Abstract
Gastric cancer (GC) is one of the most common malignancies that threatens human health. As the molecular mechanisms unerlying GC are not completely understood, identification of genes related to GC could provide new insights into gene function as well as potential treatment targets. We discovered that UGT2B15 may contribute to the pathogenesis and progression of GC using GEO data and bioinformatic analysis. Using TCGA data, UGT2B15 mRNA was found to be significantly overexpressed in GC tissues; patients with higher UGT2B15 had a poorer prognosis. It was further discovered that UGT2B15 and FOXA1 were both upregulated, and UGT2B15 and Foxa1 were positively correlated in GC. It is known that Foxa1 is a vital threshold to activate the Hippo-YAP signaling pathway. In addition, we suggest that a potential molecular mechanisms includes UGT2B15 which may upregulate Foxa1, activate the Hippo-YAP signaling pathway and contribute to the development of GC. Taken together, our findings demonstrate that UGT2B15 may be an oncogene in GC and is a promising therapeutic target for cancer treatment.
Keywords: GEO database, TCGA, UGT2B15, gastric cancer, FOXA1
Introduction
Gastric cancer (GC) is a type of malignant digestive tract tumor with a poor prognosis. GLOBOCAN 2015 reported GC as the third leading cause of cancer-related mortality in the world, with the majority of cases in developing countries (1). The pathogenesis of GC involves constant activation of protooncogenes and inactivation of key tumor suppressor genes that can lead to abnormalities in cell functions such as proliferation, differentiation and apoptosis (2–5). While surgery is the predominant treatment for GC, many patients develop advanced stage disease, or experience recurrence after surgery, and therefore, require chemotherapy and radiotherapy (6,7). Currently, due to a lack of concrete knowledge concerning the molecular basis of GC, there is no effective approach with which to predict tumorigenesis and prevent recurrence. Therefore, it is extremely important to discover the molecular mechanisms underlying tumorigenesis and progression, and to identify novel target genes for treatment.
It has been reported that mRNAs regulate many cellular processes, such as proliferation, differentiation, multi-drug resistance (MDR), and epithelial-mesenchymal transition (EMT). Some of these processes have been identified as molecular drivers of malignancy, including GC (8–11). For example, Musashi-1 protein was found to be significantly upregulated and correlated with tumor size, tumor-node-metastasis (TNM) stage, depth of invasion, lymph node metastasis and distant metastasis (12). The level of protein expression of Musashi-1 was found to be an independent prognostic indicator for the survival rate of GC patients (12). Therefore, mRNAs play a key role in the pathogenesis, progression and MDR of malignancies. Thus, the study of mRNA expression profiles is a strategy by which to understand the underlying functional mechanisms and identify biomarkers in GC.
mRNA expression microarray platforms are used to explore aberrant mRNA expression and discover differentially expressed genes (DEGs). Microarrays have been used to identify DEGs, some of which have been demonstrated to lead to tumorigenesis, progression and MDR in malignancies (13–17). At present, bioinformatic analysis is expanding as a way to better help investigators analyze mRNA expression via microarray, study complex biological networks, and identify candidate genes.
In the present study, we retrieved three mRNA profiles (GSE54129, GSE79973, GSE56807) from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). Subsequently, DEGs were identified by comparing GC with noncancerous gastric tissues. Overlapped DEGs present in the 3 mRNA expression profiles were then identified. We next performed the same DEG enrichment analyses by Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG). The subnetwork extraction algorithms were applied to analyze gene co-expression and protein-protein interactions (PPIs) by STRING and Cytoscape. By analyzing subnetworks, we found five key candidate genes, ALDH3A1, COL11A1, BGN, PGA4 and UGT2B15. While ALDH3A1, COL11A1, BGN and PGA4 have been reported to contribute to the pathogenesis and progression of GC, UGT2B15 has never been reported in GC. It was observed that significant differences in UGT2B15 were correlated with prognosis in GC in the Cancer Genome Atlas (TCGA). We further demonstrated significant differences in UGT2B15 mRNA and protein expression in GC tissues. We analyzed the relationship between UGT2B15 expression and clinicopathological characteristics and explored molecular mechanisms in GC. Our results provide an important insight with which to discover new biomarkers and prognostic markers in GC patients.
Materials and methods
Microarray data information and identification of DEGs
NCBI-GEO is a free database of microarray or gene profiles, from which gastric cancer and normal or adjacent mucosal tissue gene expression profiles for GSE54129, GSE79973 and GSE56807 were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/). Microarray data was based on Agilent GPL 570 platform (Affymetrix Human Genome U133 plus 2.0 Array), and provided 126 GC tissues and 36 non-cancerous gastric tissues. We chose these 3 datasets for integrated analysis in this study. All procedures of this study complied with the following protocol: i) The raw data of high throughput functional genomic expression of each microarray was analyzed by GEO2R software (http://www.ncbi.nlm.nih.gov/geo/). DEGs were identified by classical t-test, and statistically significant DEGs were defined using P<0.05 and logFC >2 as the cut-off criterion. ii) Overlapping DEGs were obtained by uploading the DEG profile datasets and performing integrated analysis using Funrich software (http://www.funrich.org/).
