The outer membrane of Gram-negative bacteria acts as an effective barrier against toxic compounds, and therefore compromising this structure could increase sensitivity to currently available antibiotics. In this study, we show that the Mla pathway, a system involved in maintaining the integrity of the outer membrane, is genetically and functionally different in Burkholderia cepacia complex species compared to that in other proteobacteria. Mutants in mla genes of Burkholderia cenocepacia or Burkholderia dolosa were sensitive to Gram-positive antibiotics, while this effect was not observed in Escherichia coli or Pseudomonas aeruginosa. The Mla pathway in Burkholderia species may represent an ideal genus-specific target to address their intrinsic antimicrobial resistances.
KEYWORDS: Burkholderia, Burkholderia cenocepacia, Burkholderia dolosa, antibiotic resistance, outer membrane
ABSTRACT
Antibiotic resistance is a threat to our modern society, and new strategies leading to the identification of new molecules or targets to combat multidrug-resistant pathogens are needed. Species of the genus Burkholderia, including the Burkholderia cepacia complex (Bcc), Burkholderia pseudomallei, and Burkholderia mallei, can be highly pathogenic and are intrinsically resistant to multiple classes of antibiotics. Bcc species are nonetheless sensitive to extracellular products released by Pseudomonas aeruginosa in interspecies competition. We screened for Burkholderia transposon mutants with increased sensitivity to P. aeruginosa spent medium and identified multiple mutants in genes sharing homology with the Mla pathway. Insertional mutants in representative genes of the Bcc Mla pathway had a compromised cell membrane and were more sensitive to various extracellular stresses, including antibiotics and human serum. More precisely, mla mutants in the Bcc species Burkholderia cenocepacia and Burkholderia dolosa were more susceptible to Gram-positive antibiotics (i.e., macrolides and rifampin), fluoroquinolones, tetracyclines, and chloramphenicol. Genetic complementation of mlaC insertional mutants restored cell permeability and resistance to Gram-positive antibiotics. Importantly, Bcc mla mutants were not universally weaker strains since their susceptibilities to other classes of antibiotics were unaffected. Although cell permeability of homologous mla mutants in Escherichia coli or P. aeruginosa was also impaired, they were not more sensitive to Gram-positive antibiotics or other antimicrobials as was observed in Bcc mla mutants. Together, the data suggest that the Mla pathway in Burkholderia may play a different biological role, which could potentially represent a Burkholderia-specific drug target in combination therapy with antibiotic adjuvants.
IMPORTANCE The outer membrane of Gram-negative bacteria acts as an effective barrier against toxic compounds, and therefore compromising this structure could increase sensitivity to currently available antibiotics. In this study, we show that the Mla pathway, a system involved in maintaining the integrity of the outer membrane, is genetically and functionally different in Burkholderia cepacia complex species compared to that in other proteobacteria. Mutants in mla genes of Burkholderia cenocepacia or Burkholderia dolosa were sensitive to Gram-positive antibiotics, while this effect was not observed in Escherichia coli or Pseudomonas aeruginosa. The Mla pathway in Burkholderia species may represent an ideal genus-specific target to address their intrinsic antimicrobial resistances.
INTRODUCTION
Bacteria belonging to the genus Burkholderia represent a wide range of metabolically diverse species (1). Members of the Burkholderia cepacia complex (Bcc), known to chronically colonize cystic fibrosis (CF) airways, and the level 3 pathogens Burkholderia pseudomallei and Burkholderia mallei (2) are the most common Burkholderia species associated with deadly infections in humans (1). One hallmark of Burkholderia species is their high intrinsic level of resistance to antibiotics, including to polymyxins (3), which are often used as last-resort antimicrobials against multidrug-resistant Gram-negative pathogens (4). The intrinsic resistance of Burkholderia to several classes of antibiotics is primarily attributed to the very low permeability of its outer membrane (OM), the presence of several efflux pumps, and its unique lipopolysaccharide (LPS) structure (3). For all Gram-negative bacteria, including Burkholderia, the low permeability is the result of the lipid asymmetry of the OM, with LPS at the outer leaflet and phospholipids at the inner leaflet. This particular arrangement is critical for maintaining barrier function of the OM and protection toward extracellular stresses, including antibiotics (5). Furthermore, the atypical LPS structure of Burkholderia directly confers resistance to polymyxins, including colistin (6).
As antibiotic resistance is becoming a pandemic threat, the development of innovative strategies leading to the identification of new molecules or targets against multidrug-resistant pathogens is required (7, 8). We and others have shown that Bcc species are highly susceptible to Pseudomonas aeruginosa toxins, including the secondary metabolites called phenazines (9, 10), which are antibiotics (11). Due to chemical gradients occurring in multispecies communities (12), we speculated that Bcc organisms must have intrinsic resistance mechanisms for tolerating subinhibitory concentrations of P. aeruginosa toxins that should also be required to protect against clinically relevant antibiotics.
Herein, we wanted to take advantage of the natural competitive interactions between P. aeruginosa and the Bcc to identify new drug targets in Burkholderia that are potentially associated to its “intrinsic resistome” (5). We performed a chemical genomics approach to identify hypersensitive transposon mutants of Burkholderia cenocepacia strain K56-2 to toxins/antibiotics (i.e., spent medium) of P. aeruginosa strain PA14.
RESULTS
Identification of B. cenocepacia mutants susceptible to subinhibitory concentrations of P. aeruginosa spent medium.
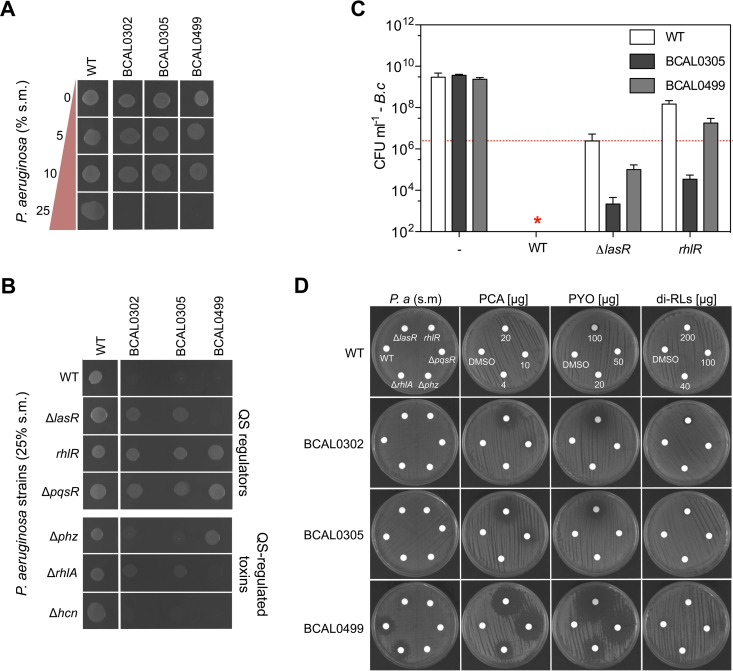
To identify genes or bacterial components associated with the intrinsic resistance of Bcc species to antimicrobials and bacterial toxins, we screened for hypersensitive Burkholderia mutants using P. aeruginosa PA14 supernatant as a source of toxins targeting sensitive B. cenocepacia K56-2 mutants. We made a small random transposon library of ∼3,800 B. cenocepacia K56-2 mutants and visually compared the growth of each mutant colony to that of their wild-type (WT) parent on agar containing spent medium (25%, vol/vol) extracted from P. aeruginosa planktonic cultures (24 h in lysogeny broth [LB]). At this concentration (25%, vol/vol), the growth of WT K56-2 colonies on agar was similar to that without Pseudomonas supernatant (Fig. 1A; also see Fig. S1 in the supplemental material). We identified 20 mutants with growth defects (24 h at 37°C and 72 h at room temperature) specific to agar containing P. aeruginosa spent medium that were not observed on LB agar alone (Table 1 and Fig. S1). Transposon (Tn) insertions affecting genes involved in regulation/signaling (BCAL0499, BCAM1948, and BCAM2426), transport (BCAL0302, BCAL0305, and BCAM2618), lipoprotein (BCAM2620), type IV secretion system (BCAM0324), hypothetical genes, and mobile elements were identified (Table 1). Although we tested a small number of Tn mutants, our mutagenesis was effective, as demonstrated by the selection of three loci with different Tn insertions in multiple neighboring genes (BCAL1079 to BCAL1082, BCAL0302 to BCAL0305, and BCAM2618 to BCAM2620) and three genes with multiple insertions (BCAL0179, BCAL0305, and BCAL3252).
FIG 1.
Sensitivity of B. cenocepacia K56-2 mutants to P. aeruginosa spent medium (s.m.). Killing of B. cenocepacia K56-2 mutants by P. aeruginosa PA14 supernatant is concentration dependent (A) and involves quorum-sensing-regulated molecules on agar (B and D) and in liquid cocultures (C). Panels A, B, and D show representative pictures from at least three experiments. Data from panel C represent means ± standard deviations (SD) from three replicates of coculture competition experiments done in large flasks (see Materials and Methods). The red line in panel C represents CFU of B. cenocepacia WT and mutant strains at time zero (∼2 × 106 CFU · ml−1). The star in panel C indicates that no B. cenocepacia CFU were recovered from the cocultures or that they were below the detection limits (∼200 CFU).
TABLE 1.
