Abstract
A new group of long terminal repeats (LTR) retrotransposons, termed terminal-repeat retrotransposons in miniature (TRIM), are described that are present in both monocotyledonous and dicotyledonous plant. TRIM elements have terminal direct repeat sequences between ≈100 and 250 bp in length that encompass an internal domain of ≈100–300 bp. The internal domain contains primer binding site and polypurine tract motifs but lacks the coding domains required for mobility. Thus TRIM elements are not capable of autonomous transposition and probably require the help of mobility-related proteins encoded by other retrotransposons. The structural organization of TRIM elements suggests an evolutionary relationship to either LTR retrotransposons or retroviruses. The past mobility of TRIM elements is indicated by the presence of flanking 5-bp direct repeats found typically at LTR retrotransposon insertion sites, the high degree of sequence conservation between elements from different genomic locations, and the identification of related to empty sites (RESites). TRIM elements seem to be involved actively in the restructuring of plant genomes, affecting the promoter, coding region and intron-exon structure of genes. In solanaceous species and maize, TRIM elements provided target sites for further retrotransposon insertions. In Arabidopsis, evidence is provided that the TRIM element also can be involved in the transduction of host genes.
Keywords: mobile elements‖transduction‖nonautonomous‖genome evolution‖sequence analysis
Transposons, also known as mobile elements or transposable elements, are discrete genetic sequences found in nearly all eukaryotic genomes. As their name indicates, transposons have the ability to move from one location of the genome to another, generating target site duplications (TSDs) after insertion. Autonomous transposons encode the enzymes necessary for their mobilization, whereas nonautonomous elements require these proteins to be provided in trans by an autonomous element (reviewed in ref. 1).
Retrotransposons are a class of transposable element that moves via an RNA intermediate that is reverse-transcribed into extrachromosomal DNA and inserted into the genome by their encoded reverse transcriptase, RNaseH and integrase enzymes (reviewed in refs. 2 and 3). This replicative mode of transposition is similar to that used by retroviruses. However, unlike the retroviruses, which are found usually in animals, retrotransposons are found in all eukaryotes, where they are the most abundant class of mobile DNA (reviewed in refs. 4 and 5). Retrotransposons are classified into two types: those with long terminal repeats (LTRs) and those without LTRs (non-LTR retrotransposons). LTR retrotransposons are subclassified further into the Ty1-copia and Ty3-gypsy groups, which differ from each other in distinct sequence features and in the order of encoded gene products (6, 7). Retrotransposons make up a large portion of many eukaryotic genomes including humans (8, 9) and a variety of plants (4, 10, 11).
Many properties of transposable elements suggest that they are parasitic or selfish DNAs (12, 13). However, like any other component of a heritable genome, transposon DNA can serve as a genomic resource for mutation and natural selection. Indeed, a more recent paradigm suggests that transposable elements may play a central role in the evolution of gene function and genome structure in eukaryotic organisms (for reviews see refs. 1, 4, 5, and 14–16). For example, transposons can contribute regulatory cis-factors (17, 18), alter transcript splicing (19, 20), promote exon shuffling (21), and facilitate chromosomal rearrangements or restructuring (22, 23), leading to changes in spatial and temporal expression patterns of host genes or even the generation of novel genes. Furthermore, retrotransposons are involved in altering the genome size of eukaryotic organisms either by increasing (10, 11) or decreasing (24–26) their copy numbers within the host genome.
The properties and abundance of transposable elements make it important to identify and characterize the whole spectrum of mobile elements that inhabit the eukaryotic genomes and to investigate their origin, interactions with host genomes, and contributions to the evolution of host genes and genomes. In this report a unique group of mobile retrotransposons is described. We provide evidence that these elements have some features typical of LTR retrotransposons but are unusually small and lack coding capacity for mobility-related proteins. We also demonstrate that they are present in both dicotyledonous and monocotyledonous plants and have been active in restructuring plant genomes.
Materials and Methods
Data Analysis.
Terminal-repeat retrotransposons in miniature (TRIM) elements were identified initially during sequence analysis of a genomic clone containing the Solanum tuberosum (potato) urease gene (C.-P.W, S. Tiller, H. V. Davies, and A.K., unpublished data) and during the analysis of the genome sequence of Arabidopsis, where they were referred to as Katydid (27).
