Bacteria have evolved several secretion strategies for polling and responding to environmental flux and insult. Of these, the type 1 secretion system (T1SS) is known to secrete an array of biologically diverse proteins—from small, <10-kDa bacteriocins to gigantic adhesins with a mass >1 MDa.
KEYWORDS: T1SS, adhesin, biofilm, LapA, retention module
ABSTRACT
Bacteria have evolved several secretion strategies for polling and responding to environmental flux and insult. Of these, the type 1 secretion system (T1SS) is known to secrete an array of biologically diverse proteins—from small, <10-kDa bacteriocins to gigantic adhesins with a mass >1 MDa. For the last several decades, T1SSs have been characterized as a one-step translocation strategy whereby the secreted substrate is transported directly into the extracellular environment from the cytoplasm with no periplasmic intermediate. Recent phylogenetic, biochemical, and genetic evidences point to a distinct subgroup of T1SS machinery linked with a bacterial transglutaminase-like cysteine proteinase (BTLCP), which uses a two-step secretion mechanism. BTLCP-linked T1SSs transport a class of repeats-in-toxin (RTX) adhesins that are critical for biofilm formation. The prototype of this RTX adhesin group, LapA of Pseudomonas fluorescens Pf0-1, uses a novel N-terminal retention module to anchor the adhesin at the cell surface as a secretion intermediate threaded through the outer membrane-localized TolC-like protein LapE. This secretion intermediate is posttranslationally cleaved by the BTLCP family LapG protein to release LapA from its cognate T1SS pore. Thus, the secretion of LapA and related RTX adhesins into the extracellular environment appears to be a T1SS-mediated two-step process that involves a periplasmic intermediate. In this review, we contrast the T1SS machinery and substrates of the BLTCP-linked two-step secretion process with those of the classical one-step T1SS to better understand the newly recognized and expanded role of this secretion machinery.
INTRODUCTION
The type 1 secretion system (T1SS) is a relatively simple yet highly conserved secretion strategy used throughout Gram-negative bacteria to translocate small substrates as well as extremely large proteins to the extracellular environment. Much of our current knowledge of T1SS has been gathered over the last several decades from studies of the T1SS toxins HlyA and CyaA of Escherichia coli and Bordetella pertussis, respectively. These toxins, and the majority of type 1 secreted proteins, belong to the repeats-in-toxin (RTX) family first described by Rodney A. Welch (1). A consensus model has emerged from these studies describing T1SS as a single-step secretion strategy that forms a transmembrane tunnel to translocate the unfolded substrate directly from the cytoplasm to the extracellular environment with no periplasmic intermediate (2–5). The glycine- and aspartate-rich nonapeptide-repeat calcium-binding RTX motifs are thought to discourage premature folding inside the cell due to low cytoplasmic calcium concentrations (<100 nM), leaving the apo-RTX motifs intrinsically disordered (6, 7). The comparatively calcium-rich extracellular environment (∼2 mM) drives a ratchet-like folding process, triggered by direct calcium binding, which enables the protein to adopt its final folded structure necessary for its biological function. In the case of CyaA, the binding of extracellular calcium also accelerates the secretion of the toxin (8).
The capacity of the T1SS to secrete diverse proteins, many of which are involved in pathogenesis within the host, has been recognized for quite some time. T1SS substrates range from small <10-kDa bacteriocins involved in bacterial warfare to gigantic >1-MDa multifunctional adhesins (9, 10), demonstrating the importance of this system to bacterial survival and adaptation. T1SS RTX adhesins are not only some of the largest bacterial proteins but also some of the most diverse, often sharing little sequence identity across closely related strains (11). These large adhesins are involved in bacterial virulence as host colonization or cell entry factors and also play roles in commensal and beneficial interactions with hosts. For example, RtxA of the intracellular pathogen Legionella pneumophila is involved in monocyte binding and entry (12). Conversely, LapA of the plant-beneficial Pseudomonas putida promotes biofilm formation on plant seeds (13).