B. cenocepacia K56-2 mutants selected for their sensitivity to P. aeruginosa spent medium on agar surface
| Mutant IDb | Tn insertion or J2315 identifiera | Description or function (gene) |
|---|---|---|
| 15G6 | BCAL0179 | Hypothetical protein |
| 36D7 | BCAL0179 | Hypothetical protein |
| 37A8 | BCAL0182 | Putative plasmid recombinase |
| 25E5 | BCAL0302 | ABC transport permease (mlaE) |
| 30G10 | BCAL0305 | ABC transport (mlaC) |
| 2C5 | BCAL0305 | ABC transport (mlaC) |
| 12C7 | BCAL0499 | Two-component response regulator |
| 14C3 | BCAL1048 | Putative alpha/beta hydrolase |
| 37B7 | BCAL1172 | Hypothetical protein |
| 28H12 | BCAL1246 | Putative malonyl coenzyme A-acyl carrier protein |
| 37H7 | (BCAL3225) | Transposase upstream of capsular polysaccharide |
| 2D10 | (BCAL3252) | Transposase |
| 35H10 | (BCAL3252) | Transposase |
| 4E2 | BCAL3336 | Purine ribonucleotide biosynthesis (purH) |
| 8H11 | (BCAM0275a) | Hypothetical protein |
| 13F4 | (BCAM0324) | Type IV secretion system (virB1) |
| 16H4 | BCAM1948 | Redox-sensitive transcriptional regulator (soxR) |
| 24C3 | (BCAM2426) | c-di-GMP phosphodiesterase |
| 17F3 | BCAM2618 | ABC transport periplasmic protein (argT) |
| 33C6 | BCAM2620 | Lipoprotein (yaeC) |
Transposon (Tn) insertion mapping was analyzed using the complete genome sequence and annotation of Burkholderia cenocepacia strain J2315 (13), which is closely related to strain K56-2 (14) and available in the Burkholderia Genome Database (15). Genes in parentheses represent the open reading frames likely affected by the Tn inserted in the intergenic region.
ID, identification.
Genes coding for proteins with diverse functions may be required by B. cenocepacia K56-2 to tolerate sublethal concentrations of P. aeruginosa toxins on an agar surface. Among the selected Tn mutants, different patterns of growth were observed on agar containing P. aeruginosa spent medium. First, the majority of the Tn mutants grew like their WT parent for the first 24 h; however, unlike WT K56-2, the opacity of their colonies faded over time (Fig. S1). On the other hand, the growth of three Tn mutants affecting genes in a locus coding for different ABC transporters (Tn mutant 25E5 [BCAL0302] and Tn mutants 2C5 and 30G10 [BCAL0305]) was severely impaired within the first 24 h, but the opacity of their colonies increased over time (Fig. S1). Lastly, we had one Tn mutant in a two-component regulatory system (Tn mutant 12C7 [BCAL0499]) that was unable to grow over a period of 96 h (Fig. S1). These three genes (BCAL0302, BCAL0305, and BCAL0499) were further characterized in this study.
Insertional mutants (i.e., plasmid insertion via single crossover recombination) in the ABC transporter genes BCAL0302 and BCAL0305, as well as in the two-component response regulator gene BCAL0499, were made in B. cenocepacia strain K56-2. Their hypersensitivity profiles to P. aeruginosa toxins (i.e., spent medium) on agar were confirmed (Fig. 1A), thereby supporting our initial genetic results. Growth of the insertional mutants (now referred to as mutant or mutants for the rest of the text) on Pseudomonas spent medium-containing agar could be rescued by either reducing the concentration of WT spent medium to 10% (vol/vol) or by adding supernatant (25%, vol/vol) extracted from the less toxic P. aeruginosa quorum-sensing (QS) regulator mutants in ΔlasR, rhlR, and ΔpqsR (9). More specifically, the three B. cenocepacia mutants could grow on agar containing supernatant extracted from rhlR and ΔpqsR mutants, while growth on agar with spent medium from the ΔlasR mutant was still toxic for BCAL0499 mutant but not for the BCAL0302 and BCAL0305 mutants (Fig. 1B). Furthermore, BCAL0305 and BCAL0499 mutants were more sensitive than WT K56-2 in liquid cocultures with the less toxic P. aeruginosa ΔlasR and rhlR mutants (Fig. 1C) demonstrating that the hypersensitivity phenotype was exhibited on agar surfaces and in liquid cultures.
Phenazines, rhamnolipids, and hydrogen cyanide (HCN) are known QS-regulated toxins involved in Bcc killing in liquid cocultures (9, 10). Growth on agar containing supernatant extracted from a P. aeruginosa PA14 HCN biosynthesis mutant (ΔhcnABC) was still blocked, suggesting that the B. cenocepacia hypersensitive mutants were susceptible to toxins other than HCN (Fig. 1B). In fact, growth of the BCAL0499 mutant was restored on agar supplemented with spent medium extracted from a phenazine-null mutant of P. aeruginosa (Δphz), while supernatant from a rhamnolipid-null mutant (ΔrhlA) partially restored growth of BCAL0302 and BCAL0305 mutants on agar (Fig. 1B). A disk diffusion assay on agar with phenazine-1-carboxylic acid (PCA) and pyocyanin (PYO), two P. aeruginosa phenazines (11), confirmed that the BCAL0499 mutant was highly susceptible to phenazines compared to WT K56-2 but was not more susceptible to dirhamnolipids (Fig. 1D). Zones of inhibition around disks containing supernatants extracted from WT P. aeruginosa PA14 or different isogenic mutants (ΔlasR, rhlR, ΔpqsR, Δphz, and ΔrhlA mutants) were only observed with BCAL0499 mutant exposed to pigmented spent media (blue-green color; PYO), namely, that from WT and the ΔrhlA mutants (Fig. 1D). Mutants BCAL0302 and BCAL0305 mutants were more sensitive to PCA and PYO than was the WT but were less sensitive than the BCAL0499 mutant (Fig. 1D). However, as suggested by their growth on ΔrhlA agar (Fig. 1B), BCAL0302 and BCAL0305 mutants were more sensitive to dirhamnolipids, as demonstrated by a small zone of inhibition with 200 μg that was not present with WT K56-2 (Fig. 1D).
Altogether, our chemical genetic screen using P. aeruginosa spent medium as a source of small molecules was demonstrated to be an effective strategy in identifying key components of B. cenocepacia K56-2 for tolerating sublethal concentrations of P. aeruginosa-derived toxins. Among those findings, we identified a two-component regulatory system and ABC transporters required for tolerating phenazines and/or rhamnolipids.
The Mla pathway plays a critical role in the intrinsic resistance of B. cenocepacia and Burkholderia dolosa to antibiotics.
To determine whether the selected B. cenocepacia K56-2 transposon mutants (Table 1) were also sensitive to clinically relevant antimicrobials, we first evaluated antibiotics that are not typically used as first line therapy against Bcc species (16) using a disk assay on LB agar.
We tested azithromycin, cefepime, imipenem, and tobramycin, representing the antibiotic classes of macrolides, cephalosporins, carbapenems, and aminoglycosides, respectively. Tn mutants in the ABC transporter genes BCAL0302 (Tn mutant 25E5) and BCAL0305 (Tn mutants 2C5 and 30G10) were sensitive to azithromycin, while the WT parent and all of the other Tn mutants were resistant (see Table S1 in the supplemental material). None of the Tn mutants were more sensitive to the other tested antibiotics than the WT (Table S1). The two-component response regulator BCAL0499 mutant was not more sensitive than the WT to any tested antibiotics (see Table S2 in the supplemental material). We then decided to only investigate further mutants/genes (i.e., BCAL0302 and BCAL0305) where a clear difference in zone of clearing was observed.
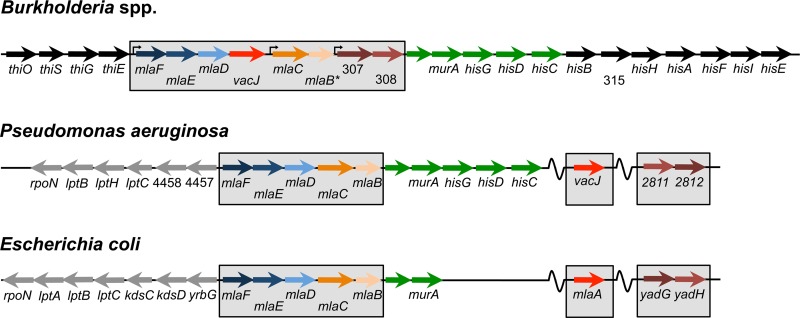
Genes BCAL0302 and BCAL0305 are part of an eight-gene locus coding for ABC transporter systems that share high sequence identities at both gene and protein levels with the Mla pathway (Fig. 2 and Table 2) found in various Gram-negative bacteria, including Escherichia coli, Shigella flexneri, and P. aeruginosa (17–19). Reciprocal BLASTP analyses showed that only MlaB from E. coli K-12 or PA4452 from P. aeruginosa strain PAO1 had no homologue in B. cenocepacia (Table 2). However, based on the genetic organization (Fig. 2) and the presence of sulfate transporter and anti-σ factor antagonist (STAS) domains in BCAL0306, MlaB, and PA4452, we determined that BCAL0306 was the most likely Burkholderia MlaB homologue (Fig. 2 and Table 2). Although genetic similarities are observed between the three species, all Burkholderia genes are in a single and continuous genetic locus, whereas mlaA and vacJ are located elsewhere in the genomes of E. coli and P. aeruginosa, respectively (Fig. 2). For the rest of the study, BCAL0302 and BCAL0305 genes will be referred to as mlaE and mlaC, respectively.
FIG 2.