In potato, a DOTPLOT (28) revealed closely spaced direct repeats within intron 6 of the urease gene. Closer inspection indicated that the direct repeats were flanked by a 5-bp TSD and contained a primer binding site (PBS) and polypurine tract (PPT) within the region between the repeats. Initial DNA-database searches with the similarity search algorithm BLAST (29) using the complete element as the query revealed the presence of similar sequences at several genomic locations in a number of solanaceous species. Independently, TRIM elements were identified during a survey of transposons in the Arabidopsis genome as part of the Arabidopsis Genome Initiative annotation effort (27). Iterative database analyses (with BLAST) by using TRIM sequences from previous searches as queries for new searches identified TRIM elements from several monocotyledonous and dicotyledonous plant species. In this search strategy all hits against the database were examined manually for typical features of TRIMs (e.g., a sequence of less than 540 bp containing direct repeats enclosing PBS and PPT motifs). Therefore, elements with some degree of truncation also were found for which the termini could not be mapped accurately because of deletions and/or sequence degeneracy. Sequence information of large insert clones was accessed from public sequence databases (GenBank, www.ncbi.nlm.nih.gov/GenBank/index.html, and the European Molecular Biology Laboratory, www.ebi.ac.uk/embl/index.html) between October 2000 and May 2001. Sequence analysis was performed by using programs of the UWGCG software package (version 10, University of Wisconsin Genetics Computer Group, www.gcg.com) or the EMBOSS program package (European Molecular Biology Open Software Suite, www.uk.embnet.org/Software/EMBOSS/). Multiple alignments were generated with PILEUP (UWGCG) or CLUSTALW (30). Graphical manipulations were carried out by using GENEDOC (31). TRIM elements were mapped onto the assembled genomic sequences of Arabidopsis accessed at the Arabidopsis Genome Initiative (released in March 2001, www.Arabidopsis.org/agi.html).
Related to Empty Sites (RESites) Analysis.
Sequences that were similar to the region immediately flanking an insertion but that did not harbor the insertion and had only one copy of the target site were defined as RESites (32). RESites can be used to delimit the termini of the insertion, confirm the size and sequence of the TSD, and provide supporting evidence for past mobility. RESites were identified by using the sequences immediately flanking an insertion as a query in a BLAST search as described (32).
Results
Identification, Characterization, and Distribution of TRIM Elements in Plants.
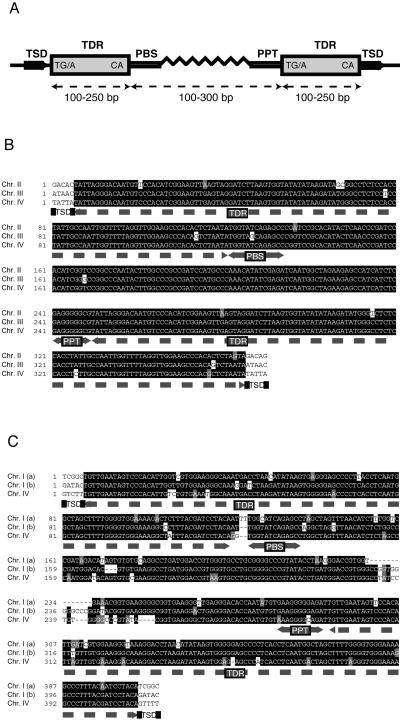
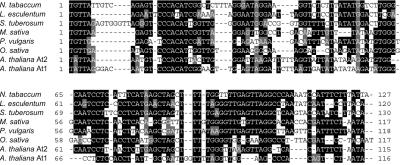
Database searches and sequence analyses revealed the presence of a family of repetitive sequences from a number of plant species that share a unique set of characteristics. Specifically, these elements are short in size (mostly ≈350 bp), have terminal direct repeats (TDRs) of 100–250 bp (on average <140 bp) that flank an internal domain containing a PBS (located immediately downstream of the 5′ terminal repeat and complementary to the methionine tRNA), and a PPT (located immediately upstream of the 3′ terminal repeat; Fig. 1A). None of the elements characterized to date encodes mobility-related proteins in the internal domain (Fig. 1). Intact elements are flanked by 5-bp direct repeats, representing the TSD that was generated after insertion. These previously uncharacterized mobile elements have been named TRIM (Fig. 1). TRIM elements are present in multiple copies and are dispersed within the host chromosomes (shown in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). TRIM elements seem ubiquitous in the plant kingdom as they have been found in both dicotyledonous and monocotyledonous species (Table 1). To date, TRIM elements from the plant species examined have similar overall lengths and structures and show some sequence similarity, especially in the TDRs (Figs. 1 and 2). For example, there is over 80 and 90% sequence identity among the TDRs of TRIM elements in Arabidopsis and rice, respectively. Furthermore, TDRs of TRIM elements from distantly related species show significant similarity (Fig. 2) ranging from 60 to 75% at the sequence identity level. In brief, TRIM elements are distinguished by their exceptionally small overall size (<540 bp) and presence in multiple copies in different locations within the host genome with highly conserved TDRs and short internal sequences (≈100–300 bp). Moreover, despite the complete lack of coding capacity for mobility-related proteins (which makes it impossible to classify TRIM elements conventionally into Ty1-copia or Ty3-gypsy groups), they are able to transpose in trans, making them the smallest active LTR retrotransposons known to date.
