Abstract
Canine parvovirus-2 (CPV-2) is the most common viral cause of gastroenteritis in puppies in Nigeria and worldwide. After its emergence in 1978, the wild-type CPV-2 was rapidly replaced by three antigenic subtypes CPV-2a, CPV-2b and CPV-2c. Subtype CPV-2a has been reported in Nigeria based on limited number of samples. As such, this study was carried out with 56 faecal samples and four CPV-2 vaccines using polymerase chain reaction and sequencing to have a better idea of CPV-2 subtypes circulating in dogs in Nigeria. 54 (96.4%) out of the 56 samples were positive for CPV-2 subtypes. CPV-2a is the predominant subtype followed by CPV-2b and CPV-2c. Also, co-existence of subtypes CPV-2a and CPV-2b; and CPV-2a and CPV-2c were found in some dogs. Sequence analysis revealed the occurrence of three mutations D413 N, N426D/E, and T440A. To our knowledge, this is the first report of the three subtypes of CPV-2 in Nigeria.
Keywords: Canine parvovirus-2, VP2, Antigenic subtypes, Polymerase chain reaction, Multiple sequence alignment, Nigeria
Canine parvoviral enteritis is endemic in Nigeria and in most parts of the world. Canine parvovirus-2 is the causative agent of canine parvoviral enteritis which is a highly contagious disease of dogs especially puppies characterized by bloody diarrhea and dehydration. The virus is a 26 nm, non-enveloped icosahedral, single-stranded DNA virus with the genome size of about 5200 nucleotides. The genome contains two non-structural proteins (NS1 and NS2) and three structural proteins (VP1, VP2 and VP3) at the 3´ and 5´ ends, respectively [14]. Canine parvovirus-2 was first described in 1978 and was presumed to have emerged from Feline Panleukopenia Virus (FPV) thorough point mutations thereby allowing the virus to replicate in dogs. Since then the virus has spread globally, two new subtype of the virus CPV-2a and CPV-2b appeared in 1979 and 1985, respectively [16] due to mutations in the VP2 region; CPV-2a harbours M87L, I101T, A300G, and D305Y whereas CPV-2b has an additional substitution, N426D. Thereafter, CPV-2c was reported in Italy in 2000, however, with retrospective studies identified in Germany in 1996 [7]. CPV-2c has an amino acid change D426E, which may probably provide this subtype with evolutionary benefits [13]. Canine parvovirus subtypes have been reported in various parts of the world. In North America, CPV 2a and 2b have been detected in the United States and canine parvovirus CPV-2c has been reported by [17] to be widespread in several states. In South America, CPV-2a and CPV-2b have been reported in dogs in Uruguay, Brazil and Argentina [12]. In Europe, all the subtypes have been detected in Italy, Germany, Portugal, Spain, France and United Kingdom [8, 15]. In Asia, all the three subtypes of the virus have also been detected in India, China and Taiwan [5]. In Africa, all