Genetic organization of the Mla pathways in different bacterial species. Comparison between Burkholderia species and P. aeruginosa strain PAO1 and E. coli K-12 strain MG1655. Colors indicate homologous genes between species based on reciprocal BLASTP and BLASTX analyses. An asterisk represents the likely mlaB homologue in Burkholderia, since no homology was shared with P. aeruginosa or E. coli homologues, but all three genes have STAS domains. Genes with numbers in Burkholderia use the J2315 identifiers, for which “BCAL” was not included on the figure. As for P. aeruginosa, the numbers are based on the PAO1 identifiers and “PA” was also not included on the figure.
TABLE 2.
Similarities between the Mla pathway genes/proteins of Burkholderia, E. coli and P. aeruginosa
| Bcca | Protein (% identity by BLASTP) |
Predicted function | |
|---|---|---|---|
| P. aeruginosab | E. coli K-12 | ||
| BCAL0301 | PA4456 (55) | MlaF (55) | ATP-binding protein |
| BCAL0302 | PA4455 (66) | MlaE (61) | Permease |
| BCAL0303 | PA4454 (47) | MlaD (48) | Periplasmic substrate-binding protein |
| BCAL0304 | VacJ (44) | MlaA (39) | Lipoprotein |
| BCAL0305 | PA4453 (24) | MlaC (26) | Periplasmic binding protein |
| BCAL0306 | PA4452 (0)c | MlaB (0)c | Protein with STAS domain |
| BCAL0307 | PA2812 (42) | YadG (42) | ATP-binding protein |
| BCAL0308 | PA2811 (35) | YadH (38) | Membrane protein |
The organization of the described ABC transporters (Fig. 2) is the same among all Bcc strains within the database (15). For simplicity, we therefore used genes or locus tags of B. cenocepacia strain J2315.
Locus tags of P. aeruginosa strain PAO1 are represented.
Very low similarity at the amino acid level, but proteins from the three species have a STAS domain.
Using the mlaE and mlaC mutants, we further demonstrated their increased sensitivity to tetracyclines (tetracycline, doxycycline, and tigecycline), chloramphenicol, macrolides (azithromycin, clarithromycin, and erythromycin), fluoroquinolones (ciprofloxacin and levofloxacin), and rifampin (Tables 3 and S2). Importantly, the sensitivity of mlaE and mlaC mutants to penicillins, cephalosporins, carbapenems, monobactams, aminoglycosides, oxazolidinones, lincosamides, and fosfomycin remained the same as that of WT K56-2 (Tables 2 and S2). These results strongly suggest that mutations in the Mla pathway did not necessarily lead to generally weak mutants susceptible to all antimicrobials, but rather to those susceptible to class-specific antibiotics.
TABLE 3.
Antibiotic resistance profile of B. cenocepacia K56-2 mla mutantsa
| Antibiotic target | Antibiotic category | Antibiotic class | Antibiotic | Antibiotic amt (μg) | Resistance profile of B. cenocepacia K56-2b |
||
|---|---|---|---|---|---|---|---|
| WT | mlaE | mlaC | |||||
| Cell wall synthesis | Beta-lactams | Penicillins | Ampicillin | 25 | R | R | R |
| Amoxicillin-clavulanate | 30 | R | R | R | |||
| Piperacillin-tazobactam | 110 | S | S | S | |||
| Cephalosporins | Cefepime | 30 | S | S | S | ||
| Cefoxitin | 30 | R | R | R | |||
| Ceftazidime | 30 | S | S | S | |||
| Ceftriaxone | 30 | S | S | S | |||
| Carbapenems | Imipenem | 10 | S | S | S | ||
| Meropenem | 10 | S | S | S | |||
| Monobactams | Aztreonam | 30 | S | S | S | ||
| Protein synthesis | 30S | Aminoglycosides | Tobramycin | 10 | R | R | R |
| Amikacin | 30 | R | R | R | |||
| Tetracyclines | Tetracycline | 30 | S | ↑S | ↑S | ||
| Doxycycline | 30 | S | ↑S | ↑S | |||
| Tigecycline | 15 | S | ↑S | ↑S | |||
| 50S | Oxazolidonones | Linezolid | 30 | R | R | R | |
| Chloramphenicol | 30 | S | ↑S | ↑S | |||
| Macrolides | Azythromycin | 15 | R | ↑↑S | ↑↑S | ||
| Clarithromycin | 15 | R | ↑↑S | ↑↑S | |||
| Erythromycin | 15 | R | ↑↑S | ↑↑S | |||
| Lincosamides | Clindamycin | 2 | R | R | R | ||
| DNA topoisomerases | Fluoroquinolones | Ciprofloxacin | 5 | S | ↑S | ↑S | |
| Levofloxacin | 5 | S | ↑S | ↑S | |||
| mRNA synthesis | Rifampin | 5 | R | ↑↑S | ↑↑S | ||
| Other | Fosfomycin | 50 | R | R | R | ||
Summary of the raw data with zones of clearing measurements (mm) is shown in Table S2.
Bacterial strains were considered resistant (R) when no zone of clearing was observed, while any clearing was recorded as sensitive (S). When the susceptibility of mlaE or mlaC mutant strains was greater than that of the WT (i.e., greater zone of clearing), their hypersensitivity was labeled as ↑S. On the other hand, mlaE or mlaC mutant strains that were sensitive to antibiotics while the WT was resistant were labeled as ↑↑S.
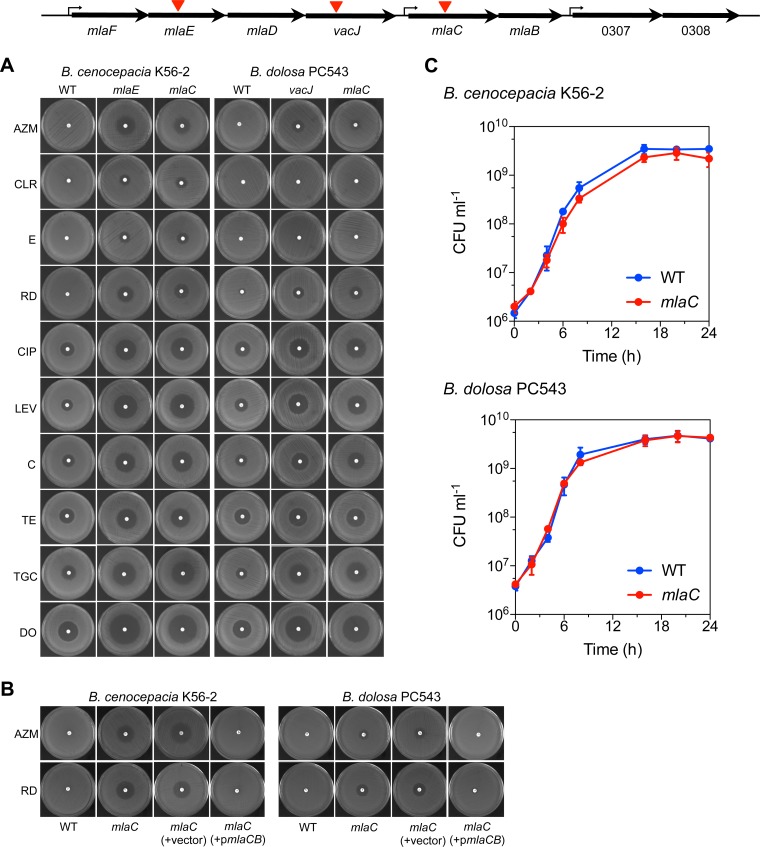
In a recent study, we identified a vacJ Tn mutant of B. dolosa strain PC543 that was unable to modify its morphotype or to swarm upon exposure to small amounts of rhamnolipids (20). Here, we showed that the vacJ Tn mutant and a newly made mlaC insertional mutant in B. dolosa PC543 had similar increased sensitivity to macrolides, tetracyclines, fluoroquinolones, and rifampin (Fig. 3A). Those results were recapitulated in cultures of B. cenocepacia K56-2 and B. dolosa PC543 mlaC mutants compared to those of the WT (Table 4). The growth in liquid culture over 24 h of the mlaC mutants was comparable to that of their respective WT parent (Fig. 3C). The lack of general growth defect by the mutants could not explain the increased sensitivity to specific antibiotics. The mlaCB genes are predicted to be transcribed as a 2-gene operon (Fig. 2 and 3) (15). Consequently, genetic complementation of the mlaC mutants in B. cenocepacia K56-2 and B. dolosa PC543 was carried out with a plasmid containing the mlaCB genes under the control of their native promoter. Complementation of the mlaC mutants restored the resistance to rifampin and azithromycin to WT level in both B. cenocepacia K56-2 and B. dolosa PC543 (Fig. 3B). These data genetically demonstrate that the Mla pathway participates in the intrinsic resistance of Burkholderia to Gram-positive antibiotics.
FIG 3.
The impact of the Bcc Mla pathway on antibiotic resistance. (A) Disk diffusion assay demonstrating the increased susceptibility of the mlaC and mlaE insertional mutants or the vacJ Tn mutant of B. cenocepacia K56-2 or B. dolosa PC543, indicated by the red triangles in the Mla genetic region map inset, to 10 antibiotics representing 5 different classes (macrolide, rifampin, fluoroquinolone, chloramphenicol, and tetracycline). (B) Genetic complementation of the mlaC insertional mutants with pHERD26T-mlaCB (pmlaCB) or with the empty pHERD26T (vector) using the Gram-positive antibiotics azithromycin and rifampin. (C) mlaC does not impact growth of B. cenocepacia K56-2 or B. dolosa PC543 in large shaken LB cultures. Panels A and B show representative pictures from at least three experiments. Data from panel C represent means ± SD from three replicates. AZM, azithromycin; CLR, clarithromycin; E, erythromycin; RD, rifampin; CIP, ciprofloxacin; LEV, levofloxacin; C, chloramphenicol; TE, tetracycline; TGC, tigecycline; and DO, doxycycline.