Figure 1.
Schematic diagram of the general structure of TRIM elements. (A) The archetypal TRIM contains the following sequence features: TSD, TDRs (flanking shaded boxes), PBS, and PPT. Start and end bases of TDRs are given in the flanking boxes. The possible length of different elements is given underneath (maximum length <540 bp). (B) Multiple alignment of selected TRIM elements (Katydid-At1) elements from A. thaliana. The alignment was created with CLUSTALW by using default parameters. The BOXSHADE program was used for shading. Structural features (abbreviations as described for A) are marked underneath. The elements in chromosomes II (Chr. II, gi 6598569, position 33,850–33,481), III (Chr. III, gi 7629988, position 45,624–45,993), and IV (Chr. IV, gi 3309259, position 95,888–95,519) are shown. (C) Multiple alignment of selected TRIM elements from rice (Oryza sativa). Alignment and abbreviations are as described above. The elements from chromosomes I (Chr. I(a), gi 10800055, position 29,385–29,802), I (Chr. I (b), gi 9711848, position 74,966–74,558), and IV (Chr. IV, gi 5777612, position 77,595–77,183) are shown.
Table 1.
TRIM elements found in plant sequences
| Organism | Intact* | Solo LTR | Truncated† | Total | EST‡ |
|---|---|---|---|---|---|
| Brassicaceae | |||||
| Arabidopsis thaliana | 21 | 5 | 17 | 43 | 1 |
| Solanaceae | |||||
| Solanum tuberosum | 3 | 2 | 3 | 8 | 4 |
| Solanum ochranthum | 1 | 0 | 0 | 1 | 0 |
| Lycopersicon pennellii | 0 | 0 | 1 | 1 | 1 |
| Lycopersicon esculentum | 3 | 0 | 7 | 10 | 3 |
| Nicotiana tabacum | 1 | 0 | 0 | 1 | 0 |
| Fabaceae | |||||
| Lotus japonicus | 0 | 0 | 2 | 2 | 1 |
| Medicago sativa | 1 | 0 | 0 | 1 | 0 |
| Medicago trunculata | 0 | 7 | 5 | 11 | 9 |
| Phaseolus vulgaris | 1 | 1 | 0 | 2 | 0 |
| Glycine max | 0 | 0 | 1 | 1 | 1 |
| Poaceae | |||||
| Oryza sativa | 8 | 4 | 4 | 16 | 1 |
| Zea mays | 1 | 0 | 1 | 2 | 0 |
Elements with small internal deletions were counted as intact.
Elements for which one or both termini could not be resolved.
Each EST represents a TRIM insertion in a different gene (duplicate ESTs for the same gene were not counted).
Figure 2.
Multiple alignment of selected 5′ TDR sequences of TRIM elements from different species. Sequences from Nicotiana tabacum (gi 9392606, position 3,821–3,947), Lycopersicon esculentum (gi 4220970, position 38–157), S. tuberosum (gi14599414, position 1,522–1,651), M. sativa (gi 19642, position 8,088–8,204), P. vulgaris (gi 2576326, position 1,878–1,761), O. sativa (gi 10800055, position 29,390–29506), A. thaliana Katydid-At2 (gi 7649355, position 68,877–68,994), and A. thaliana Katydid-At1 (gi 6598490, position 7722–7607) are shown. The alignment was created with CLUSTALW (gap opening parameter, 10; gap extension parameter, 1). The program BOXSHADE was used for the shading.