Gene ontology and pathway enrichment analysis
Cytoscape (http://www.cytoscape.org/) is an open source software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other data. Gene Ontology (GO) and Pathway Enrichment Analysis for overlapping DEGs was analyzed using Bingo and ClueGo in Cytoscape software, with P<0.05 as the cut-off criterion.
Protein-protein interaction network and seed candidate genes
i) Overlapped DEGs were uploaded into STRING, and DEG-encoded proteins and protein-protein interaction network (PPI) were constructed, and then the results were downloaded in table TSV format data. ii) The tabular data obtained above was uploaded into Cytoscape software, which was used to construct protein-interaction relationship sub-networks and analyze the interaction relationship of the candidate DEG-encoding proteins in GC, and obtained seed candidates genes by calculating node degree.
Expression of candidate genes
TCGA has an interactive web server for analyzing RNA sequencing expression data from 9,736 tumors and 8,587 normal samples from the TCGA and the GTEx projects, using a standard processing pipeline. It provides customizable functions such as tumor/normal differential expression analysis, profiling according to cancer types or pathological stages, patient survival analysis, similar gene detection, correlation analysis and dimensionality reduction analysis. We downloaded the mRNA profile and clinical data with Cancer Browser (https://genome-cancer.ucsc.edu/).
Analysis of overall survival time
To analyze the prognostic value of UGT2B15, GC patients were divided into 2 groups: The upregulated expression and the downregulated expression group using mean UGT2B15 expression as cut-off in the TCGA data. The 2 patient cohorts were compared on a Kaplan-Meier survival plot and the log-rank P-value was calculated.
Immunohistochemical assays
The study included 32 gastric cancer patients, containing 16 males and 16 females, aged 61.1±12.2 years, recruited between August 2012 and September 2014 in Shenzhen People's Hospital (Shenzheng, China). Written informed consent was obtained from all patients prior to surgery, and the study was approved by our Institutional Ethics Committee. Sections were deparaffinized and rehydrated in steps through a series of graded ethanol and distilled water, and then treated with 3% H2O2 in methanol for 30 min to block endogenous peroxidase activity. The sections were rinsed for 5 min in PBS twice, and incubated with 10% normal goat serum for 30 min to block non-specific antibody binding. After washing, the samples were incubated with primary anti-rabbit UGT2B15 (cat. no. ab154864; Abcam, Cambridge, MA, USA) at 1:500 dilution at 4°C overnight, washed in PBS 3 times, then incubated with a secondary antibody goat anti-rabbit IgG (cat. no. ab6789; Abcam). Later, the sections were stained with DAB according to the manufacturer's protocols, mounted and photographed using a digitalized microscope camera (Nikon Corp., Tokyo, Japan).