TABLE 4.
Impact of mlaC on the antibiotic sensitivity of B. cenocepacia and B. dolosa in liquid cultures
| Antibiotic | MIC (μg · ml−1) for: |
|||
|---|---|---|---|---|
|
B. cenocepacia K56-2 |
B. dolosa PC543 |
|||
| WT | mlaC | WT | mlaC | |
| Azythromycin | 256 | 16 | >256 | 64 |
| Erythromycin | >256 | 16 | >256 | 128 |
| Rifampin | 32 | 1 | 32 | 4 |
| Ciprofloxacin | 2 | 0.5 | 2 | 0.5 |
| Chloramphenicol | 16 | 4 | 32 | 8 |
| Tetracycline | 8 | 2 | 4 | 2 |
| Ceftazidime | >256 | >256 | >256 | >256 |
| Tobramycin | >256 | >256 | >256 | >256 |
Notably, our data demonstrate that mutations in the Mla pathway render Bcc sensitive to typical Gram-positive antibiotics, such as macrolides and rifampin. We rescreened the Tn library in B. cenocepacia K56-2 and the 34 B. dolosa PC543 Tn mutants, which were previously selected for their inability to differentiate morphologically upon exposure to rhamnolipids (20) and identified BCAM2829 and bamC as genes also required for azithromycin and rifampin resistance (Fig. S2). BCAM2829 is a VacJ-like lipoprotein (VacJ2), while BamC is a predicted lipoprotein that participates in the assembly and insertion of outer membrane β-barrel proteins in E. coli (21). At the amino acid level, VacJ2 is 54% identical to the VacJ lipoprotein of the Mla pathway.
Together, our data demonstrate that the Mla pathway is critical for the intrinsic antibiotic resistance to macrolides, rifampin, fluoroquinolones, tetracyclines, and chloramphenicol in both B. cenocepacia and B. dolosa. On the other hand, the antibiotic resistance profile of the two-component regulatory system mutant BCAL0499 was similar to that of its WT parent, demonstrating that this mutation primarily affected tolerance to small molecules like phenazines but not to antibiotics (Table S2). Although vacJ2 and bamC were directly selected with azithromycin, our original chemical genetic screen using P. aeruginosa supernatant directly led to the identification of the Mla pathway as a major determinant of the Bcc intrinsic resistome.
Bcc mutants in the Mla pathway have a defective cell permeability.
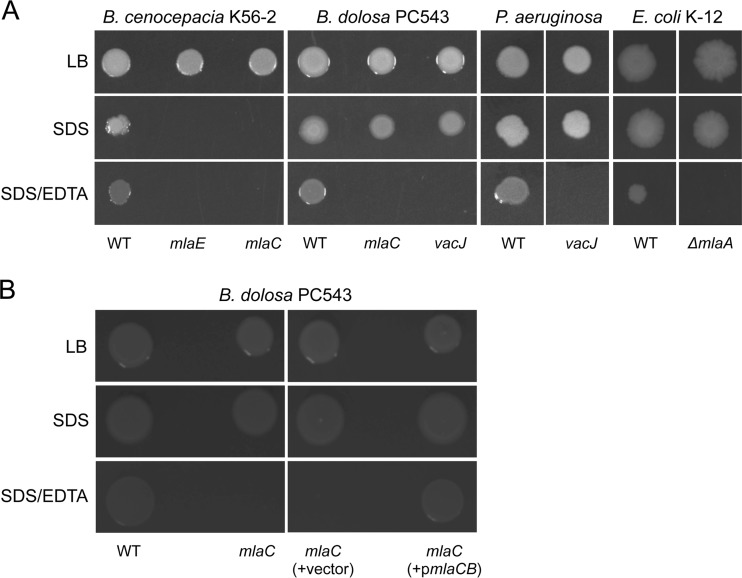
Studies in E. coli have shown that the Mla pathway is involved in lipid homeostasis in the outer leaflet and is consequently responsible for maintaining the barrier function of the outer membrane (OM) in Gram-negative bacteria (18, 22–24). Consistent with those findings, the sensitivity of Bcc mla or vacJ mutants to Gram-positive antibiotics, such as macrolides and rifampin (Fig. 3A), would also suggest a potential outer membrane defect.
As an indicator of cell permeability, we performed an SDS-EDTA sensitivity assay to determine whether the Mla pathway played a role in the OM permeability of members of the Bcc. By chelating cations, EDTA destabilizes LPS interactions, and bacterial death occurs upon exposure to the detergent SDS when the OM is compromised. B. cenocepacia K56-2 mutants (i.e., mlaC and mlaE mutants) were highly sensitive to SDS (0.025%, wt/vol) (Fig. 4A). In B. dolosa PC543 background, the mlaC mutant and the vacJ Tn mutant were resistant to 0.025% (wt/vol) SDS, but unlike their WT parent, they were unable to grow on SDS-containing agar in the presence of EDTA (Fig. 4A). Genetic complementation of the mlaC mutant with a plasmid containing the mlaCB genes restored the ability of the mutant strain to grow on SDS-EDTA agar like its WT PC543 parent (Fig. 4B). The vacJ2 Tn mutant had an intermediate sensitivity to SDS-EDTA, with a 1.5-log reduction in survival compared to that of its WT K56-2 parent. On the other hand, the SDS-EDTA sensitivity of the bamC Tn mutant in B. dolosa PC543 could not be evaluated, since it was highly sensitive to SDS like B. cenocepacia K56-2 mla mutants (data not shown).
FIG 4.
Mutants in the Mla pathway have impaired permeability. (A) Outer membrane permeability was assessed by the SDS-EDTA sensitivity assay using representative mla or vacJ mutants in different species, including B. cenocepacia K56-2, B. dolosa PC543, P. aeruginosa PA14, and E. coli K-12 (strain BW25113). (B) Genetic complementation of the mlaC insertional mutant in B. dolosa PC543 with pHERD26T-mlaCB (pmlaCB) or with the empty pHERD26T (vector). The growth or absence of growth from 2 μl of undiluted cultures spotted on agar is represented. The final concentrations of SDS and EDTA were 0.025% (vol/vol) and 0.55 mM, respectively. Pictures were taken after 48 h at 37°C.
As suggested by the SDS-EDTA sensitivity results (Fig. 4A), mutations in the Mla pathway disrupt the OM permeability of the Bcc, P. aeruginosa, and E. coli, but they do not have the same effect on antibiotic susceptibility. For instance, mla or vacJ mutants in E. coli or P. aeruginosa were not more sensitive to macrolides, rifampin, fluoroquinolones, chloramphenicol, and tetracyclines (see Tables S3 and S4 in the supplemental material). Altogether, our data strongly suggest that mutations in the Burkholderia Mla pathway or in vacJ2 increase OM permeability (Fig. 4A) and likely enhance susceptibility to several classes of antibiotics (Fig. 3 and Table 3).
Bcc mutants in the Mla pathway have impaired motility and colony development on agar.
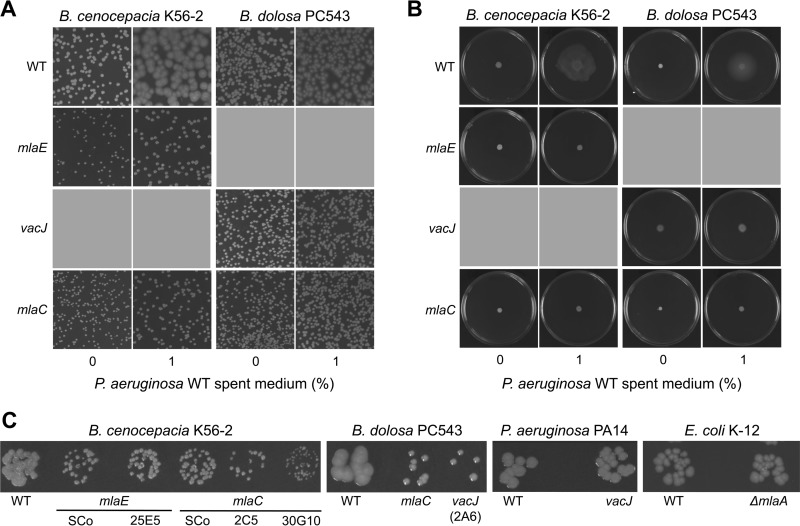
We previously showed that B. dolosa PC543 and B. cenocepacia K56-2 exhibited different colony morphotypes and swarming behaviors when exposed to P. aeruginosa spent medium or rhamnolipids (20). As a vacJ Tn mutant failed to demonstrate these phenotypes (20), we next showed that mlaC and mlaE mutants also did not differentiate morphologically (Fig. 5A) and could not swarm (Fig. 5B) upon exposure to P. aeruginosa supernatant-containing rhamnolipids. In addition, colonies of mlaC, mlaE, or vacJ mutants were smaller in size compared to those of their respective WT parents (Fig. 5C). Interestingly, homologue mutants of vacJ in P. aeruginosa PA14 or ΔmlaA in E. coli K-12 did not have smaller colonies like those observed in B. cenocepacia K56-2 or B. dolosa PC543 (Fig. 5C). Data on antibiotic resistance, motility, and colony size tend to show that the Mla pathway has a different impact in the biology of Burkholderia species compared to that in P. aeruginosa and E. coli.