In Arabidopsis thaliana, a total of 43 TRIM elements were identified (Table 1) and subdivided into three groups on the basis of overall sequence similarity: Katydid-At1, -At2, and -At3. Thirty-one copies of Katydid-At1 are present including 12 truncated elements (i.e., termini could not be mapped accurately). Katydid-At2 is present in seven copies including three truncated elements, whereas there are five copies of Katydid-At3, of which two are truncated. Katydid-At1 elements have TDRs of 116 bp (Fig. 1B). The left and right TDRs are nearly identical in many elements. Because LTRs are identical initially after element insertion, the high sequence identity between the TDRs suggests that Katydid-At1 elements were recently, and are perhaps still, active (33). The fact that three RESites were identified (Fig. 3) and the high nucleotide similarity between some Katydid-At1 elements (up to 98%, Fig. 1B) also attest to recent mobility. In contrast, similarity between members or the TDRs of Katydid-At2 or -At3 was lower, indicating that they may be ancient insertions (Table 2). Katydid-At2 elements have TDRs similar to Katydid-At1 in size (≈115 bp) but have only limited similarity in sequence (Fig. 2). Although one Katydid-At3 element shared structural characteristics with other TRIM elements, two members had unusual features (Table 2). For example, one element (gi 4263586) has terminal inverted repeats rather than TDRs. Because a perfect 5-bp flanking TSD can be identified, it is possible that this unique element was mobilized with this structure. The other element is truncated at one end and yet still appears to have a 5-bp TSD. Structural rearrangements were observed also within other Arabidopsis Katydid elements. Although most Katydid-At1 elements have TDRs, “solo” and “triple” direct repeat structures were identified (Fig. 6, which is published as supporting information on the PNAS web site). Solo and triple direct repeat elements are also immediately flanked by a 5-bp TSD. Solo direct repeats were generated most likely by illegitimate recombination between TDRs of an intact Katydid-At1 element (10, 24, 25). The middle repeat of the triple direct repeats-containing element is unlikely to be a solo direct repeat, because flanking TSDs are not present. Within the triple repeats the TDRs share 93% with each other but showed lower (89 and 90%) similarity with the middle direct repeat (gi 3150006). It currently is unclear as to the precise mechanisms that generated these unusual TRIM elements.
Figure 3.
Identification of RESites for TRIM elements (Katydid-At1). Sequences harboring the Katydid-At1 insertion (depicted by the black boxes with arrowheads) are shown above the corresponding RESites. The positions on clone and gi numbers are indicated. Target sequences and TSDs are underlined. Insertion in gi 3702730 is a solo LTR.
Unlike other Arabidopsis retrotransposons, which cluster in the pericentromeric and centromeric regions where gene density is low, Katydid-At1 is dispersed throughout the genome (Table 2). In addition to the elements mined from nuclear sequences, we have identified a Katydid-At1 insertion in the mitochondrial genome of Arabidopsis (gi 1785729). Similar to most elements found in the mitochondrial genome of Arabidopsis (34), this Katydid-At1 element is truncated and degenerate. Transposable elements make up ≈4% of the total mitochondrial DNA and are predominately retrotransposon fragments that originated from reverse transcripts of nuclear genomic elements (34). Similarly, we consider that the mitochondrial Katydid-At1 element is likely to be of nuclear origin, because it shares sequence similarity with nuclear insertions and no other such elements could be identified in mitochondrial genomes.
In rice, TRIM elements from different genomic locations are highly similar, having nearly identical TDRs and highly conserved internal domains (Fig. 1C). These features of TRIM elements attest to the recent activity of these elements in the rice genome. At the date of this analysis (March 2001), 16 rice TRIM elements could be found in the sequence database. There were eight complete and three truncated elements, and five were present only as solo LTRs (Tables 1 and 2). Similar to Arabidopsis Katydid elements, the rice elements also are located on a variety of chromosomes, and some of the insertions are near or within putative genes and overlap with predicted ORFs (e.g., gi 9711848 and 5777612 in Table 2). However, only one expression sequence tag (EST, coding for a protein with similarity to leucine-rich repeat-like proteins, gi 4715547) with partial similarity to a TDR of a rice TRIM was identified. In maize, only one complete TRIM element has been found to date. This element possesses unusually long TDRs of ≈230 bp and a short internal domain of 72 bp. However, similar to TRIM elements from other plants, this element contains a PBS and a PPT motif and 5-bp TSDs.
In contrast to A. thaliana and rice, only limited genomic sequence data are available for members of the family of solanaceous and leguminous species (Table 1). In both groups ancient TRIM insertions were found (Table 2, gi 9392606) including truncated elements (Table 2, gi 19200 and 170430), solo LTRs (Table 2, gi 21590 and 8747823), and more complex structures, probably generated by several insertion-deletion events involving more than one element (Table 2, gi 9251206 and 10764221). However, TRIM elements with well conserved sequences were identified also (Table 2, gi 9562370 and 19642), indicating that TRIM elements have a long evolutionary history in these plants and that some still may be active.
Retrotransposons Nested Within TRIM Elements.
The TRIM in intron 6 of one of the alleles of the urease gene from potato (S. tuberosum, cv. Désirée (gi 14599411) contains an insertion of a truncated LTR retrotransposon of the Ty1-copia group (Table 2). In the wild species Solanum ochranthum, the TRIM in intron 6 of urease sustained a long nuclear interspersed element insertion, a non-LTR retrotransposon 30 bp downstream of the Ty1-copia insertion found in potato (gi 14599445, Table 2). In both cases the insertion was located within the internal sequence of the TRIM. Another case for TRIM elements being targeted by Ty1-copia retrotransposons was found in the maize genome. In this case, a complete element of ≈9 kb inserted within the 3′ TDR of a TRIM element (Table 2).