Statistical analysis
Gene and protein expression was analyzed by t-test. Data are presented as mean ± SD. All statistical analyses were conducted using GraphPad Prism version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Identification of DEGs and bioinformatic analysis
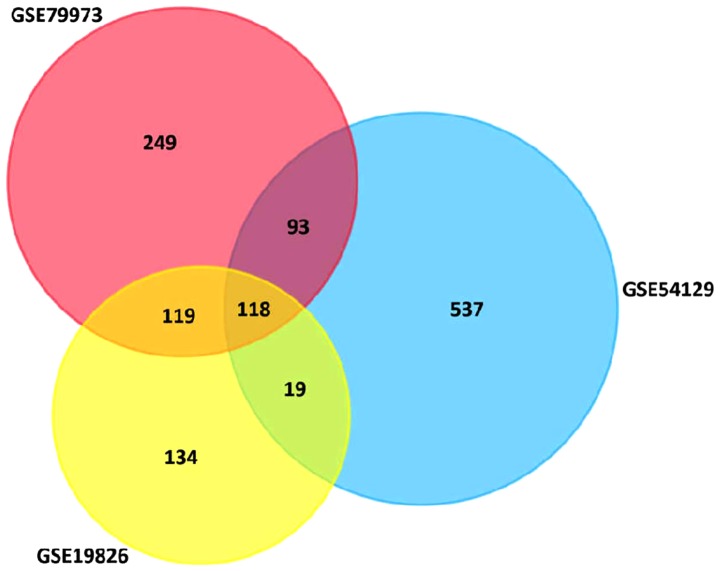
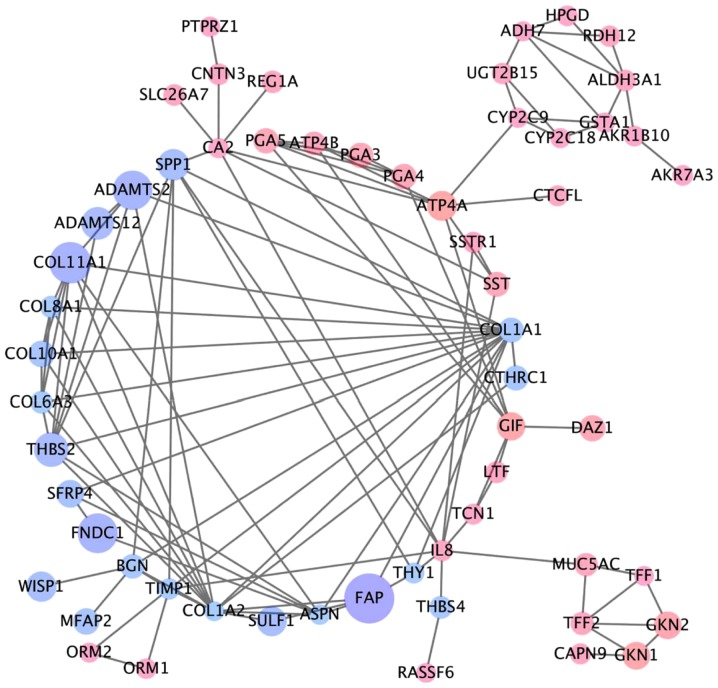
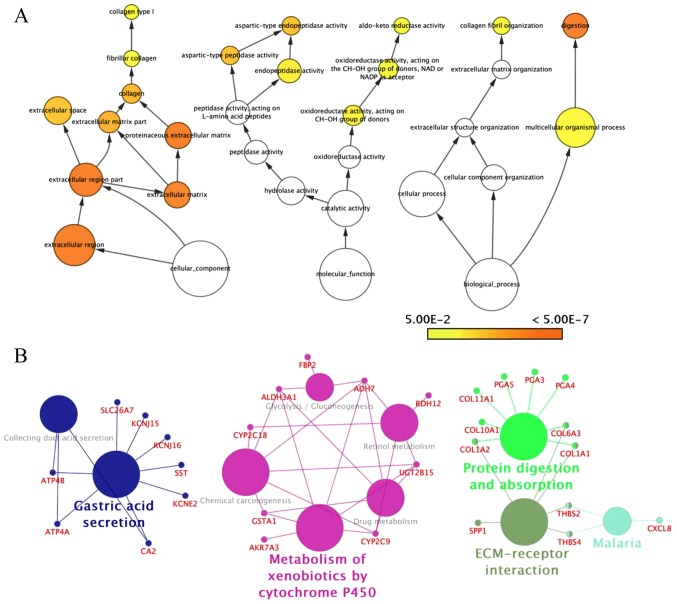
NCBI-GEO is a free database of microarray/gene profiles, from which GC cancer and normal or adjacent mucosal tissue gene expression profiles for GSE54129, GSE79973 and GSE56807 were obtained. The microarray data of GSE54129 contained 111 GC tissues and 21 normal GC tissues. GSE79973 and GSE56807 data contained 10 and 5 pairs of GC tissues and matched paraneoplastic tissues, respectively. We obtained 579 DEGs, including 189 upregulated DEGs and 390 downregulated DEGs in GSE79973, 677 DEGs including 166 upregulated and 511 downregulated DEGs in GSE54129, and 390 DEGs including 247 upregulated DEGs and 143 downregulated DEGs in GSE19826 (FC >2.0, P<0.05). The overlap between the 3 profiles was illustrated in a Venn diagram (Fig. 1). A total of 118 overlapping genes were in the common region, containing 87 upregulated and 31 downregulated DEGs (Table I). A PPI network included 110 nodes and 104 edges of the 118 DEGs, but 58 DEGs did not fall into the DEG PPI network through STRING (data not shown). We proposed PPI network complex for further analysis using Cytotype. This identified 5 central node genes, ALDH3A1, BGN, COL11A1, PGA3 and UGT2B15 with the filtering of degree >4 criteria (Fig. 2). These results revealed enrichment of DEGs in: Cellular process and cellular component organization within the biological process group; catalytic activity and hydrolase activity within the molecular function group; and extracellular region, extracellular matrix and extracellular region within the cellular component group using Bigo in Cytoscape (Fig. 3A). Analysis of the DEG functional and signaling pathway enrichment was conducted using ClueGo in Cytoscape to reveal that the DEGs are part of important pathways in GC, including metabolism of xenobiotics by cytochrome P450, chemical carcinogenesis, drug metabolism, ECM interaction, and protein digestion and absorption (Fig. 3B).
Figure 1.
Identification of DEGs and bioinformatic analysis. (A) Identification of 118 overlapped DEGs from the 3 cohort profile data sets (GSE79973, GSE19826 and GSE54129). Different color areas represent different datasets. DEGs were identified using a classical t-test and statistical significance defined by P<0.05 and [logFC]>2 as cut-off criterion. DEGs, differentially expressed genes.