FIG 5.
Bcc mla or vacJ mutants have impaired colony morphotype, colony size, and motility. (A and B) Colony morphotype (A) and swarming motility (B) upon exposure to P. aeruginosa PA14 spent medium of different mla or vacJ mutant representative strains in B. cenocepacia K56-2 or B. dolosa PC543. (C) mla or vacJ mutants have smaller colonies on LB agar compared to WT in B. cenocepacia K56-2 and B. dolosa PC543 but not in P. aeruginosa PA14 or E. coli K-12 K-12 (strain BW25113). Tn mutant numbers are labeled below mlaE, mlaC, or vacJ. All pictures were taken after 48 h at 37°C. SCo, single crossover mutant.
The Bcc Mla pathway is required for host innate resistance.
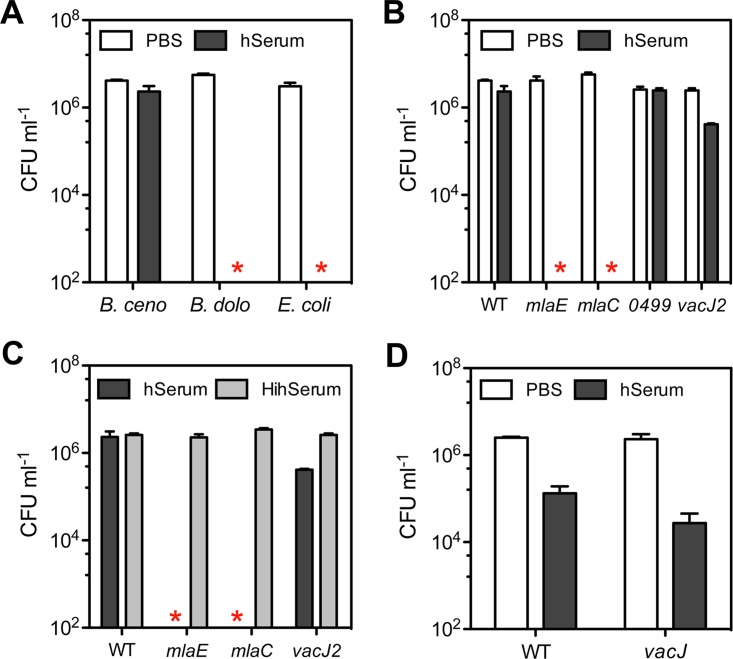
Some studies have demonstrated that the Mla pathway is important for resisting the bactericidal effect of human serum in various bacterial pathogens (19, 25–27). To determine whether a similar role exists in the Bcc, we collected and pooled serum from five healthy donors (4 females and 1 male) and tested the killing activity of the serum over a period of 2 h at a final concentration of 30%.
Prior to evaluating Bcc mutants in the Mla pathway, we first evaluated serum resistance of WT B. cenocepacia K56-2 and B. dolosa PC543 in comparison to that of the sensitive E. coli K-12 strain BW25113. B. cenocepacia K56-2 was resistant to human serum, as previously reported (28, 29), while B. dolosa PC543 was as sensitive as E. coli K-12 (Fig. 6A). Consequently, mlaC and vacJ mutants in B. dolosa PC543 were not evaluated due to the high serum sensitivity of WT PC543. Mutants mlaE and mlaC in B. cenocepacia K56-2 were highly sensitive to human serum, as no viable colonies could be recovered after 2 h (Fig. 6B). On the other hand, the two-component regulatory system mutant BCAL0499 was resistant, while the vacJ2 Tn mutant selected for its sensitivity to azithromycin was slightly sensitive to serum, with a close to 1-log-unit reduction in survival (Fig. 6B). Heat inactivation of the serum restored bacterial viability of mlaE, mlaC, and vacJ2 mutants to WT levels (Fig. 6C) suggesting that heat-labile components of human serum, such as complement, are likely involved in the killing activity. Although P. aeruginosa PA14 was more sensitive to serum than was B. cenocepacia K56-2, the impact of a compromised Mla pathway on serum resistance was greater in B. cenocepacia (Fig. 6B) compared to that in P. aeruginosa (Fig. 6D).
FIG 6.
The role of the Mla pathway in serum resistance. (A) Resistance to 30% human serum (hSerum) over a period of 2 h was evaluated for B. cenocepacia K56-2 (B. ceno), B. dolosa PC543 (B. dolo), and E. coli K-12 strain BW25113 (E. coli). (B) Resistance to 30% hSerum of the mlaC, mlaE, and BCAL0499 insertional mutants and the vacJ2 Tn mutant in comparison to WT B. cenocepacia K56-2. (C) The killing activity of 30% heat-inactivated human serum (HihSerum) was tested with mlaC and mlaE insertional mutants, the vacJ2 Tn mutant, and WT B. cenocepacia K56-2 and compared to that of hSerum. (D) Resistance to 30% hSerum of a vacJ mutant compared to its WT P. aeruginosa PA14 parent strain. Stars represent the absence of recovered CFU or below the detection limits (∼200 CFU). All data represent means ± SD from three replicates.
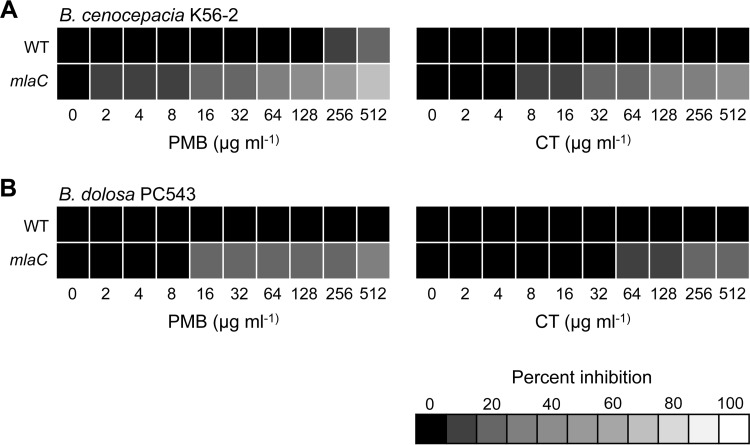
In addition to serum and/or complement, cationic antimicrobial peptides (AMPs) represent another nonspecific innate host response toward pathogens. Here, we evaluated the impact on the resistance to AMPs of the mlaC mutation in both B. cenocepacia K56-2 and B. dolosa PC543. We used polymyxin B (PMB) and colistin as proxies of host AMP susceptibility since the resistance profile of commensal and pathogenic bacteria to PMB or colistin is similar to that to mammalian-derived AMPs (30). Burkholderia species are highly resistant to AMPs (6), and disruption of mlaC did not lead to complete killing in both B. cenocepacia and B. dolosa but rather led to a dose-dependent growth inhibition for both PMB and colistin (Fig. 7). Although WT B. dolosa PC543 is intrinsically more resistant to PMB than WT B. cenocepacia K56-2, the reduced growth of the mlaC mutant had the same trend in both genetic backgrounds in the presence of increasing concentrations of PMB or colistin (Fig. 7).
FIG 7.
The impact of the Mla pathway on resistance to polymyxins. MIC determination in liquid cultures (LB) for WT and mlaC mutants in B. cenocepacia K56-2 (A) and B. dolosa PC543 (B) genetic background against polymyxin B (PMB) and colistin (CT).
Altogether, our results suggest that the Bcc Mla pathway is likely required to effectively tolerate the bactericidal action of nonspecific host innate immune responses, such as complement and AMPs.
DISCUSSION
The intrinsic resistance of many Gram-negative bacteria to antibiotics involves several mechanisms, including low OM permeability and multidrug efflux pumps (5, 31). A large proportion of essential genes in Burkholderia species are involved in maintaining the integrity of the cell envelope, representing potential new drug targets (32). Here, we applied a simple chemical genetic screen to identify components of the intrinsic resistome of Burkholderia to subinhibitory concentrations of P. aeruginosa toxins. Among the determinants identified as being required to tolerate Pseudomonas toxins and antibiotics in B. cenocepacia K56-2 is the Mla pathway, a system shown to maintain the lipid asymmetry of the OM in E. coli and consequently its primary function as a barrier to toxic compounds (18, 23, 33). Although the Mla pathway is not essential in B. cenocepacia K56-2 (32), it was proposed to be essential in B. cenocepacia strain J2315 (34). Even though these differences could be attributed to strain-specific factors, it further demonstrates that the Mla pathway plays an essential role in the biology of Burkholderia and could be used as a potential drug target.
Our study showed that the genetic organization (Fig. 2) and phenotypes associated with the Mla pathways, such as antibiotic resistance, differed greatly between bacterial species. For example, although mla or vacJ mutants in E. coli or P. aeruginosa have increased cell permeability (18, 19, 35), they are not more sensitive to Gram-positive antibiotics (see Tables S3 and S4 in the supplemental material) (18), as described in this study for Bcc species (Fig. 3 and Table 3). Similar to the Bcc, vacJ mutants in Actinobacillus pleuropneumoniae and Haemophilus parasuis are more sensitive to various antibiotics, including tilmicosin (27, 36), a macrolide antibiotic used in veterinary medicine. Together, these differences in antibiotic susceptibility suggest that factors other than cell permeability that would be species specific might be required by Gram-negative bacteria to be resistant to Gram-positive antibiotics. Supporting this possibility, synergy between efflux pumps and OM diffusion, which is antibiotic and species specific, occurs in Gram-negative bacteria (31, 37). In E. coli, MlaA interacts with the OmpF/C porins (22), demonstrating that Mla proteins can physically interact with other systems in the OM that are not necessarily involved in transport of phospholipids. Could the Mla pathway interact with efflux pumps in Burkholderia species and therefore reduce antibiotic efflux in the absence of Mla proteins, which would then be necessary for proper anchoring of pump-associated proteins in the OM?