Insertion Within Genes.
TRIM elements contribute to coding and untranslated regions of plant genes. However, in many cases this notion is based on the observation that TRIM elements overlap with sequences predicted to contain a gene by computer-assisted intron/exon assignment. Although experimental evidence confirming the presence of a gene at predicted locations is often still lacking, the frequent presence of TRIM elements in ESTs from species of the Solanaceae and Fabaceae (Tables 1 and 2) demonstrates that insertion into transcribed coding sequences is common at least in some species. In some cases, ESTs provided evidence for the insertion of TRIM elements into coding sequences with similarity to known genes, for example glycolate oxidase in tomato (gi 10902684), a nucleic acid binding domain/leucine-rich repeat-like protein in potato (gi 9562370), or a P450-like protein in Medicago trunculata (gi 10520041). In solanaceous and leguminous species TRIM elements were found also in promoter regions and introns of genes (Table 2). For example, in potato and S. ochranthum elements were found in intron 6 of the urease gene (gi 14599411, gi 14599414, and gi 14599445) in Medicago sativa in intron 9 of the nodulin-25 gene (gi 19642), in potato in the promoter region of the proteinase inhibitor II gene (gi 21553), and in Phaseolus vulgaris a TRIM solo terminal repeat was found in the promoter region of the chalcone isomerase gene (gi 20981, Table 2). In contrast, TRIM elements are found rarely in ESTs from Arabidopsis and rice, although they often overlap with computer-predicted genes (Table 2).
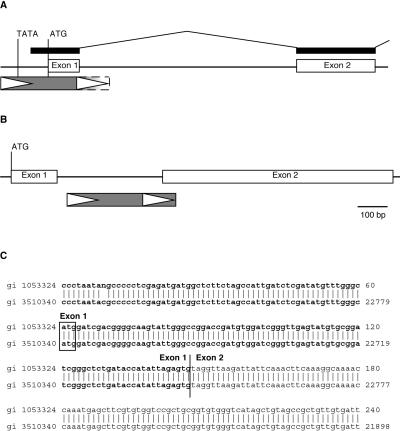
Two examples of Katydid-At1 elements altering host genes in Arabidopsis are shown in Fig. 4. In the first case (Fig. 4 A and C) a Katydid-At1 element (gi 3510340, position 22,675–22,946) overlaps with the transcription and possible translation start site and the first exon-intron boundary of a gene encoding a cytochrome P450-like protein (gi 3510340, position 19,519–22,694). That the gene is transcribed and correctly spliced is indicated by the existence of two ESTs corresponding to this gene (gi 5841198 and 1053954). The insertion of Katydid-At1, probably at the 5′ terminus of a preexisting gene, introduced a transcription start site (a TATA motif within the terminal repeat is present ≈40 bp upstream of transcription start). The element is truncated in the 3′-terminal repeat ≈20 bp downstream of the first exon-intron boundary. Thus, the first intron was either created de novo, or the 5′ boundary of an already existing intron was replaced because of the insertion of the element. Additionally, the potential use of a translation start site within the region between the two TDRs introduces 28 amino acids encoded by the element to the N terminus of the protein. Fig. 4B shows a case in which a Katydid-At1 element appears to have created a new intron within a gene, although no EST data to support this hypothesis are available.
Figure 4.
Examples of TRIM (Katydid-At1) contributions to gene structures. The gray boxes with white arrowheads depict Katydid-At1 elements. Open boxes represent exons. The predicted start of translation (ATG) and TATA box are indicated. (A) The Katydid-At1 element in gi 3510340 (position 22,673–22,946) contributes sequences for the promoter, first exon, and first exon-intron boundary of a gene encoding a cytochrome P450-like gene. This insertion is truncated. Corresponding identical EST sequences (gi 5841198 and gi 1053954) are represented by thick black bars. Thin lines connecting thick black bars represent a putative intron that is not found in the EST. (B) The Katydid-At1 found on gi 7209738 contributes a splice site to a predicted cytochrome P450 gene. The scale is as described for A. (C) Sequence alignment of Katydid-At1 from A showing the putative translation start codon (boxed) and exon boundaries.
Transduction of Host Genes.
