induces breast cancer cell apoptosis by directly targeting cyclin D1 (38). In the present study, it was revealed that let-7a, as a novel miRNA, directly suppresses cyclin D1-associated signals, which effectively inhibits cell growth and induces cell apoptosis. As downstream factors of cyclin D1, Bcl-2 was decreased, whereas Bax was increased, in let-7a-treated A549 and H1299 cells. These phenomena may be associated with the inhibitory effect of let-7a on lung cancer cell proliferation. The regulation of Bcl-2 promotes the mitochondria to release cytochrome c, which further induces cell apoptosis through caspase signals (39). The current results demonstrated that let-7a could induce lung adenocarcinoma cell apoptosis by reducing Bcl-2 and upregulating cleaved caspase-3, −8 and −9.
Although it was revealed in the present study that let-7a inhibits the malignant behavior of lung cancer cells by targeting cyclin D1, the mechanisms of let-7a remain to be elucidated. The regulatory networks between miRNAs and mRNAs indicate that their interactions are complex. Therefore, it is necessary to assess the roles of other miRNAs in the regulation of cyclin D1 in lung adenocarcinoma. Further investigations are warranted in order to extend these findings, and in vivo studies are required to provide more convincing data about the roles of cyclin D1 and let-7a in lung adenocarcinoma (31).
In summary, the present study demonstrates novel roles of let-7a in lung cancer. Let-7a significantly inhibits cell proliferation and enhances apoptosis of lung adenocarcinoma cells by directly regulating cyclin D1 signals. The current findings suggest that let-7a may be a novel therapeutic target in patients with lung cancer.
Acknowledgements
We thank Lixia Zhang (Binzhou Medical University) for their help of electron microscopy and flow cytometry analysis.
Glossary
Abbreviations
- miRNAs
microRNAs
- NSCLC
non-small cell lung cancer
- 3′ UTR
3′ untranslated region
- qRT-PCR
real-time polymerase chain reaction
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- FLNa
filamin A
Funding
The present study was supported by the National Natural Science Foundation of China (nos. 31371321, 81772281 and 81502536), the Shandong Science and Technology Committee (nos. 2017GSF221011, ZR2016CL09 and ZR2015HL060), the Health and Family Planning Commission of Shandong Province (no. 2014WS0186, 2015WS0499), and the Shandong Province Taishan Scholar Project (no. ts201712067).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
SYX and CZ conceived and designed the study. WZ, JXH, RMH, QZ, JQG, YJL, NX, LYL and PYW performed the experiments. WZ, JXH and SYX wrote the paper. CZ and SYX reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The experiments were performed according to the relevant guidelines of the Code of Ethics of the World Medical Association for experiments involving humans and the Medical Ethics Committee of Binzhou Medical University. Written informed consent was obtained from all patients prior to the collection of lung tissue samples.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Geyik E, Igci YZ, Pala E, Suner A, Borazan E, Bozgeyik I, Bayraktar E, Bayraktar R, Ergun S, Cakmak EA, et al. Investigation of the association between ATP2B4 and ATP5B genes with colorectal cancer. Gene. 2014;540:178–182. doi: 10.1016/j.gene.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 2.Ozcan O, Kara M, Yumrutas O, Bozgeyik E, Bozgeyik I, Celik OI. MTUS1 and its targeting miRNAs in colorectal carcinoma: Significant associations. Tumour Biol. 2016;37:6637–6645. doi: 10.1007/s13277-015-4550-4. [DOI] [PubMed] [Google Scholar]
- 3.Ye L, Wang H, Liu B. miR-211 promotes non-small cell lung cancer proliferation by targeting SRCIN1. Tumour Biol. 2016;37:1151–1157. doi: 10.1007/s13277-015-3835-y. [DOI] [PubMed] [Google Scholar]