Table I.
| Genes | |
|---|---|
| Downregulated | INHBA, COL10A1, FNDC1, FAP, THBS2, CST1, SULF1, SFRP4, SPP1, CEMIP, IGF2BP3, MIR675///H19, KLK10, ADAMTS2, CTHRC1, COL11A1, ASPN, PDLIM7, BGN, WISP1, CXCL8, THBS4, COL1A1, COL1A2, MFAP2, ADAMTS12, COL8A1, TIMP1, RARRES1, COL6A3, THY1 |
| Upregulated | AKR7A3, GATA5, DUOX2, RASSF6, B4GALNT3, REG1A, SCIN, TCN1, CNTN3, SPINK7, OTC, CTSE, CYP2C18, ORM2///ORM1, PTPRZ1, SH3RF2, ATP4B, VSTM2A, CYP2C9, LINC00982, ACER2, RFX6, CA2, FAM3B, CAPN13, ADTRP, SCNN1B, TRIM74///TRIM73, C6orf58, SST, UGT2B15, TMED6, CLDN18, MFSD4A, C16orf89, LINC00675, SULT1C, DAZ4///DAZ2///DAZ3///DAZ1, ANXA10, KCNJ15, UPK1B, CAPN9, HPGD, GATA6-AS1, MAP7D2, VSIG2, SSTR1, SLC26A7, KCNJ16, RDH12, PSCA, FUT9, PIK3C2G, TPCN2, CA9, KIAA1324, LINC00261, ADGRG2, SCGB2A1, ALDH3A1, FBP2, MUC5AC, SLC26A9, MAL, TFF1, ADH7, TFF2, PGC, GSTA1, MSMB, AKR1B10, CXCL17, ESRRG, LTF, DPCR1, KRT20, PSAPL1, KCNE2, SOSTDC, LIPF, GIF, PGA4///PGA3///PGA5, ATP4A, GKN1, GKN2, CWH43, VSIG1 |
A total of 118 overlapping genes were in the common region, containing 87 upregulated and 31 downregulated DEGs. DEGs, differentially expressed genes.
Figure 2.
Protein-protein interaction analysis. Protein-protein interaction (PPI) network constructed for the DEGs using Cytoscape. Blue nodes indicate genes with lowered expression and red nodes indicate overexpressed genes. Lines represent interaction of the protein with other proteins in the network. PPI, protein-protein interactions; DEGs, differentially expressed genes.
Figure 3.
DEG GO and pathway analysis. (A) GO term enrichment analysis for DEGs. The node color represents the significance of the GO terms and the node size represents the number of genes in that category. (B) KEGG pathway enrichment analysis for DEGs. The big nodes represent the different pathways and the small nodes represent the DEGs. DEGs, differentially expressed genes; GO, Gene ontology.
Validation of the relationship between expression of UGT2B15 and clinical characteristics in GC using TCGA data
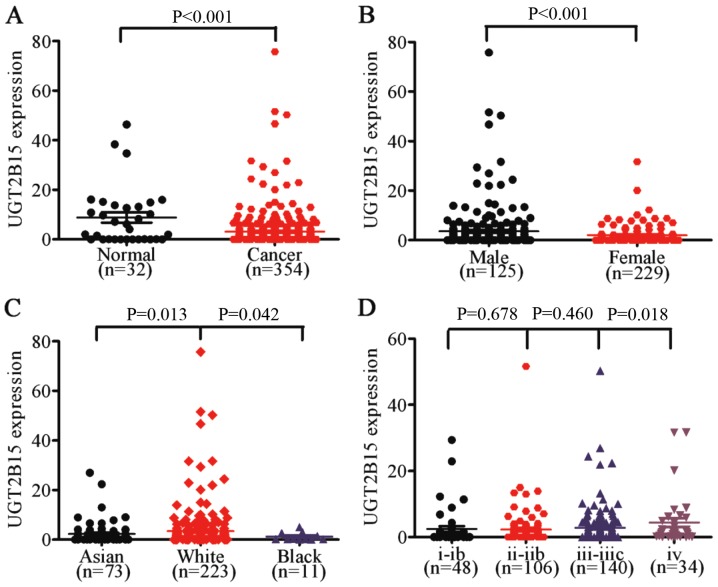
In order to validate the correlation between UGT2B15 expression and clinical characteristics of GC, we downloaded clinical data and UGT2B15 mRNA expression data from the TCGA dataset containing 354 GC and 32 normal samples (data not shown). The results showed that UGT2B15 was upregulated in GC tissue compared to that found in the non-cancerous gastric tissue (P<0.001) (Fig. 4A). We further discovered that UGT2B15 was overexpressed in male GC patients (P<0.001) compared to female GC patients, and that expression of UGT2B15 in Caucasian GC patients was significantly higher than that in Asian or black GC patients (P=0.013 and P=0.042, respectively) (Fig. 4B and C). We then demonstrated that UGT2B15 was upregulated in stage IV GC tissues (P=0.018) (Fig. 4D).