A Katydid-At1 element located in the Arabidopsis genome contains an ORF (gi 3241926) nearly identical to a putative cellular gene annotated as a nonsense-mediated mRNA decay (NMD) trans-acting factor but that actually is more similar to the yeast gene sen1. NMD is responsible for the degradation of prematurely terminated mRNAs (35), whereas SEN1 is involved in the endonucleolytic cleavage of pre-tRNAs (36). Both NMD and SEN1 contain a large domain involved in degrading ribonucleotides (35). The ORF linked to the Katydid-At1 elements lacks part of the first and second exons and does not contain intron sequences (Fig. 5). This structure most likely was generated by the recombination of a Katydid element with an mRNA of NMD during the transposition process. The presence of this ORF within Katydid-At1 is very reminiscent of the product of cellular gene transduction by retroviruses leading to the formation of oncoviruses (37). Previously, cellular gene transduction was documented for another plant retroelement, the maize Bs1 element (37–41). Although the cellular NMD (c-NMD) and transduced NMD (r-NMD, where r- refers to retrotransposon) both are located on chromosome 5, they are not linked tightly, nor are there other Katydid-At1 elements near c-NMD in the A. thaliana (Columbia ecotype) genomic sequence. This precludes the possibility that transduction resulted from a read-through transcript.
Figure 5.
TRIM (Katydid-At1) involvement in the transduction of a cellular gene. (A) Diagram illustrating the region of similarity (93%) between the cellular NMD gene (c-NMD) and the transduced version (r-NMD). The intron sequences on c-NMD (bent thin lines) are absent in r-NMD. Exons are represented by black boxes, and Katydid-At1 sequences and LTRs are represented by gray boxes and arrowheads, respectively. The gi numbers and positions on clones are indicated. (B) Amino acid sequence alignment of r-NMD and c-NMD. Identical residues are shaded in gray.
Discussion
Computer-assisted data mining of transposable elements has proven to be the most efficient and informative route to the understanding of not only mobile element evolution but also their role in eukaryotic gene and genome evolution. Although many mined elements reinforce traditional paradigms concerning sequence- and structure-based transposon classification, unusual elements occasionally are mined that force a re-evaluation of element type definition. TRIM elements are one such type of element. Their size (<540 bp) is considerably less than that of typical LTR retrotransposons (5–15 kb), which makes them the smallest TDR-containing mobile elements discovered to date. Despite their small size and lack of coding capacity, TRIM elements are obviously mobile as indicated by several observations: (i) the high sequence similarity between elements from different genomic locations (Fig. 1 B and C), (ii) the wide distribution within the individual genomes (Table 2), and (iii) the identification of RESites (Fig. 3).
Despite their small size, TRIM elements have some typical features of LTR retroelements: possessing TDRs, a 5-bp duplication of the target site sequence, and PBS and PPT motifs, which are essential for the mobilization of LTR retrotransposons. Furthermore, the solo TDRs of TRIM elements are reminiscent of solo LTRs and presumably arose by a similar mechanism (10, 24, 25). Unlike LTR retrotransposons, however, TRIM elements lack mobility-related coding sequences such as gag and pol domains (Fig. 1; refs. 3 and 4). A full-length TRIM with coding capacity for mobility-related proteins has not yet been discovered, although the sequencing of the Arabidopsis genome is nearly complete. The lack of an autonomous Arabidopsis TRIM element may merely reflect ecotype distribution or stochastic loss. In any case, TRIM elements probably are mobilized in trans by other retroelements. The mobility of elements lacking coding capacity is well documented. For example, defective or nonautonomous DNA transposons (e.g., class II elements) can be mobilized by functional autonomous element counterparts (1). The short nuclear interspersed element Alu is the most abundant element in the human genome, yet similar to other short nuclear interspersed elements, it has no coding capacity (42). The Bs1 elements of the Ty1-copia group retrotransposons in maize have been active recently but lack a pol domain (43). Recently human long nuclear interspersed elements were shown to mobilize transcripts not derived from long nuclear interspersed element sequences (44). Furthermore, several retroviral vectors have been designed that lack coding capacity and only transpose in the presence of mobility-related proteins in trans (45). The abundance of streamlined nonautonomous elements such as short nuclear interspersed elements and TRIMs in eukaryotic genomes may reflect a unique evolutionary relationship with their autonomous counterparts and the host genome.
A TRIM-like structure found in barley (gi 9623334) may have been generated by a combination of transposition and unequal recombination events among two identical LTR retrotransposons (25). Although this model would not necessarily account for the presence of PBS and PPT, it suggests a mechanism by which the TRIM progenitor might have evolved originally from LTR retroelements. In any case, because retroelement classification relies on internal domains such as gag and pol and no TRIM-related retroelement containing these domains could be identified, we cannot be certain whether TRIM elements are related to Ty1-copia or Ty3-gypsy-like retrotransposons or retroviruses or perhaps originated independently.