Burkholderia species harbor several efflux pumps, of which only a few have been partially characterized (38). The BpeAB-OprB efflux pump in Burkholderia pseudomallei strain 1026b is known to impact multiple antibiotics (i.e., macrolides, tetracyclines, fluoroquinolones, chloramphenicol, and rifampin) (39), and the mla mutants in B. cenocepacia and B. dolosa were found to be more sensitive to these antibiotics. A BpeAB-OprB homologue (BCAL2020 to BCAL2022) in B. cenocepacia strain J2315 is required for resistance to macrolides and fluoroquinolone but also to tobramycin and aztreonam (40), two antibiotics for which mla mutants in B. cenocepacia K56-2 were unaffected (Table 3). These differences could be strain specific, as was previously described in B. pseudomallei (39, 41), or involved more than one efflux pump systems if they are indeed interacting with Mla proteins. Interestingly, another copy of the bpeAB-oprB gene (BCAS0764 to BCAS0766) is present in B. cenocepacia and a few other Bcc species but not in B. dolosa (15). If this pump interacts with the Mla pathway, it might explain the high sensitivity of mla mutants to SDS in B. cenocepacia K56-2 but not in B. dolosa PC543. Whether efflux pumps truly interact with the Mla pathway in Burkholderia or in other species remains an unanswered question that would require further investigation. If true, it may help to elucidate differences in antibiotic resistance between species in mla null strains.
In most species, the organization of the mla genes is discontinuous, with the mlaFEDCB genes located together, while mlaA or vacJ is located elsewhere on the genome (Fig. 2). In Burkholderia, the mla genes are all continuous, with the mlaA or vacJ homologue located between mlaD and mlaC. Besides this difference, two additional genes, BCAL0307 and BCAL0308, are located downstream of mlaFEDACB (Fig. 2), suggesting that they may be part of the Mla pathway and possibly interacting with the other Mla proteins. Supporting this possibility, the BCAL0308 homologue protein in E. coli, YadH, was recently shown to interact and form a protein complex with MlaA, and it participates in phospholipid trafficking in order to maintain OM lipid asymmetry (33). Like the ΔmlaA mutant, a yadH mutant was hyperpermeable and not more sensitive to erythromycin (33), which is in agreement with our data (see Table S3 in the supplemental material). Although we do not have any experimental evidence in Burkholderia, we can speculate that BCAL0307 and BCAL0308 are possibly interacting with Mla proteins like YadH in E. coli.
In addition to OM permeability and antibiotic resistance, genetic studies in several species have shown that the Mla pathway also affects biofilm formation, outer membrane vesicle formation, cell morphology, survival, and pathogenesis in animals (17, 19, 27, 35, 36, 42–44). Here, we showed that it also affects swarming motility (Fig. 5B) as well as colony size (Fig. 5C), demonstrating the pleiotropic role of the Mla pathway in the biology of Burkholderia. In addition to the phenotypes stated above, we also showed that mlaC or mlaE was fully required for serum resistance, while vacJ2 was partially required in the serum-resistant strain B. cenocepacia K56-2 (Fig. 6). These results are in agreement with those of other studies, in which vacJ or mla genes conferred various levels of protection against human serum components (e.g., complement) in different species, including P. aeruginosa, Haemophilus influenzae, Actinobacillus pleuropneumoniae, B. pseudomallei, and Burkholderia thailandensis (19, 25–27). Capsule polysaccharides and extracellular polysaccharides (EPS) are bacterial factors that can interfere with the complement cascade (45), but the presence of EPS was not associated with serum resistance in Bcc strains (29). Although mechanisms of serum resistance in the Bcc are not fully understood, a few genetic studies have identified determinants conferring resistance to serum killing in B. cenocepacia K56-2, such as a complete LPS structure (28) and a trimeric autotransporter (46). The impaired growth of the mla mutants in the presence of PMB or colistin (Fig. 7) might suggest a potential LPS defect, since the loss of lipid A-core oligosaccharide heptoses and not that of O antigen was shown to reduce PMB resistance (47). However, the mla mutants were still viable with PMB or colistin at concentrations up to 512 μg · ml−1, which suggests that the LPS structure was minimally affected or that other unknown resistance mechanisms were still intact to maintain a high level of polymyxin resistance, considering that the MIC of enteropathogens toward polymyxins is typically below 1 μg · ml−1 (30).
Overall, our genetic study suggests that the Mla pathway may represent a Burkholderia-specific target to potentiate current antibiotics (48–50). The fact that the Mla pathway is present and is genetically similar in more than 675 Burkholderia sequenced strains (15), including the level 3 pathogens B. mallei and B. pseudomallei, for which the functionality seems similar to that of the Bcc (25), increases the value of developing a combination therapy against these multidrug-resistant pathogens by exploiting the Mla pathway. Therefore, targeting the Mla pathway with an antibiotic adjuvant (e.g., nonantibiotic molecule) in combination with macrolides, for example, should enhance Burkholderia killing and avoid enhanced killing of commensal bacteria, such as E. coli. In addition to characterizing the mode of action of potential nonantibiotic molecules used in combination therapy, Bcc mla mutant strains could also be used as a high-throughput screening tool to find new antimicrobial compounds that could be further developed for treating Burkholderia infections.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are described in Table S5 in the supplemental material. Bacterial strains were routinely grown in LB-Miller broth or on 1.5% LB-Miller agar (EMD Chemicals Inc., Gibbstown, NJ), supplemented with antibiotics when appropriate and incubated at 37°C. M9 minimal medium (Becton, Dickinson and Company [BD], Sparks, MD) containing 0.5% Casamino Acids (M9CAA) was used for swarming motility assays, as previously described (20). Antibiotics were added to the culture medium of Pseudomonas aeruginosa background strains, when appropriate, at the following concentrations: tetracycline (Tet) at 150 μg · ml−1 and gentamicin (Gm) at 50 μg · ml−1. Trimethoprim (Tp) was added at 100 μg · ml−1 for strains of the Bcc and Escherichia coli. Gm and kanamycin (Km) were used at 10 and 50 μg · ml−1, respectively, for E. coli. Tet was added at 5 μg · ml−1 for E. coli, while between 50 to 150 μg · ml−1 was added for Burkholderia mlaC mutants carrying Tetr plasmids (see “Cloning of malCB and complementation of the mlaC insertional mutants,” below). Bacterial stocks were frozen and stored at −80°C in 10% skim milk (BD). Phenazine-1-carboxylic acid (PCA) was purchased from Princeton BioMolecular Research (catalogue no. PBMR030094; Princeton, NJ) and dissolved in dimethyl sulfoxide (DMSO) at a final concentration of 2 mg · ml−1. Pyocyanin (PYO) was purchased from Sigma-Aldrich (catalogue no. P0046; St. Louis, MO) and dissolved in DMSO at a final concentration of 10 mg · ml−1. All chemicals used in this study were purchased from Sigma-Aldrich (St. Louis, MO).
Extraction of bacterial spent medium.
Bacterial spent medium was extracted as previously described (9, 20). Briefly, bacterial cells from overnight cultures (4 ml of LB medium in borosilicate glass test tubes) were spun down (2 min at 8,000 rpm) and washed once in phosphate-buffered saline (PBS). Flasks containing 25 ml of culture medium were inoculated with 80 μl of washed cells (1:300 dilution) and incubated for 24 h at 37°C with shaking (175 rpm). Bacterial cells were subsequently spun down, and the resulting supernatant was filtered using 0.2-μm filters (Millipore) and stored at 4°C.
Genetic manipulations.
Restriction enzymes, T4 DNA ligase, and Phusion high-fidelity DNA polymerase were obtained from New England BioLabs (Ipswich, MA).
(i) Construction of single crossover insertional mutants in B. cenocepacia K56-2 and B. dolosa PC543.
Insertional mutants in mlaE (BCAL0302), mlaC (BCAL0305), and BCAL0499 were carried out using the suicide vectors pMQ87Tp-mlaE, pMQ87Tp-mlaC, and pMQ87Tp-BCAL0499, respectively (Table S5). These vectors were created using the primer pairs BCAL0302_5L-3L, BCAL0305_5L-3L, and BCAL0499_5L-3L, respectively (see Table S6 in the supplemental material). An internal DNA fragment (∼500 to 700 bp) from the gene to be mutated was PCR amplified from B. cenocepacia strain K56-2 and subsequently cloned into pMQ87Tp (linearized with SmaI) via homologous recombination in Saccharomyces cerevisiae, as previously described (51). To take advantage of the efficient homologous recombination of S. cerevisiae, PCR primers were 60 bp long each, with 40 bp matching to the DNA sequence flanking each side of the SmaI site used to linearize pMQ87Tp. In addition to a bacterial origin of replication, this plasmid also carries an origin of replication for S. cerevisiae. Auxotrophy to uracil is used as a selection marker in yeast for the recombined plasmid. The resulting plasmid in yeast was isolated and then transformed into E. coli DH5α cells, for which newly extracted plasmid was sequenced (MOBIX Lab, McMaster University, Hamilton, ON, Canada), using the universal primers M13 forward and reverse (see Table S6 in the supplemental material) for verification. Bcc strains were then conjugated by triparental mating, using pRK2013 as the mobilizing plasmid (52). Transconjugants were selected on Pseudomonas isolation agar (PIA; BD) containing 100 μg · ml−1 of Tp.