Figure 4.
UGT2B15 expression in different patient populations. (A) UGT2B15 overexpression in GC tissues compared to normal tissues (P<0.001). (B) Higher expression of UGT2B15 in male GC patients than female GC patients (P<0.01). (C) Higher expression of UGT2B15 in Caucasian (White) GC patients compared to Asian and Black GC patients (P=0.013, P=0.043). (D) UGT2B15 is upregulated in stage V GC (P=0.018). GC, gastric cancer.
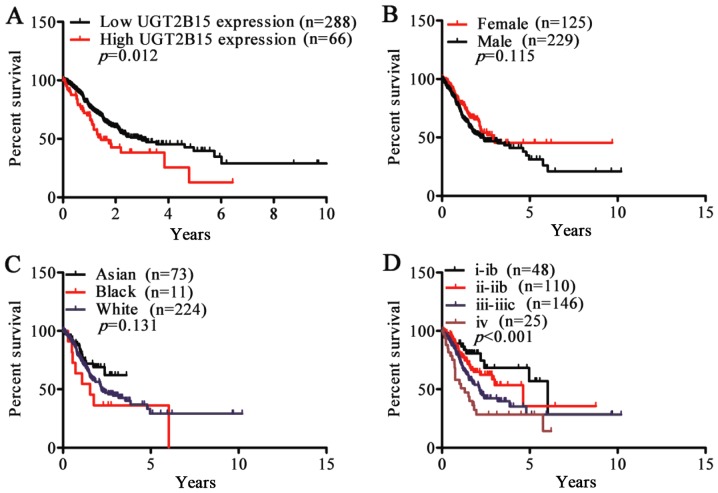
To validate overexpression of UGT2B15 as a prognostic factor in GC, the following analysis was performed. Using the mean ratio of relative UGT2B15 expression as the cut-off, 354 GC patients were classified into 2 groups, high-expression (n=66, >0.7) and low-expression (n=288, <0.7). Kaplan-Meier survival analysis and log-rank tests showed that patients with higher UGT2B15 expression had shorter survival time than those with lower UGT2B15 expression (P=0.012) (Fig 5A). However, although we found significant differences in UGT2B15 expression among different sex and races, the differences in survival curves of sex and race were not statistically significant (P=0.115, P=0.131) (Fig. 5B and C). Consistent with our results showing higher expression of UGT2B15 in stage IV disease, patients with stage IV tended to have poor prognosis (P<0.001) (Fig. 5D).
Figure 5.
Kaplan-Meier analysis of survival time in patients with different clinical characteristics. (A) Higher UGT2B15 expression is associated with poorer prognosis (P=0.012). (B) There is no significant difference in survival time between female and male GC patients (P=0.015). (C) The differences in survival curves in relation to race were not statistically significant (D) Stage IV GC patients had shorter survival time (P<0.001). GC, gastric cancer.
UGT2B15 positively correlates with Foxa1
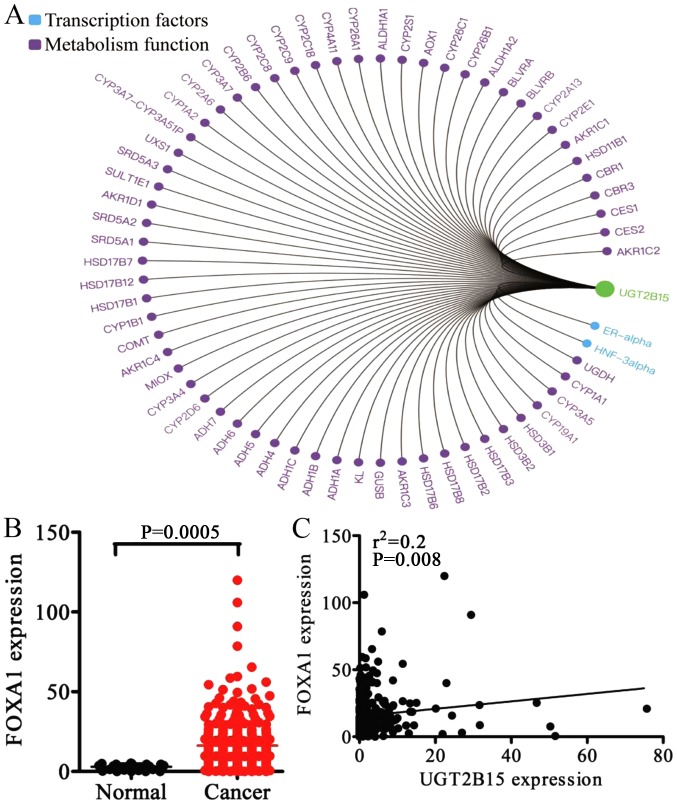
To explore the molecular mechanism of UGT2B15, Gene-Cloud Biotechnology Information (GCBI) analysis was performed to predict the function of UGT2B15; this showed that UGT2B15 had two transcription factors, ER-α and HNF-3a (FOXA1), and numerous metabolism function genes (Fig. 6A). It has been demonstrated that UGT2B15 regulates ER-α in breast cancer; however, the regulatory and functional relationships between UGT2B15 and Foxa1 remain unknown.