three subtypes have been reported to be circulating in dogs in Morocco [1], whereas only CPV-2a and CPV-2b have been reported in South Africa and Mozambique [9, 10]. Canine parvovirus was first detected molecularly by [6] in 2012 in Nigeria. Subsequently, [2, 9] then reported the presence of only CPV-2a subtype. In this study, we explored further CPV-2 subtypes circulating in dogs in Nigeria and CPV-2 vaccines commonly used to vaccinate dogs in Nigeria. Therefore, a total 56 rectal swabs were collected from dogs with cases of gastroenteritis at the Veterinary Teaching Hospital, University of Ibadan between 2012 and 2017. Four canine parvovirus vaccines commonly used in Nigeria were also tested to know the subtypes of the virus they contain. Total DNA was extracted from the samples and the vaccines using DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) following the manufacturer’s instructions. A pair of primers: VP2F: 5′-GCGCAAACAGATGAAAATCA-3′ and VP2R: 5′-CCTTTCCACCAAAAATCTGAG-3′ used in this study were designed using Primer 3 from JustBio (www.justbio.com/hosted-tools.html. Polymerase chain reaction (PCR) amplification was carried out under the following conditions: 94 °C for 2 min for initial denaturation, 35 cycles of 95 °C for 30 s, 46.7 °C for 1 min, 72 °C for 1 min, and final extension at 72 °C for 5 min. The PCR reaction was carried out in triplicate to identify samples containing multiple subtypes of the virus (co-infection). Seven of the amplified DNA fragments from the samples and that of the four vaccines were sequenced and deposited at the GenBank. These nucleotide sequences of the partial VP2 gene of the seven Nigerian and the vaccine CPV-2 genomes were compared with other published CPV-2 sequences already available in the GenBank database using BLAST search via the National Center of Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequence alignment of the partial CPV-2 VP2 gene sequences from the seven Nigerian, vaccines CPV-2 sequences, and the following CPV-2 VP2 sequences retrieved from the GenBank KX192421 (NG36_2a), KX192419 (NG40_2a), HQ602991 (NGR1-10_2a), HQ602995 (NGR15-10_2a), EU659118-USA (Ref_2a), KF385391 (Italy_2a), GU139566 (India_2a), MG583676 (China_2a), KP096430 (Brazil_2a), HQ602977 (South Africa_2a), KR559892-USA (Ref_2b), KF373569 (Italy_2b), FJ005261 (Germany_2b), EU118267(India_2b), KT074338 (China_2b), KP096449 (Brazil_2b), HQ602970 (South Africa_2b), M38245-USA_2c (Ref_2c), JQ429285 (Italy_2c), FJ005204 (Germany_ 2c), KX425920 (India_2c), GU380305 (China_2c), KP096451 (Brazil_2c) and JX416842.1 (Canine Adenovirus-2 E3 (CAV-2) gene as the outgroup) was carried out with clustal W algorithm. Phylogenetic tree was generated using the maximum likelihood method coupled with the Kimura 2-parameter model with bootstrap analysis of 1000 replicates. Phylogenetic and molecular evolutionary analyses were conducted using MEGA, version 7.0.