The presence of TRIM elements in dicotyledonous and monocotyledonous species indicates that these elements probably are ubiquitous in the plant kingdom. A comparison of the TDR sequences of TRIM elements from seven different plant species indicated significant sequence similarities (Fig. 2; also see Results). This significant sequence similarity among 5′ TDRs is surprising given that LTRs typically share little sequence similarity between members of Ty1-copia or Ty3-gypsy elements in different plant species (see refs. 1 and 4 and references therein). The relative conservation of TDRs of TRIM elements probably reflects selective constraints resulting in the TDRs only containing the minimal set of cis-factors necessary for the retrotransposition process. The high level of conservation in structure and size of TRIM elements from a wide range of plant species together with clear indications of mobility strongly suggest that TRIM elements are a form of streamlined LTR retrotransposon.
TRIM Elements Contribute to the Restructuring of the Host Genomes.
Retrotransposons have been reported to contribute to the coding regions of genes in plants (see refs. 4 and 14 and references therein). Recent data from the Human Genome Sequence has also revealed many cases of bona fide and predicted proteins that contain L1 and Alu sequences (9, 46). Similar to other LTR retrotransposons, there are indications that TRIM elements have been involved intimately in reshaping genomes and in gene evolution. TRIM elements are found dispersed throughout the genome in Arabidopsis and rice (Table 2). Because TRIM elements are comparatively small in size, it can be argued that they are less likely to cause serious disruptions of promoter and intron integrity compared with insertions of the much larger LTR retrotransposons. Alternatively, some LTR retrotransposons have been suggested to sustain deletions immediately after insertion near genes and thereby lessening the impact of transposition events (14, 19, 47). TRIM elements also might be able to contribute more effectively to ORFs of genes (Figs. 4 and 6), especially if the element itself already contains long ORFs (e.g., the Katydid-At1 element on gi 3241926). Apart from the potential direct influence of TRIM elements on genes, they seem to act indirectly as target sites for both LTR and non-LTR retrotransposons in Solanum plants and maize (Table 2). It has been reported previously that LTR retrotransposons act as target sites for the insertions of other retrotransposons in maize (10, 33) and barley (24, 25). Nested insertions may reflect the propagation of these elements into genomic regions where retrotransposons already have been established and thus may limit the deleterious effects of retrotransposon activity on host fitness (4, 10).
The ability of TRIM elements to transduce a host gene was documented for the Katydid-At1 element in A. thaliana (Fig. 5). Together with the maize Bs1 element, these are the only plant examples of retrotransposon-mediated transduction, a phenomenon thought to be limited to the acquisition of proto-oncogenes in viruses (37–40). Recent data from the human genome-sequencing project revealed many examples of long nuclear interspersed elements, specifically L1 elements, with transduced genomic sequences located in their 3′-flanking region (21, 46, 48, 49). Approximately 15–21% of these L1 elements harbor downstream sequences between 30 and 970 bp in length, meaning that ≈1% of the human genome is the result of transduced sequences. Thus, transduction may account in part for the expansion of host genomes and may facilitate exon shuffling, and amplified gene sequences may serve as the raw material for the evolution of new genes.
In conclusion, TRIM elements are the smallest known LTR retrotransposons identified to date and are important players in plant genome evolution. Moreover, like other LTR retrotransposons, they have been involved actively in the restructuring of plant genomes by acting as target sites for retrotransposon insertions by altering host gene structure and by also being capable of transducing host genes. Although only plant TRIM elements have been reported here, it is anticipated that TRIM-like elements would be found in other eukaryotic organisms.
Supplementary Material
Acknowledgments
We thank Nabil Elrouby and John Jones for critical reading of an earlier version of this manuscript. A.K. and C-P.W. acknowledge the funding of the Scottish Executive & Environmental Rural Affairs Department (SEERAD), and C-P.W. also acknowledges the European Commission Training and Mobility of Researchers program. This work was funded in part by grants from the National Science and Engineering Research Council (NSERC) of Canada (to T.B.) and a David Stewart McGill Majors Fellowship (to Q.H.L.).