Figure 6.
UGT2B15 may be regulated and positively correlated with FOXA1. (A) UGT2B15 regulates transcription factor FOXA1 and ER-α. Green nodes represent transcription factors, blue nodes represent metabolism function. (B) Foxa1 was overexpressed in GC tissues compared to normal tissues (P<0.005). (C) Pearson assay showing that UGT2B15 is positively correlated with Foxa1 (r2=0.2, P=0.008). GC, gastric cancer.
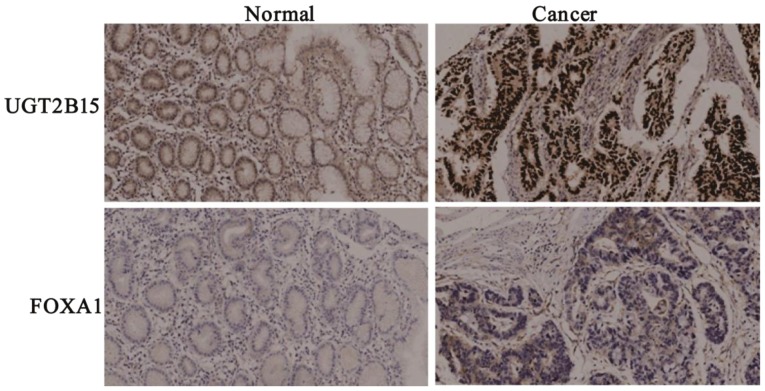
Clinical data and FOXA1 mRNA expression data were obtained from the TCGA dataset, providing 354 GC tissues and 32 non-cancerous gastric tissues (data not shown). Results showed that FOXA1 was upregulated in GC tissues compared to non-cancerous gastric tissues (P=0.0005) (Fig. 6B). The Pearson correlation coefficient confirmed that UGT2B15 and FOXA1 were positively correlated in GC tissues (R=0.2, P=0.008) (Fig. 6C). Immunohistochemistry was performed on 32 pairs of GC and adjacent non-tumorous tissues to confirm UGT2B15 and Foxa1 protein expression. Our results demonstrated the upregulation of both UGT2B15 protein and Foxa1 protein in GC tissues (Fig. 7).
Figure 7.
UGT2B15 and FOXA1 protein expression. Protein expression of UGT2B15 and Foxa1 was examined by immunohistochemistry in 32 paired GC tissues and normal gastric tissues. UGT2B15 and Foxa1 protein expression was significantly higher in GC tissues. GC, gastric cancer.
Discussion
GC is one of most common malignant tumors worldwide, particularly in East Asian countries such as China. Most patients with GC are diagnosed at the advanced stage and present with a relatively poor prognosis for overall survival. As is well recognized, GC results from the accumulation of multiple molecular alterations in cells (1).
Therefore, identification of genes with oncogenic potential or tumor suppressor activity may be of great value for diagnosis and treatment. In the present study, we investigated prognostic and predictive genes based on date available from public databases, and established and validated one prognostic signature, UGT2B15.
UGT2B15 is one of several functional members of the UGT2B subfamily. This protein is expressed primarily in the liver along with several extra-hepatic tissues, including prostate and breast cancer, and plays an important role in the glucuronidation of androgenic steroids (18,19). Hwang et al discovered polymorphisms of UGT2B15 that contributed to individual variations in drug and hormone metabolism in Korean as well as other ethnic populations (20). It has been reported that UGT2B15 may lead to progression and drug resistance in cancer. In addition, it has been found that UGT2B15 is downregulated in prostate cancer, and is negatively correlated with castration-resistant prostate cancer and lymph node metastases (19). However, Pfeiffer et al demonstrated UGT2B15 upregulation in prostate cancer that led to castration resistance (21). It has also been shown that UGT2B15 can reduce tamoxifen therapeutic efficacy and contribute to the development of acquired resistance to tamoxifen treatment via overexpression of UGT2B15 in breast cancer cells (18). However, Sutiman et al reported that UGT2B15 appeared to be of minor importance in breast cancer patients receiving tamoxifen (22). Therefore, the function of UGT2B15 is unclear in cancer.
In the present study, we first demonstrated UGT2B15 overexpression in GC tissues; moreover, it was upregulated in male, Caucasian and stage IV GC patients. Furthermore, patients with overexpressed UGT2B15 or stage IV GC cancer had significantly poorer overall survival time; however, no significant difference in overall survival time was detected in different races or sex. GO analysis and pathway analysis demonstrated UGT2B15 enrichment in extracellular region, chemical carcinogenesis, and drug metabolism. These results also suggest that UGT2B15 plays a role in progression and drug metabolism.