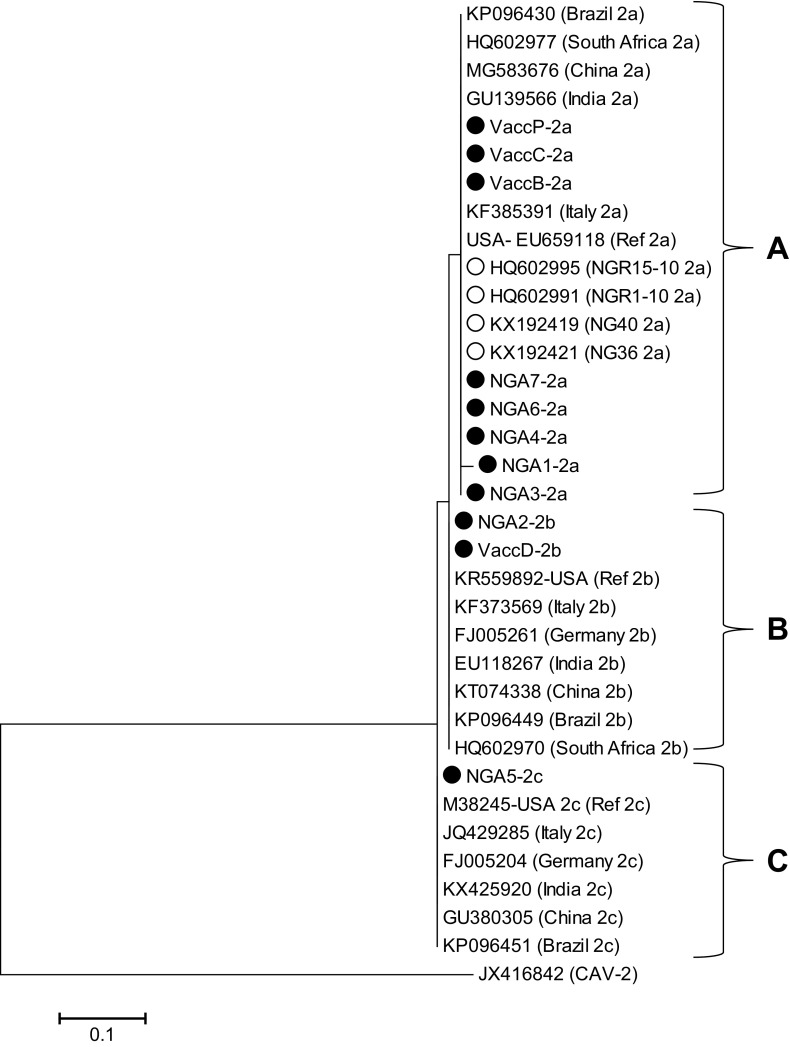
Partial sequences of VP2 gene were amplified from the extracted DNA. 54 (96.4%) out of the 56 tested faecal samples were PCR positive for CPV-2. BLAST search revealed that sequence analysis carried out on all the selected seven positive samples and four vaccines were correspondent to CPV-2 published sequences. The seven CPV-2 detected genomes were designated as NGA1, NGA2, NGA3, NGA4, NGA5, NGA6 and NGA7 and vaccine sequences VaccB, VaccC, VaccD and VaccP have been deposited at the GenBank. Multiple sequence alignments of the nucleotides and the deduced amino acid sequences of the eleven sequences from this study (seven from CPV-2 positive samples and four from the vaccines) and six sequences retrieved from the GenBank NG36_2a, NG40_2a, NGR-10_2a, Ref_2a, Ref_2b and Ref_2c evidenced three non-synonymous mutations. As depicted in Fig. 1a, at position 1239 guanine is substituted by adenine (GAT → AAT). This mutation results in amino acid change D413 N due to a transition in the first position of the codon (Fig. 1b). This change was observed in in the Nigerian and vaccine CPV-2 sequences NGA2_2b, VaccD_2b and Ref_2c as well as in the sequences retrieved from the GenBank but implication of this mutation is not yet known. At positions 1278–1280, a change in the first codon (AAT → GAT) results in the substitution N426D and a change in the last codon (AAT → GAA) results in N426E which are CPV-2b and CPV-2c, respectively. As such from this study, five of the samples (NGA1, NGA3, NGA4, NGA6 and NGA7 and three vaccines (VaccB, VaccC and VaccP) are CPV-2a; one sample (NGA-2) and vaccine (VaccD) are CPV-2b whilst only NGA5 is CPV-2c. Therefore, this study revealed the picture of CPV-2 subtypes circulating in dogs in Nigeria and in the vaccines commonly used in vaccinating dogs in Nigeria. Although, CPV-2a detected in VaccB, VaCCC and VaccP could be wild-type CPV-2 because most vaccines still contain the wild-type CPV-2 [18], and because we amplified and sequenced a fragment of CPV-2 which did not include substitutions M87L, I101T, A300G, and D305Y which distinguish wild-type CPV-2 from the subtype CPV-2a [7]. At position 1320 (Fig. 1a), a change in the first codon (ACA → GCA) results in T440A (Fig. 1b) which is in an exposed region comprising amino acids 267–498 of the VP2 protein, known as the large GH loop. This region exhibits the greatest variability amongst parvoviruses and is considered as the main antigenic site of the virus [3]. Mutation T440A has also been reported [11] and they proposed that viruses evolve to escape the immune system via antigenic drift, and this mutation may be due to vaccine immune pressure and may be responsible for vaccine failure. To corroborate these reports, this mutation was observed in