(ii) Cloning of mlaCB and complementation of the mlaC insertional mutants.
A 1,367-bp DNA fragment containing the mlaCB genes and the upstream region was PCR amplified from B. cenocepacia K56-2 genomic DNA using Phusion high-fidelity DNA polymerase with the primers mlaCB-For and mlaCB-Rev (Table S6). The PCR product was then subcloned into pCR2.1 using the TA cloning kit (Invitrogen). Since the Phusion enzyme does not leave 3′ A overhangs, an extra extension cycle of 10 min at 72°C with Taq DNA polymerase (Invitrogen) was performed prior to the subcloning step into E. coli DH5α cells. The resulting plasmid, pCR2.1-mlaCB, was then sequenced (MOBIX Lab), using the universal primers M13 forward and reverse (Table S6) for verification. Then, a 1,321-bp KpnI-EcoRI fragment from pCR2.1-mlaCB was cloned into pHERD26T (53) digested with the same enzymes in the opposite direction of the arabinose-inducible promoter to generate pHERD26T-mlaCB. E. coli strains carrying pHERD26T and pHERD26T-mlaCB were then conjugated into Burkholderia strains by triparental mating, using pRK2013 as the mobilizing plasmid (52). Tet was used at a concentration of 50 μg · ml−1 in mlaC mutants of B. cenocepacia K56-2 and B. dolosa PC543 carrying pHERD26T, while 150 μg · ml−1 Tet was used with pHERD26T-mlaCB.
(iii) Transposon mutant libraries in B. cenocepacia K56-2.
Random transposon mutagenesis was performed as previously described (54, 55). Briefly, plasmid pSCrhaBout (55) was conjugated into B. cenocepacia K56-2 by triparental mating, using pRK2013 as the mobilizing plasmid (52). Transconjugants were selected on LB agar plates containing the required antibiotics (100 and 50 μg · ml−1 of Tp and Gm, respectively). Transposon mutants were transferred to 96-well microtiter plates containing 100 μl per well of LB (with Tp at 100 μg · ml−1) and allowed to grow at 37°C for 16 to 20 h. Glycerol was then added to each well at a final concentration of approximately 15% before plates were sealed and stored at −80°C.
(iv) Screening of the transposon mutant libraries.
The transposon library (40 plates) was used to identify mutants with impaired ability to grow upon exposure to sublethal concentrations (25%, vol/vol) of P. aeruginosa (strain PA14) metabolites. Briefly, microtiter plates containing 100 μl per well of LB (with Tp at 100 μg · ml−1) were inoculated directly from each plate of the frozen library using a 96-pin replicator and incubated with shaking (175 rpm) at 37°C overnight. Overnight cultures were then transferred with a 96-pin replicator onto LB agar plates containing filtered spent medium from P. aeruginosa (strain PA14) at a final concentration of 25% and incubated for 24 h at 37°C and room temperature for an extra 72 h. Growth of transposon mutants was monitored daily and those impaired in growth on spent medium agar were retested at least 2 more times.
(v) Mapping of transposon insertion sites.
Transposon insertion mapping was performed as previously described (54, 55). Briefly, genomic DNA was extracted from overnight cultures of selected transposon mutants by using the Promega Wizard genomic DNA purification kit (Promega, Madison, WI), digested with NotI or XhoI restriction enzymes (NEB), self-ligated using T4 DNA ligase (Rapid DNA ligation kit, Roche, Germany), and transformed into E. coli DH5α competent cells. Transformants were selected on LB agar plates containing 100 μg · ml−1 of Tp. Plasmids from these respective transformants were extracted and sequenced (MOBIX Lab) using primer 824 (55). Sequences obtained were then compared using BLAST to sequences from the complete genome of B. cenocepacia strain J2315 (13) at the Burkholderia Genome Database (www.burkholderia.com) (15).
Monoculture and coculture growth.
Competitive experiments and bacterial growth curves were performed as previously described (9). Briefly, overnight cultures were spun down (2 min at 8,000 rpm) and washed once in phosphate-buffered saline (PBS) and used for inoculating liquid cultures in shaken flasks.
(i) Monoculture and coculture.
Flasks containing 25 ml of culture medium were inoculated with 80 μl of washed cells (1:300 dilution) and incubated for 24 h at 37°C with shaking (175 rpm).
(ii) Bacterial survival quantification.
Viable cell counts were used to determine bacterial growth or survival of both monoculture and coculture growth. Bacterial cultures were serially diluted and plated on agar-containing medium with a detection limit of 200 CFU · ml−1. To accurately quantify B. cenocepacia viable counts in cocultures, LB agar was supplemented with Gm (50 μg · ml−1) to counter select P. aeruginosa.
Antibiotic susceptibility assays.
Antibiotic sensitivity was performed using the disk diffusion assay and the broth microdilution method.
(i) Disk diffusion.
LB agar plates were entirely streaked with cotton swabs that were previously immersed into LB medium containing test bacteria that were diluted to an optical density at 600 nm (OD600) of 0.1 from an overnight culture. Antibiotic disks or disks containing Pseudomonas toxins with concentrations stated in the text (i.e., PCA, PYO, or dirhamnolipids) were then deposited on inoculated agar and incubated at 37°C for 24 to 48 h.
(ii) Broth microdilution method.
MIC values of selected antibiotics was determined by macrodilutions in LB as previously described (56), with minor modifications. Briefly, overnight cultures were diluted to an OD600 of 0.1 and added to 96-well plates at a final dilution of 1:200 (vol/vol). The final volume per well was 100 μl, including the tested antibiotic serially diluted. Inoculated plates were sealed with a sticky, breathable membrane (catalogue no. 9123-6100; Breathe-Easy, USA Scientific) and incubated for 24 h at 37°C with shaking (175 rpm). The last dilution without visual growth was considered the MIC value.
Colony morphotype assay.
Morphology assays were performed as previously described (20). Briefly, bacterial cells from overnight cultures were pelleted for 2 min at 8,000 rpm and washed with PBS, and a final 10−6-fold dilution was plated on agar-containing spent medium at a final concentration of 1% (vol/vol) and incubated at 37°C for 24 to 48 h.
Swarming motility assays.
Assays were performed as previously described (20). Briefly, 2 μl of an overnight culture was spotted in the middle of swarm plates allowed to dry for 1 h at room temperature and then incubated at 37°C for 24 to 48 h. Swarm plates were made of M9CAA supplemented with agar at 0.5% (wt/vol) supplemented or not supplemented with 1% (vol/vol) bacterial spent medium.
SDS-EDTA sensitivity assay.
Susceptibility to SDS-EDTA was performed as previously described (18), with a few modifications. Briefly, 2-μl aliquots overnight cultures in LB were spotted on agar and incubated at 37°C for 48 h. Growth of spotted colonies on regular LB agar was then compared to growth on agar containing 0.55 mM EDTA, 0.025% SDS, or 0.025% SDS–0.55 mM EDTA.
Serum bactericidal assay.
Heparinized blood was collected from 5 healthy volunteers (4 females and 1 male), as approved by the Hamilton Integrated Research ethics board. Once coagulated, collecting tubes were centrifuged for 10 min, and human serum (hSerum) was frozen at −80°C. Resistance to hSerum was performed as previously described (57), with a few modifications. Prior to use, sera from all volunteers were pooled equally and diluted with PBS to a final concentration of 30% (vol/vol). Briefly, overnight cultures in LB were washed in PBS and 1 μl (∼1 × 106 to 3 × 106 CFU · ml−1) was added to 100 μl of 30% hSerum and incubated for 2 h at 37°C. Incubation in PBS was performed in parallel as the negative control. Serum was heat inactivated (HihSerum) at 65°C for 30 min. Viable bacteria at time zero and after 2 h were serially diluted and spotted on agar for enumeration.
Supplementary Material
ACKNOWLEDGMENTS
We thank Laura Rossi for critical reading of the manuscript.
This study was supported by funding from Cystic Fibrosis Canada to M.G.S. S.P.B. was the recipient of a postdoctoral research fellowship from Cystic Fibrosis Canada and M.G.S. is Canada Research Chair in Interdisciplinary Microbiome Research.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00156-18.