- 4.Castro D, Moreira M, Gouveia AM, Pozza DH, De Mello RA. MicroRNAs in lung cancer. Oncotarget. 2017;8:81679–81685. doi: 10.18632/oncotarget.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan C, Wang D, Zhang Y, Yu W. MicroRNA-1284 inhibits cell viability and induces apoptosis of ovarian cancer cell line OVCAR3. Oncol Res. 2016;24:429–435. doi: 10.3727/096504016X14685034103518. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu WL, Chang JM, Chong IW, Hung YL, Chen YH, Huang WT, Kuo HF, Hsieh CC, Liu PL. Curcumin inhibits LIN-28A through the activation of miRNA-98 in the lung cancer cell line A549. Molecules. 2017;22:E929. doi: 10.3390/molecules22060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong Z, Dong Z, Yang L, Chen X, Gong Z. Inhibition of proliferation of human lung cancer cells by green tea catechins is mediated by upregulation of let-7. Exp The Med. 2012;4:267–272. doi: 10.3892/etm.2012.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan L, Gong Z, Zhong Z, Dong Z, Liu Q, Le Y, Guo J. Lin-28 reactivation is required for let-7 repression and proliferation in human small cell lung cancer cells. Mol Cell Biochem. 2011;355:257–263. doi: 10.1007/s11010-011-0862-x. [DOI] [PubMed] [Google Scholar]
- 10.Braga TV, Evangelista FC, Gomes LC, Araujo S, Carvalho MD, Sabino AP. Evaluation of MiR-15a and MiR-16-1 as prognostic biomarkers in chronic lymphocytic leukemia. Biomed Pharmacother. 2017;92:864–869. doi: 10.1016/j.biopha.2017.05.144. [DOI] [PubMed] [Google Scholar]
- 11.Ru Y, Chen XJ, Zhao ZW, Zhang PF, Feng SH, Gao Q, Gao SG, Feng XS. CyclinD1 and p57kip2 as biomarkers in differentiation, metastasis and prognosis of gastric cardia adenocarcinoma. Oncotarget. 2017;8:73860–73870. doi: 10.18632/oncotarget.18008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X, Sun F, Luo S, Zhao W, Yang T, Zhang G, Gao M, Lu R, Shu Y, Mu W, et al. let-7a is an antihypertrophic regulator in the heart via targeting calmodulin. Int J Biol Sci. 2017;13:22–31. doi: 10.7150/ijbs.16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He M, Xue Y. MicroRNA-148a suppresses proliferation and invasion potential of non-small cell lung carcinomas via regulation of STAT3. Onco Targets Ther. 2017;10:1353–1361. doi: 10.2147/OTT.S123518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang R, Yang C, Ma X, Wang Y, Luo D, Huang C, Xu Z, Liu P, Yang L. MiR-let-7a inhibits cell proliferation, migration, and invasion by down-regulating PKM2 in gastric cancer. Oncotarget. 2016;7:5972–5984. doi: 10.18632/oncotarget.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirali S, Aghaei M, Shabani M, Fathi M, Sohrabi M, Moeinifard M. Adenosine induces cell cycle arrest and apoptosis via cyclinD1/Cdk4 and Bcl-2/Bax pathways in human ovarian cancer cell line OVCAR-3. Tumour Biol. 2013;34:1085–1095. doi: 10.1007/s13277-013-0650-1. [DOI] [PubMed] [Google Scholar]
- 16.Li ZCJ, Yu Q, Wu X, Pan A, Li L. Evaluation of CCND1 amplification and CyclinD1 expression: Diffuse and strong staining of CyclinD1 could have same predictive roles as CCND1 amplification in ER positive breast cancers. Am J Transl Res. 2016;8:142–153. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YY, Ren T, Cai YY, He XY. MicroRNA let-7a inhibits the proliferation and invasion of nonsmall cell lung cancer cell line 95D by regulating K-Ras and HMGA2 gene expression. Cancer Biother Radiopharm. 2013;28:131–137. doi: 10.1089/cbr.2012.1307. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Wang D, Gu C, Liu X, Pei W, Li J, Cao Y, Jiao Y, Tong J, Nie J. Down-regulation of let-7 microRNA increased K-ras expression in lung damage induced by radon. Environ Toxicol Pharmacol. 2015;40:541–548. doi: 10.1016/j.etap.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhang YX, Yan YF, Liu YM, Li YJ, Zhang HH, Pang M, Hu JX, Zhao W, Xie N, Zhou L, et al. Smad3-related miRNAs regulated oncogenic TRIB2 promoter activity to effectively suppress lung adenocarcinoma growth. Cell Death Dis. 2016;7:e2528. doi: 10.1038/cddis.2016.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin-induced cytotoxicity via regulating microRNA let-7a in nasopharyngeal carcinoma. J Biochem Mol Toxicol. 2017;31:7. doi: 10.1002/jbt.21904. [DOI] [PubMed] [Google Scholar]