seven out of the 10 Nigerian CPV-2 subtypes sequences employed in multiple sequence alignment but were not observed in the vaccine sequences which is suspicious of reversion of vaccine virus to virulence. Isolates NGA1 (CPV-2a), NGA2 (CPV-2b) and NGA4 (CPV-2a) and NGA5 (CPV-2c) were from dogs with the case file numbers 2014-187 and 2016-484, respectively (Table 1). This indicates co-infection of subtypes within the same animal. The co-infection/co-existence of CPV-2a and N/D426E subtype, in the same animal has previously been reported [4]. These reports confirm that the infection of one animal with several parvoviruses may occur. Phylogenetic analysis using maximum likelihood method inferred from partial VP2 sequences showed that the five Nigerian sequences (NGA1, NGA3, NGA4, NGA6 and NGA7) and vaccine sequences VaccP, VaccC and VaccB belonged to cluster A as other CPV-2a from other parts of the world; NGA-2 and VaccD belongs to cluster B as well as other CPV-2b sequences retrieved from the GenBank while NGA5 belongs to cluster C which contained CPV-2c from other parts of the world (Fig. 1b). This showed that the Nigerian CPV-2 sequences clustering into their respective subtypes, and it further confirms the classification of the virus into their respective subtypes. In conclusion, this study revealed that all three CPV-2 subtypes are circulating in dogs in Nigeria. Continual monitoring and molecular characterization of CPV-2 especially full length of VP2 of clinical samples and vaccines are imperative to recognize mutations that can induce vaccine failure and mechanisms responsible for CPV-2 evolution (Fig. 2).
Fig. 1.
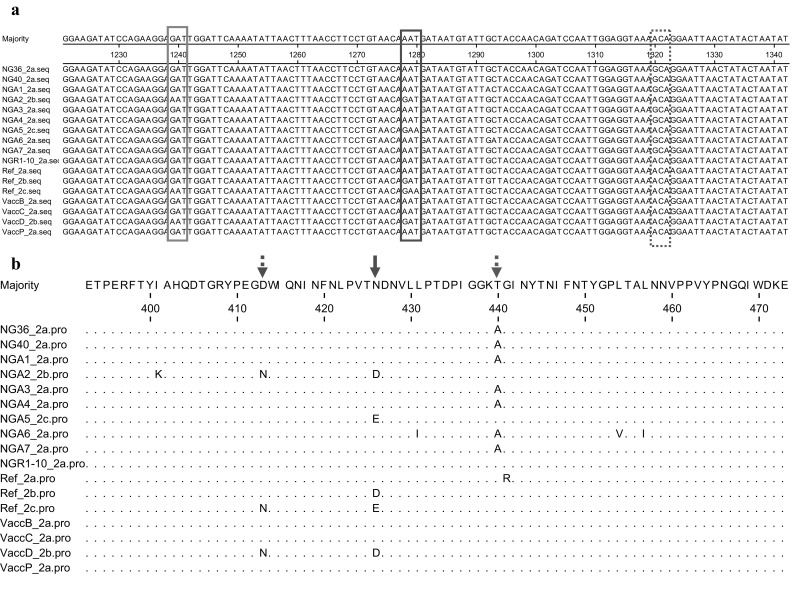
a Multiple sequence alignment of the nucleotide sequences of partial VP2 gene of CPV-2 of Nigerian and vaccine sequences with CPV-2 sequences retrieved from the GenBank. The VP2 of Nigerian and vaccine CPV-2 sequences here analyzed is represented by the sequence of samples NGA1, NGA2, NGA3, NGA4, NGA5, NGA6 and NGA7; VaccB, VaccC, VaccD and VaccP. Reference strains are: CPV-2a (Ref_2a, EU659118); CPV-2b (Ref_2b, KR559892) and CPV-2c (Ref_2c, M38245). The sites of mutations of VP2 gene in the Nigerian strains are at positions 1239-1241, 1278-1280 and 1320-1322 in the boxes. b Multiple sequence alignment of the deduced amino acid sequences of VP2 protein of CPV-2 subtypes. The VP2 of Nigerian and vaccines CPV-2 sequences here analyzed are represented by the sequence of sample NGA1, NGA2, NGA3, NGA4, NGA5, NGA6 and NGA7; VaccB, VaccC, VaccD and VaccP. Reference strains are: CPV-2a (Ref_2a, EU659118); CPV-2b (Ref_2b, KR559892) and CPV-2c (Ref_2c, M38245). Dots indicate position where the VP2 sequences are identical to that of the consensus sequence. The sites of mutations of VP2 protein in the Nigerian and vaccines sequences are at positions 413, 426 and 440
Table 1.