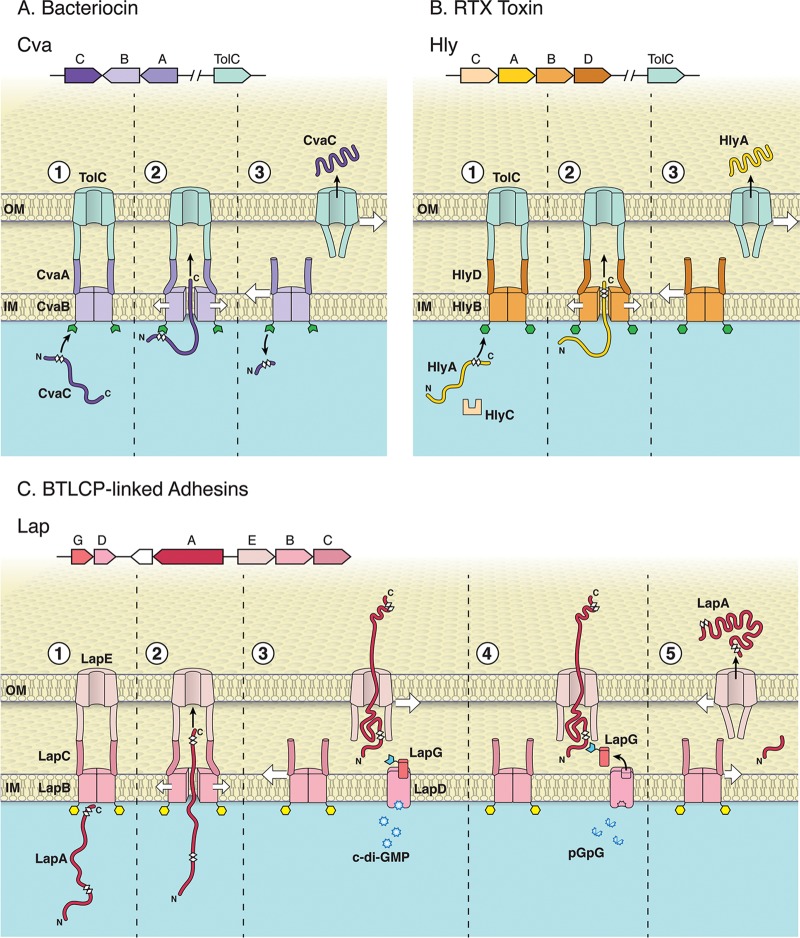
The transport of secreted substrates via the T1SS requires a tripartite translocation apparatus (Fig. 1A and B) composed of an inner membrane ATP-binding cassette (ABC) transporter, a membrane fusion protein (MFP), and outer membrane (OM) pore, TolC (14, 15). Substrate-induced assembly of the nanomachine forms a transmembrane-spanning tunnel that secretes an unfolded cytoplasmic substrate directly into the extracellular environment (16). A secretion signal located at the N terminus (bacteriocins) or C terminus (all other known substrates) directs the unfolded substrate to its specific ABC transporter/MFP partner (17–19). Importantly, the ABC transporter/MFP inner membrane complex is typically dedicated to secreting a single T1SS substrate, while the outer membrane pore can be promiscuous (20, 21). As described above, the T1SS was believed to have no periplasmic intermediate.
FIG 1.