To confirm the biological effects of UGT2B15 in GC, we discovered that UGT2B15 can regulate transcription factors ER-α and HNF-3a (Foxa1) using Gene-Cloud Biotechnology information (GCBI). It has been confirmed that UGT2B15 regulates ER-α in breast cancer (18); however, the regulatory and functional relationships between UGT2B15 and Foxa1 remain unknown. We discovered that FOXA1 mRNA was upregulated in GC, and was positively correlated with expression of UGT2B15 in GC tissues with TCGA data. It has been demonstrated that FOXA1 acts as an oncogene and has roles in tumorigenesis, progression, and drug resistance, in ovarian, breast and GC (23–25). For example, overexpression of Foxa1 promoted GC cell proliferation and metastasis, via a mechanism involving Foxa1 activation of the Hippo-YAP signaling pathway by upregulation of YAP1 in GC (25). However, we demonstrated that UGT2B15 and Foxa1 protein were overexpressed in GC tissues by IHC. This implies that UGT2B15 regulates Foxa1 expression contributing to the development of GC through activating the Hippo-YAP signaling pathway.
In conclusion, this study suggests that UGT2B15 has an oncogenic role, and may be a promising therapeutic target for GC treatment. Further investigation is needed to elucidate the biological mechanisms of UGT2B15 in GC in vivo and in vitro.
Acknowledgements
The authors thank Mr. He Jie for his technical support and ‘World of Plainness’ authored by Yao Lu for the motivation.
Funding
The present study was supported by the Natural Science Foundation of Hunan Province (no. 2017JJ3270), the Three Engineering Training Funds in Shenzhen (no. SYLY201718 and SYJY201801) and the Technical Research and Development Project of Shenzhen (no. JCYJ20150403101028164).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YB, JY and DeL conceived and designed the study. DiL, XC, NW, MY, AL and SW acquired the data. GH, BZ, YY, DiL, HL, WZ and WX analyzed the data. DiL, PL and CM drafted the manuscript and were involved in the conception of the study. SP, PC and LJ performed the statistical analysis. JY and DeL obtained the data. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The study was approved by the Shenzhen People's Hospital Institutional Ethics Committee (Shenzhen, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Yan C, Zhu M, Huang T, Yu F, Jin G. Genome-wide association studies identified loci contribute to phenotypic variance of gastric cancer. Gut. 2017 doi: 10.1136/gutjnl-2017-315230. [DOI] [PubMed] [Google Scholar]
- 3.Huang LY, Wang X, Cui XF, Li H, Zhao J, Wu CC, Min L, Zhou Z, Wan L, Wang YP, et al. IRTKS is correlated with progression and survival time of patients with gastric cancer. Gut. 2017 doi: 10.1136/gutjnl-2016-313478. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Xu W. Neutrophils diminish T-cell immunity to foster gastric cancer progression: The role of GM-CSF/PD-L1/PD-1 signalling pathway. Gut. 2017;66:1878–1880. doi: 10.1136/gutjnl-2017-313923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gan L, Meng J, Xu M, Liu M, Qi Y, Tan C, Wang Y, Zhang P, Weng W, Sheng W, et al. Extracellular matrix protein 1 promotes cell metastasis and glucose metabolism by inducing integrin beta4/FAK/SOX2/HIF-1alpha signaling pathway in gastric cancer. Oncogene. 2017;37:744–755. doi: 10.1038/onc.2017.363. [DOI] [PubMed] [Google Scholar]
- 6.Brower V. Apatinib in treatment of refractory gastric cancer. Lancet Oncol. 2016;17:e137. doi: 10.1016/S1470-2045(16)00138-8. [DOI] [PubMed] [Google Scholar]
- 7.Bang YJ, Im SA, Lee KW, Cho JY, Song EK, Lee KH, Kim YH, Park JO, Chun HG, Zang DY, et al. Randomized, double-blind phase II trial with prospective classification by ATM protein level to evaluate the efficacy and tolerability of olaparib plus paclitaxel in patients with recurrent or metastatic gastric cancer. J Clin Oncol. 2015;33:3858–3865. doi: 10.1200/JCO.2014.60.0320. [DOI] [PubMed] [Google Scholar]
- 8.Reyna DE, Garner TP, Lopez A, Kopp F, Choudhary GS, Sridharan A, Narayanagari SR, Mitchell K, Dong B, Bartholdy BA, et al. Direct activation of BAX by BTSA1 overcomes apoptosis resistance in acute myeloid leukemia. Cancer Cell. 2017;32:490–505. doi: 10.1016/j.ccell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buttner R, Gosney JR, Skov BG, Adam J, Motoi N, Bloom KJ, Dietel M, Longshore JW, Lopez-Rios F, Penault-Llorca F, et al. Programmed death-ligand 1 immunohistochemistry testing: A review of analytical assays and clinical implementation in non-small-cell lung cancer. J Clin Oncol. 2017;35:3867–3876. doi: 10.1200/JCO.2017.74.7642. [DOI] [PubMed] [Google Scholar]