Co-existence of multiple CPV-2 subtypes in the same animal
| Dog case file number/vaccine | Subtype 2a | Subtype 2b | Subtype 2c |
|---|---|---|---|
| 2014-187 | + (NGA1) | + (NGA2) | – |
| 2014-329 | + (NGA3) | – | – |
| 2016-484 | + (NGA4) | – | + (NGA5) |
| 2016-063 | + (NGA6) | – | – |
| 2013-456 | + (NGA7) | – | – |
| VaccB | + | – | – |
| VaccC | + | – | – |
| VaccD | – | + | – |
| VaccP | + | – | – |
Fig. 2.
Phylogenetic analysis of CPV-2 based on VP2 gene nucleotide sequences. Phylogenetic tree was constructed via multiple alignments of nucleotide sequence of VP2 gene from Nigeria CPV-2 and vaccine sequences. Canine adenovirus-2 E3 gene was used as the outgroup. The tree was analyzed by maximum likelihood method with bootstrapping (1000). CPV-2 clusters A, B and C are labeled. Bar, 0.2 nucleotide substitutions per site. CPV-2 sequences from this study have black circles whereas Nigeria CPV-2 sequences retrieved from the GenBank are have white circles
Acknowledgements
This work was supported in part by the AFSCAN grant. The author(s) received no financial support for the authorship and/or publication of this article.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Amrani N, Desario C, Kadiri A, Cavalli A, Berrada J, Zro K, Sebbar G, Colaianni ML, Parisi A, Elia G, Buonavoglia C, Malik J, Decaro N. Molecular epidemiology of canine parvovirus in Morocco. Infect Genet Evol. 2016;41:201–206. doi: 10.1016/j.meegid.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Apaah TT, Daly JM, Tarlinton RE. Canine parvovirus (CPV-2) variants circulating in Nigerian dogs. VetRecOpen. 2016;3:e000198. doi: 10.1136/vetreco-2016-000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battilani M, Ciulli S, Tisato E, Prosperi S. Genetic analysis of canine parvovirus isolates (CPV-2) from dogs in Italy. Virus Res. 2002;83:149–157. doi: 10.1016/S0168-1702(01)00431-2. [DOI] [PubMed] [Google Scholar]
- 4.Battilani M, Gallina L, Vaccari F, Morganti L. Co-infection with multiple variants of canine parvovirus type 2 (CPV-2) Vet Res Commun. 2007;31(Suppl. 1):209–212. doi: 10.1007/s11259-007-0007-6. [DOI] [PubMed] [Google Scholar]
- 5.Chiang SY, Wu HY, Chiou MT, Chang MC, Lin CN. Identification of a novel canine parvovirus type 2c in Taiwan. Virol J. 2016;13(1):160. doi: 10.1186/s12985-016-0620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chollom SC, Fyaktu EJ, Okwori AEJ, Agada GOA, Hashimu G, Akele RY, Voumangai EI, Dash T, Egah DZ. Molecular detection of canine parvovirus in Jos, Nigeria. J Vet Med Anim Health. 2013;5:57–59. [Google Scholar]