Overview of three classes of ABC transporters. The T1SS consists of an ABC transporter, MFP, and OM protein. Unfolded T1SS substrates interact with their cognate inner membrane ABC transporter/MFP complex and then recruit and open an OM pore. The inner membrane complex dissociates from TolC following secretion, and TolC returns to the closed state. Variations among these T1SS types are detailed below. The N- and C-terminal portions of the substrates are indicated in the figure, and the white diamonds illustrate the glycine-rich regions of each substrate. (A) Secretion of the bacteriocin CvaC (colisin V); (top) operonic structure of the genes encoding CvaC, CvaB (ABC transporter), and CvaA (MFP) and distally encoded TolC OM pore in E. coli; (bottom) the CvaB protein contains an N-terminal secretion signal with a canonical double glycine motif recognized by CvaB (step 1). CvaB contains a calcium-dependent N-terminal C39 peptidase (green open hexagon) that cleaves CvaC C-terminal to the double glycine site (thereby activating the protein [step 2]) during transport to the extracellular environment. The CvaBA-TolC complex disassembles following secretion of CvaC into the extracellular environment; the closing of the TolC aperture after the secretion complex disassembles is illustrated here and in the subsequent panels (step 3). (B) Secretion of the RTX toxin HlyA; (top) operonic structure of the genes encoding HlyC (accessory acyltransferase that acylates and activates HlyA), HlyA (RTX toxin), HlyB (ABC transporter), HlyD (MFP), and the distally encoded TolC OM pore in E. coli; (bottom) N terminus of HlyB contains a catalytically inactive C39-like domain ([CLD] green hexagon). The CLD of HlyB binds unfolded glycine-rich RTX motifs on the C terminus of HlyA (step 1). The RTX motifs (final ∼50 to 60 aa in HlyA) that engage HlyB, and are ultimately necessary to recruit TolC for secretion, are N-terminal to the secretion signal (step 2). The HlyBD-TolC complex disassembles following secretion of HlyA into the extracellular environment (step 3). (C) Secretion of the BTLCP-linked RTX adhesin LapA in P. fluorescens Pf0-1; (top) operonic structure of the genes encoding LapG (periplasmic calcium-dependent BTLCP that cleaves LapA), LapD (c-di-GMP-sensing effector that sequesters LapG when c-di-GMP is bound), LapA (BTLCP-linked RTX adhesin), LapE (TolC-like OM pore), LapB (ABC transporter), and LapC (MFP); (bottom) LapA secretion requires LapB, which has a CLD distinct from HylB (yellow hexagon [step 1]). LapA is secreted in a C-terminal to N-terminal direction (step 2), but secretion of LapA is stalled during translocation, leaving LapA threaded through LapE with its cleavable N-terminal retention domain localized in the periplasm and multiple adhesive repeats exposed at the cell surface (step 3). LapA is fixed at the cell surface as a biofilm-promoting secretion intermediate when c-di-GMP levels are high and LapD has bound this dinucleotide. c-di-GMP-bound LapD sequesters LapG (open blue hexagon [step 3]), protecting LapA from LapG proteolysis. When c-di-GMP levels are reduced via conversion of this signal to the linear molecule pGpG, LapG is released from LapD (note empty LapG binding site), LapG can cleave LapA at a canonical dialanine site within the retention module (step 4). The cleaved LapA is released from the cell surface, and the unoccupied LapE is now free to interact with substrate-engaged LapBC (step 5). Because the CLD of LapB is distinct from the CLD of HlyB and the C39 domain of CvaB, its role in LapA secretion is unclear. Given the relatedness of these transport systems, the CLD of LapB may bind the glycine-rich regions found at the N or C terminus of LapA (see step 1). (Copyright William Scavone, Kestrel Studio; reproduced with permission.)
In contrast to this T1SS single-step model, recent work on LapA of Pseudomonas fluorescens Pf0-1, a giant, c-di-GMP-regulated, cell surface-associated type 1-secreted RTX adhesin, suggests a different approach, wherein the secretion substrate is tethered to the cell surface via a cognate outer membrane pore component. This adhesin can be released from the cell surface; thus, its transport out of the cell can be considered a two-step secretion strategy (Fig. 1C). This review aims to summarize and draw on classical T1SS studies to highlight and contrast a recently appreciated subclass of T1SS transporters and substrates and to update current thinking about the range of functions performed by the T1SS machinery. Although T1SS adhesins are found throughout Gram-negative bacteria, we have intentionally focused on those substrates linked with bacterial transglutaminase-like cysteine proteinases (BTLCPs) and their unique two-step secretion strategy. For a general review of type 1-secreted adhesins and other large RTX proteins, such as multifunctional autoprocessing repeats-in-toxin (MARTX) toxin proteins, please see other recent reviews (9, 22).