REFERENCES
- 1.Eberl L, Vandamme P. 2016. Members of the genus Burkholderia: good and bad guys. F1000Res 5:1007. doi: 10.12688/f1000research.8221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan JR. 2016. Biosecurity and bioterrorism, 2nd ed. Butterworth-Heinemann, Oxford, UK. [Google Scholar]
- 3.Rhodes KA, Schweizer HP. 2016. Antibiotic resistance in Burkholderia species. Drug Resist Updat 28:82–90. doi: 10.1016/j.drup.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivas P, Rivard K. 2017. Polymyxin resistance in Gram-negative pathogens. Curr Infect Dis Rep 19:38. doi: 10.1007/s11908-017-0596-3. [DOI] [PubMed] [Google Scholar]
- 5.Cox G, Wright GD. 2013. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol 303:287–292. doi: 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Loutet SA, Valvano MA. 2011. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front Microbiol 2:159. doi: 10.3389/fmicb.2011.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piddock LJ. 2012. The crisis of no new antibiotics–what is the way forward? Lancet Infect Dis 12:249–253. doi: 10.1016/S1473-3099(11)70316-4. [DOI] [PubMed] [Google Scholar]
- 8.Wright GD. 2015. Solving the antibiotic crisis. ACS Infect Dis 1:80–84. doi: 10.1021/id500052s. [DOI] [PubMed] [Google Scholar]
- 9.Bernier SP, Workentine ML, Li X, Magarvey NA, O'Toole GA, Surette MG. 2016. Cyanide toxicity to Burkholderia cenocepacia is modulated by polymicrobial communities and environmental factors. Front Microbiol 7:725. doi: 10.3389/fmicb.2016.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smalley NE, An D, Parsek MR, Chandler JR, Dandekar AA. 2015. Quorum sensing protects Pseudomonas aeruginosa against cheating by other species in a laboratory coculture model. J Bacteriol 197:3154–3159. doi: 10.1128/JB.00482-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price-Whelan A, Dietrich LE, Newman DK. 2006. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol 2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 12.Bernier SP, Surette MG. 2013. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol 4:20. doi: 10.3389/fmicb.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varga JJ, Losada L, Zelazny AM, Kim M, McCorrison J, Brinkac L, Sampaio EP, Greenberg DE, Singh I, Heiner C, Ashby M, Nierman WC, Holland SM, Goldberg JB. 2013. Draft Genome Sequences of Burkholderia cenocepacia ET12 Lineage Strains K56-2 and BC7. Genome Announc 1:e00841-1. doi: 10.1128/genomeA.00841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ, Ranganathan SC, Waters VJ, Ratjen FA. 2014. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, gram-negative bacteria, and multiple infections. Ann Am Thorac Soc 11:1120–1129. doi: 10.1513/AnnalsATS.201402-050AS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter CD, Cooley BJ, Needham BD, Fisher CR, Trent MS, Gordon V, Payne SM. 2014. The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect Immun 82:660–669. doi: 10.1128/IAI.01057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munguia J, LaRock DL, Tsunemoto H, Olson J, Cornax I, Pogliano J, Nizet V. 2017. The Mla pathway is critical for Pseudomonas aeruginosa resistance to outer membrane permeabilization and host innate immune clearance. J Mol Med (Berl) 95:1127–1136. doi: 10.1007/s00109-017-1579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernier SP, Hum C, Li X, O'Toole GA, Magarvey NA, Surette MG. 2017. Pseudomonas aeruginosa-derived rhamnolipids and other detergents modulate colony morphotype and motility in the Burkholderia cepacia complex. J Bacteriol 199:e00171-17. doi: 10.1128/JB.00171-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noinaj N, Gumbart JC, Buchanan SK. 2017. The beta-barrel assembly machinery in motion. Nat Rev Microbiol 15:197–204. doi: 10.1038/nrmicro.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abellon-Ruiz J, Kaptan SS, Basle A, Claudi B, Bumann D, Kleinekathofer U, van den Berg B. 2017. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol 2:1616–1623. doi: 10.1038/s41564-017-0046-x. [DOI] [PubMed] [Google Scholar]
- 23.Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. 2017. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285. doi: 10.1016/j.cell.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thong S, Ercan B, Torta F, Fong ZY, Wong HY, Wenk MR, Chng SS. 2016. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. Elife 5:e19042. doi: 10.7554/eLife.19042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim J. 2015. The characterization of the lipoprotein VacJ in Burkholderia pseudomallei and Burkholderia thailandensis. PhD dissertation. London School of Hygiene and Tropical Medicine, University of London, London, UK. [Google Scholar]
- 26.Nakamura S, Shchepetov M, Dalia AB, Clark SE, Murphy TF, Sethi S, Gilsdorf JR, Smith AL, Weiser JN. 2011. Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog 7: e1001247. doi: 10.1371/journal.ppat.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie F, Li G, Zhang W, Zhang Y, Zhou L, Liu S, Wang C. 2016. Outer membrane lipoprotein VacJ is required for the membrane integrity, serum resistance and biofilm formation of Actinobacillus pleuropneumoniae. Vet Microbiol 183:1–8. doi: 10.1016/j.vetmic.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Ortega X, Hunt TA, Loutet S, Vinion-Dubiel AD, Datta A, Choudhury B, Goldberg JB, Carlson R, Valvano MA. 2005. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J Bacteriol 187:1324–1333. doi: 10.1128/JB.187.4.1324-1333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zlosnik JE, Gunaratnam LC, Speert DP. 2012. Serum susceptibility in clinical isolates of Burkholderia cepacia complex bacteria: development of a growth-based assay for high throughput determination. Front Cell Infect Microbiol 2:67. doi: 10.3389/fcimb.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. 2015. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnamoorthy G, Leus IV, Weeks JW, Wolloscheck D, Rybenkov VV, Zgurskaya HI. 2017. Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. mBio 8:e01172–17. doi: 10.1128/mBio.01172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gislason AS, Turner K, Domaratzki M, Cardona ST. 2017. Comparative analysis of the Burkholderia cenocepacia K56-2 essential genome reveals cell envelope functions that are uniquely required for survival in species of the genus Burkholderia. Microb Genom 3. doi: 10.1099/mgen.0.000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babu M, Bundalovic-Torma C, Calmettes C, Phanse S, Zhang Q, Jiang Y, Minic Z, Kim S, Mehla J, Gagarinova A, Rodionova I, Kumar A, Guo H, Kagan O, Pogoutse O, Aoki H, Deineko V, Caufield JH, Holtzapple E, Zhang Z, Vastermark A, Pandya Y, Lai CC, El Bakkouri M, Hooda Y, Shah M, Burnside D, Hooshyar M, Vlasblom J, Rajagopala SV, Golshani A, Wuchty S, Saier J FGM, Uetz P, Moraes TF, Parkinson J, Emili A. 2018. Global landscape of cell envelope protein complexes in Escherichia coli. Nat Biotechnol 36:103–112. doi: 10.1038/nbt.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong YC, Abd El Ghany M, Naeem R, Lee KW, Tan YC, Pain A, Nathan S. 2016. Candidate essential genes in Burkholderia cenocepacia J2315 identified by genome-wide TraDIS. Front Microbiol 7:1288. doi: 10.3389/fmicb.2016.01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L, Gao X, Wei J, Chen L, Zhao X, Li B, Duan K. 2012. PA2800 plays an important role in both antibiotic susceptibility and virulence in Pseudomonas aeruginosa. Curr Microbiol 65:601–609. doi: 10.1007/s00284-012-0196-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhao L, Gao X, Liu C, Lv X, Jiang N, Zheng S. 2017. Deletion of the vacJ gene affects the biology and virulence in Haemophilus parasuis serovar 5. Gene 603:42–53. doi: 10.1016/j.gene.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Krishnamoorthy G, Wolloscheck D, Weeks JW, Croft C, Rybenkov VV, Zgurskaya HI. 2016. Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob Agents Chemother 60:7372–7381. doi: 10.1128/AAC.01045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podnecky NL, Rhodes KA, Schweizer HP. 2015. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol 6:305. doi: 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mima T, Schweizer HP. 2010. The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob Agents Chemother 54:3113–3120. doi: 10.1128/AAC.01803-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, Fondi M, Perrin E, Decorosi F, Viti C, Giovannetti L, Leoni L, Fani R, Riccardi G, Mahenthiralingam E, Buroni S. 2011. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS One 6:e18902. doi: 10.1371/journal.pone.0018902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan YY, Tan TM, Ong YM, Chua KL. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob Agents Chemother 48:1128–1135. doi: 10.1128/AAC.48.4.1128-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuccui J, Easton A, Chu KK, Bancroft GJ, Oyston PC, Titball RW, Wren BW. 2007. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect Immun 75:1186–1195. doi: 10.1128/IAI.01240-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roier S, Zingl FG, Cakar F, Schild S. 2016. Bacterial outer membrane vesicle biogenesis: a new mechanism and its implications. Microb Cell 3:257–259. doi: 10.15698/mic2016.06.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki T, Murai T, Fukuda I, Tobe T, Yoshikawa M, Sasakawa C. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol 11:31–41. doi: 10.1111/j.1365-2958.1994.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 45.Miajlovic H, Smith SG. 2014. Bacterial self-defence: how Escherichia coli evades serum killing. FEMS Microbiol Lett 354:1–9. doi: 10.1111/1574-6968.12419. [DOI] [PubMed] [Google Scholar]
- 46.Mil-Homens D, Fialho AM. 2012. A BCAM0223 mutant of Burkholderia cenocepacia is deficient in hemagglutination, serum resistance, adhesion to epithelial cells and virulence. PLoS One 7:e41747. doi: 10.1371/journal.pone.0041747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loutet SA, Flannagan RS, Kooi C, Sokol PA, Valvano MA. 2006. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J Bacteriol 188:2073–2080. doi: 10.1128/JB.188.6.2073-2080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD. 2011. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol 7:348–350. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 49.Stokes JM, MacNair CR, Ilyas B, French S, Cote JP, Bouwman C, Farha MA, Sieron AO, Whitfield C, Coombes BK, Brown ED. 2017. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat Microbiol 2:17028. doi: 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright GD. 2016. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol 24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 51.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernier SP, Nguyen DT, Sokol PA. 2008. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect Immun 76:38–47. doi: 10.1128/IAI.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardona ST, Mueller CL, Valvano MA. 2006. Identification of essential operons with a rhamnose-inducible promoter in Burkholderia cenocepacia. Appl Environ Microbiol 72:2547–2555. doi: 10.1128/AEM.72.4.2547-2555.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hacek DM, Dressel DC, Peterson LR. 1999. Highly reproducible bactericidal activity test results by using a modified National Committee for Clinical Laboratory Standards broth macrodilution technique. J Clin Microbiol 37:1881–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeShazer D, Brett PJ, Woods DE. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol 30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.