Abbreviations
- TSD
target site duplications
- LTR
long terminal repeat
- TRIM
terminal-repeat retrotransposons in miniature
- PBS
primer binding site
- PPT
polypurine tract
- RESite
related to empty site
- TDR
terminal direct repeat
- EST
expressed sequence tag
- gi
NCBI GenInfo identifier
- NMD
nonsense-mediated mRNA decay
Footnotes
References
- 1.Kunze R, Saedler H, Lonnig W E. Adv Bot Res. 1997;27:331–470. [Google Scholar]
- 2.Boeke J D, Corces V G. Ann Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- 3.Boeke J D, Stoye J P. In: Retroviruses. Varmus H, Hughes S, Coffin J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 343–345. [PubMed] [Google Scholar]
- 4.Kumar A, Bennetzen J L. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 5.Kidwell M G, Lisch D. Trends Ecol Evol. 2000;15:95–99. doi: 10.1016/s0169-5347(99)01817-0. [DOI] [PubMed] [Google Scholar]
- 6.Doolittle W F, Feng D F, Johnson M S, McClure M A. Q Rev Biol. 1989;64:1–29. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 7.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smit A F. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 9.Li W-H, Gu Z, Wang H, Nekrutenko A. Nature (London) 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- 10.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 11.Pearce S R, Harrison G, Li D, Heslop-Harrison J S, Kumar A, Flavell A J. Mol Gen Genet. 1996;250:305–315. doi: 10.1007/BF02174388. [DOI] [PubMed] [Google Scholar]
- 12.Doolittle W F, Sapienza C. Nature (London) 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 13.Orgel L E, Crick F H C. Nature (London) 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 14.Wessler S R, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 15.McDonald J F, Matyunina L V, Wilson S, Jordan I K, Brown N J, Miller W J. Genetica (The Hague) 1997;100:3–13. [PubMed] [Google Scholar]
- 16.Kazazin H H, Moran J V. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 17.Lister C, Jackson D, Martin C. Plant Cell. 1993;5:1541–1553. doi: 10.1105/tpc.5.11.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R. J Biol Chem. 1998;273:891–897. doi: 10.1074/jbc.273.2.891. [DOI] [PubMed] [Google Scholar]
- 19.Marillonnet S, Wessler S R. Plant Cell. 1997;9:967–978. doi: 10.1105/tpc.9.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodbeck D, Amherd R, Callearts P, Hintermann E, Mayer U A, Affolter M. Cell Biol. 1998;17:621–633. doi: 10.1089/dna.1998.17.621. [DOI] [PubMed] [Google Scholar]
- 21.Moran J V, DeBerardinis R J, Kazazian H H. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal A, Eastman Q M, Schatz D G. Nature (London) 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 23.Végh Z, Vincze E, Kadirov R, Toth G, Kiss G B. Plant Mol Biol. 1990;15:295–306. doi: 10.1007/BF00036915. [DOI] [PubMed] [Google Scholar]
- 24.Vicient C M, Suoniemi A, Anamthawat-Johnson K, Tanskanen J, Beharav A, Nevo E, Schulman A H. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirasu K, Schulman A H, Lahaye T, Schulze-Lefert P. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrov D A. Trends Genet. 2001;17:23–28. doi: 10.1016/s0168-9525(00)02157-0. [DOI] [PubMed] [Google Scholar]
- 27.Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 28.Maizel J V, Lenk R P. Proc Natl Acad Sci USA. 1981;78:7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholas K B, Nicholas H B, Jr, Deerfield D W I. EMBNEW NEWS. 1997;4:14. [Google Scholar]
- 32.Le Q H, Wright S, Yu Z, Bureau T. Proc Natl Acad Sci USA. 2000;97:7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SanMiguel P, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 34.Unseld M, Marienfeld J R, Brandt P, Brenneicke A. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 35.Jacobson A, Peltz S W. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 36.DeMarini D J, Winey M, Ursic D, Webb F, Culbertson M R. Mol Cell Biol. 1992;12:2154–2164. doi: 10.1128/mcb.12.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varmus H E. Annu Rev Genet. 1984;18:553–612. doi: 10.1146/annurev.ge.18.120184.003005. [DOI] [PubMed] [Google Scholar]
- 38.Bureau T E, White S E, Wessler S R. Cell. 1994;77:479–480. doi: 10.1016/0092-8674(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 39.Jin Y-K, Bennetzen J L. Plant Cell. 1994;6:1177–1186. doi: 10.1105/tpc.6.8.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmgren M G. Plant Mol Biol. 1994;25:137–140. doi: 10.1007/BF00023232. [DOI] [PubMed] [Google Scholar]
- 41.Elrouby, N. & Bureau, T. (2001) J. Biol. Chem., in press. [DOI] [PubMed]
- 42.Deininger P L. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 619–636. [Google Scholar]
- 43.Jin Y-K, Bennetzen J L. Proc Natl Acad Sci USA. 1989;86:6235–6239. doi: 10.1073/pnas.86.16.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esnault C, Maestre J, Heidmann T. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 45.Kim H S, Kim S, Robbins P D. Adv Virus Res. 2000;55:545–663. doi: 10.1016/s0065-3527(00)55017-9. [DOI] [PubMed] [Google Scholar]
- 46.International Human Genome Sequencing Consortium. Nature (London) 2001;409:860–921. [Google Scholar]
- 47.White S E, Habera L E, Wesller S R. Proc Natl Acad Sci USA. 1994;91:11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickeral O K, Makalowski W, Boguski M S, Boeke J D. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodier J L, Ostertag E M, Kazazian H H. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.