- 10.Ge W, Zhao K, Wang X, Li H, Yu M, He M, Xue X, Zhu Y, Zhang C, Cheng Y, et al. iASPP is an antioxidative factor and drives cancer growth and drug resistance by competing with Nrf2 for keap1 binding. Cancer Cell. 2017;32:561–573. doi: 10.1016/j.ccell.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Sidaway P. Breast cancer: LAG3 expression indicates favourable outcomes. Nat Rev Clin Oncol. 2017;14:712. doi: 10.1038/nrclinonc.2017.162. [DOI] [PubMed] [Google Scholar]
- 12.Shou Z, Jin X, He X, Zhao Z, Chen Y, Ye M, Yao J. Overexpression of Musashi-1 protein is associated with progression and poor prognosis of gastric cancer. Oncol Lett. 2017;13:3556–3566. doi: 10.3892/ol.2017.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimada Y, Okumura T, Sekine S, Moriyama M, Sawada S, Matsui K, Yoshioka I, Hojo S, Yoshida T, Nagata T, et al. Expression analysis of iPS cell-inductive genes in esophageal squamous cell carcinoma by tissue microarray. Anticancer Res. 2012;32:5507–5514. [PubMed] [Google Scholar]
- 14.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–240. [PubMed] [Google Scholar]
- 15.Vendrell JA, Thollet A, Nguyen NT, Ghayad SE, Vinot S, Bieche I, Grisard E, Josserand V, Coll JL, Roux P, et al. ZNF217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res. 2012;72:3593–3606. doi: 10.1158/0008-5472.CAN-11-3095. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y, Zheng S, Torossian A, Speirs CK, Schleicher S, Giacalone NJ, Carbone DP, Zhao Z, Lu B. Role of insulin-like growth factor-1 signaling pathway in cisplatin-resistant lung cancer cells. Int J Radiat Oncol Biol Phys. 2012;82:e563–572. doi: 10.1016/j.ijrobp.2011.06.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY, Wang XW. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Hu DG, Selth LA, Tarulli GA, Meech R, Wijayakumara D, Chanawong A, Russell R, Caldas C, Robinson JL, Carroll JS, et al. Androgen and estrogen receptors in breast cancer coregulate human UDP-glucuronosyltransferases 2B15 and 2B17. Cancer Res. 2016;76:5881–5893. doi: 10.1158/0008-5472.CAN-15-3372. [DOI] [PubMed] [Google Scholar]
- 19.Grosse L, Paquet S, Caron P, Fazli L, Rennie PS, Belanger A, Barbier O. Androgen glucuronidation: An unexpected target for androgen deprivation therapy, with prognosis and diagnostic implications. Cancer Res. 2013;73:6963–6971. doi: 10.1158/0008-5472.CAN-13-1462. [DOI] [PubMed] [Google Scholar]
- 20.Hwang MS, Lee SJ, Kim WY, Jeong HE, Shin JG. Genetic variations in UDP-glucuronosyltransferase 2B15 in a Korean population. Drug Metab Pharmacokinet. 2013;29:105–109. doi: 10.2133/dmpk.DMPK-13-SC-054. [DOI] [PubMed] [Google Scholar]
- 21.Pfeiffer MJ, Smit FP, Sedelaar JP, Schalken JA. Steroidogenic enzymes and stem cell markers are upregulated during androgen deprivation in prostate cancer. Mol Med. 2011;17:657–664. doi: 10.2119/molmed.2010.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutiman N, Lim JS, Muerdter TE, Singh O, Cheung YB, Ng RC, Yap YS, Wong NS, Ang PC, Dent R, et al. Pharmacogenetics of UGT1A4, UGT2B7 and UGT2B15 and their influence on Tamoxifen disposition in Asian breast cancer patients. Clin Pharmacokinet. 2016;55:1239–1250. doi: 10.1007/s40262-016-0402-7. [DOI] [PubMed] [Google Scholar]
- 23.Wang LL, Xiu YL, Chen X, Sun KX, Chen S, Wu DD, Liu BL, Zhao Y. The transcription factor FOXA1 induces epithelial ovarian cancer tumorigenesis and progression. Tumour Biol. 2017;39:1010428317706210. doi: 10.1177/1010428317706210. [DOI] [PubMed] [Google Scholar]
- 24.Davis DG, Siddiqui MT, Oprea-Ilies G, Stevens K, Osunkoya AO, Cohen C, Li XB. GATA-3 and FOXA1 expression is useful to differentiate breast carcinoma from other carcinomas. Hum Pathol. 2016;47:26–31. doi: 10.1016/j.humpath.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Ren H, Zhang P, Tang Y, Wu M, Zhang W. Forkhead box protein A1 is a prognostic predictor and promotes tumor growth of gastric cancer. Onco Targets Ther. 2015;8:3029–3039. doi: 10.2147/OTT.S91035. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.