- 7.Decaro N, Desario C, Amorisco F, Losurdo M, Elia G, Parisi A, Ventrella G, Martella V, Buonavoglia C. Detection of a canine parvovirus type 2c with a non-coding mutation and its implications for molecular characterisation. Vet J. 2013;196(3):555–557. doi: 10.1016/j.tvjl.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 8.Decaro N, Desario C, Amorisco F, Losurdo M, Elia G, Parisi A, Ventrella G, Martella V, Buonavoglia C. Detection of a canine parvovirus type 2c with a non-coding mutation and its implications for molecular characterisation. Vet J. 2013;196:555–557. doi: 10.1016/j.tvjl.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 9.Dogonyaro BB, Bosman A-M, Sibeko KP, Venter EH, van Vuuren M. Genetic analysis of the VP2-encoding gene of canine parvovirus strains from Africa. Vet Microbiol. 2013;165:460–465. doi: 10.1016/j.vetmic.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 10.Figueiredo J, Miranda C, Souto R, Silva E, Fafetine J, Thompson G. Genetic characterization of canine parvovirus type 2 subtypes in Maputo, Mozambique. Arch Microbiol. 2017;199(4):543–549. doi: 10.1007/s00203-016-1320-7. [DOI] [PubMed] [Google Scholar]
- 11.Geng Y, Guo D, Li C, Wang E, Wei S, Wang Z, Yao S, Zhao X, Su M, Wang X, Wang J, Wu R, Feng L, Sun D. Co-circulation of the rare CPV-2c with unique Gln370Arg substitution, new CPV-2b with unique Thr440Ala substitution, and new CPV-2a with high prevalence and variation in Heilongjiang Province, Northeast China. PLoS One. 2015;10(9):e0137288. doi: 10.1371/journal.pone.0137288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grecco S, Iraola G, Decaro N, Alfieri A, Alfieri A, Gallo Calderón M, da Silva AP, Name D, Aldaz J, Calleros L, Marandino A, Tomás G, Maya L, Francia L, Panzera Y, Pérez R. Inter- and intracontinental migrations and local differentiation have shaped the contemporary epidemiological landscape of canine parvovirus in South America. Virus Evol. 2018;4(1):vey011. doi: 10.1093/ve/vey011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapil S, Cooper E, Lamm C, Murray B, Rezabek G, Johnston L, 3rd, Campbell G, Johnson B. Canine parvovirus types 2c and 2b circulating in North American dogs in 2006 and 2007. J Clin Microbiol. 2007;45:4044–4047. doi: 10.1128/JCM.01300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miranda C, Thompson G. Canine parvovirus: the worldwide occurrence of antigenic variants. J Gen Virol. 2016;97(9):2043–2057. doi: 10.1099/jgv.0.000540. [DOI] [PubMed] [Google Scholar]
- 15.Miranda C, Parrish CR, Thompson G. Epidemiological evolution of canine parvovirus in the Portuguese domestic dog population. Vet Microbiol. 2016;183:37–42. doi: 10.1016/j.vetmic.2015.11.037. [DOI] [PubMed] [Google Scholar]
- 16.Parrish C, Have P, Foreyt WJ, et al. The global spread and replacement of canine parvovirus strains. J Gen Virol. 1988;69:1111–1116. doi: 10.1099/0022-1317-69-5-1111. [DOI] [PubMed] [Google Scholar]
- 17.Pérez R, Bianchi P, Calleros L, Francia L, Hernández M, Maya L, Panzera Y, Sosa K, Zoller S. Recent spreading of a divergent canine parvovirus type 2a (CPV-2a) strain in a CPV-2c homogenous population. Vet Microbiol. 2012;155:214–219. doi: 10.1016/j.vetmic.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Truyen U. Evolution of canine parvovirus–a need for new vaccines? Vet Microbiol. 2006;117:9–13. doi: 10.1016/j.vetmic.2006.04.003. [DOI] [PubMed] [Google Scholar]