- 21.Wang PY, Sun YX, Zhang S, Pang M, Zhang HH, Gao SY, Zhang C, Lv CJ, Xie SY. Let-7c inhibits A549 cell proliferation through oncogenic TRIB2 related factors. FEBS Lett. 2013;587:2675–2681. doi: 10.1016/j.febslet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Cheng G, Wang X, Li Y, He L. let-7a-transfected mesenchymal stem cells ameliorate monocrotaline-induced pulmonary hypertension by suppressing pulmonary artery smooth muscle cell growth through STAT3-BMPR2 signaling. Stem cell Res Ther. 2017;8:34. doi: 10.1186/s13287-017-0480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan YC, Zhu YS, Mei PJ, Sun SG, Zhang H, Chen HF, Chen C, Miao FA. Cullin1 regulates proliferation, migration and invasion of glioma cells. Med Oncol. 2014;31:227. doi: 10.1007/s12032-014-0227-x. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Wang J, Zhao H, Wang J, Tony To S. The role of Myc and let-7a in glioblastoma, glucose metabolism and response to therapy. Arch Biochem Biophys. 2015;580:84–92. doi: 10.1016/j.abb.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The 8th edition lung cancer stage classification. Chest. 2016 doi: 10.1016/j.chest.2016.10.010. doi:10.1016/j.chest.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Nakashima Y, Yao T, Hirahashi M, Aishima S, Kakeji Y, Maehara Y, Tsuneyoshi M. Nuclear atypia grading score is a useful prognostic factor in papillary gastric adenocarcinoma. Histopathology. 2011;59:841–849. doi: 10.1111/j.1365-2559.2011.04035.x. [DOI] [PubMed] [Google Scholar]
- 27.Kadota K, Suzuki K, Kachala SS, Zabor EC, Sima CS, Moreira AL, Yoshizawa A, Riely GJ, Rusch VW, Adusumilli PS, et al. A grading system combining architectural features and mitotic count predicts recurrence in stage I lung adenocarcinoma. Mod Pathol. 2012;25:1117–1127. doi: 10.1038/modpathol.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinosa AM, Alfaro A, Roman-Basaure E, Guardado-Estrada M, Palma Í, Serralde C, Medina I, Juárez E, Bermúdez M, Márquez E, et al. Mitosis is a source of potential markers for screening and survival and therapeutic targets in cervical cancer. PLoS One. 2013;8:e55975. doi: 10.1371/journal.pone.0055975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giordano TJ. The argument for mitotic rate-based grading for the prognostication of adrenocortical carcinoma. Am J Surg Pathol. 2011;35:471–473. doi: 10.1097/PAS.0b013e31820bcf21. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Hao Z, Fan R, Zou X, Jin H, Pan Y, He L, Du R, Gao L, Liu D, et al. CIAPIN1 inhibits gastric cancer cell proliferation and cell cycle progression by downregulating CyclinD1 and upregulating P27. Cancer Biol Ther. 2007;6:1539–1545. doi: 10.4161/cbt.6.10.4684. [DOI] [PubMed] [Google Scholar]
- 31.Zhong Z, Yeow WS, Zou C, Wassell R, Wang C, Pestell RG, Quong JN, Quong AA. CyclinD1/cyclin-dependent kinase 4 interacts with filamin A and affects the migration and invasion potential of breast cancer cells. Cancer Res. 2010;70:2105–2114. doi: 10.1158/0008-5472.CAN-08-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho KJ, Song J, Oh Y, Lee JE. MicroRNA-let-7a regulates the function of microglia in inflammation. Mol Cell Neurosci. 2015;68:167–176. doi: 10.1016/j.mcn.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Jeong HC, Kim EK, Lee JH, Lee JM, Yoo HN, Kim JK. Aberrant expression of let-7a miRNA in the blood of non-small cell lung cancer patients. Mol Med Rep. 2011;4:383–387. doi: 10.3892/mmr.2011.430. [DOI] [PubMed] [Google Scholar]
- 34.Zhao BW, Zhou LF, Liu YL, Wan SM, Gao ZX. Evolution of Fish Let-7 MicroRNAs and their expression correlated to growth development in Blunt Snout Bream. Int J Mol Sci. 2017;18:E646. doi: 10.3390/ijms18030646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter T, Pines J. Cyclins and cancer. II: Cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 36.Seiler R, Thalmann GN, Rotzer D, Perren A, Fleischmann A. CCND1/CyclinD1 status in metastasizing bladder cancer: A prognosticator and predictor of chemotherapeutic response. Mod Pathol. 2014;27:87–95. doi: 10.1038/modpathol.2013.125. [DOI] [PubMed] [Google Scholar]
- 37.Ghallab NA, Kasem RF, El-Ghani SF, Shaker OG. Gene expression of miRNA-138 and cyclin D1 in oral lichen planus. Clin Oral Investig. 2017;21:2481–2491. doi: 10.1007/s00784-017-2091-5. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Wei J, Mei Z, Yin Y, Li Y, Lu M, Jin S. Suppressing role of miR-520a-3p in breast cancer through CCND1 and CD44. Am J Transl Res. 2017;9:146–154. [PMC free article] [PubMed] [Google Scholar]
- 39.Gómez-Crisóstomo NP, López-Marure R, Zapata E, Zazueta C, Martínez-Abundis E. Bax induces cytochrome c release by multiple mechanisms in mitochondria from MCF7 cells. J Bioenerg Biomembr. 2013;45:441–448. doi: 10.1007/s10863-013-9508-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.