OVERVIEW OF T1SS MACHINERY AND SUBSTRATE SECRETION
The T1SS is a tripartite transport complex composed of an inner membrane ABC transporter and an MFP, as well as an outer membrane pore. ABC transporters are found in all domains of life and play diverse roles in importing and exporting molecules, big and small, into or out of the cell or cellular compartments (23). Minimally, ABC transporters contain two transmembrane domains (TMDs) and two nucleotide binding domains (NBDs) (24). For the T1SS, the 2TMB-2NBD requirement is satisfied when a single protein containing both a TMD and a NBD dimerizes in the inner membrane. Cross-linking experiments demonstrated that the ABC transporter HlyB forms a stable complex with the MFP HlyD, even in the absence of its HlyA secretion substrate (Fig. 1B, step 3) (16). The transmembrane-spanning HlyBD-TolC secretion complex is initiated when HlyBD binds the unfolded C-terminal secretion signal of the HlyA toxin, and this HlyBD-TolC complex disassembles following secretion (16). One-step secretion of unfolded HlyA commences from the C terminus to the N terminus (25). Although TolC is not encoded near the hlyCABD genes, this outer membrane protein is required for the secretion of HlyA (15).
The outer membrane component, TolC, is a trimeric pore protein. Each TolC monomer is composed of three domains: a 40-Å long β-barrel domain that is inserted in the outer membrane, a 100-Å periplasmic α-helical domain, and a periplasmic mixed α/β equatorial domain (26). The α helices of TolC form an aperture that facilitates the opening and closing of the membrane pore. When closed, the aperture diameter is only 3.9 Å, which prohibits the secretion of even small solutes. TolC mutants reflecting the open state have an aperture diameter of approximately 20 Å (27). In E. coli, TolC can also form a complex with AcrAB, an MFP and resistance-nodulation-division (RND) antiporter pump (28), as well as with the bacteriocin transporter inner membrane complex, CvaAB (21). Additionally, TolC of Rickettsia typhi was shown to secrete the ankyrin repeat-containing protein, RARP-1, which is translocated to the periplasm by Sec translocase (29). Thus, TolC-like proteins participate in the secretion of a variety of substrates.
Until recently, there were two known subclasses of T1SS substrates: small bacteriocins containing cleaved N-terminal leader peptides and a large group of diverse uncleaved substrates containing C-terminal secretion signals. Bacteriocins secreted by the T1SS in Gram-negative bacteria belong to the class II subfamily of bacteriocins. These small modified peptides of <10 kDa, which have been extensively characterized in Gram-positive bacteria, contain N-terminal double glycine (GG) motifs that are recognized by the C39 peptidase domain of the ABC transporter (Fig. 1A). During secretion, the C39 peptidase domain of the ABC transporter cleaves the bacteriocin near an N-terminal GG motif, activating the immature bacteriocin and enabling the secretion of the substrate downstream of this GG motif. Importantly, this functional C39 peptidase domain is critical for bacteriocin activation.
Recent work from the Schmitt lab demonstrated that HlyB and many other T1SS ABC transporters contain additional N-terminal domains that strongly resemble the C39 peptidase domain, found N-terminally fused to bacteriocin ABC transporters such as CvaB of E. coli (30). In HlyB, the C39 domain is catalytically inactive as it lacks the critical cysteine residue (C39) and was thus called the C39-like domain (CLD) (30). Instead of binding an N-terminal GG motif like the C39 peptidase, the HlyB-CLD binds the unfolded glycine-rich RTX motifs located near the C-terminal secretion signal of HlyA (Fig. 1B). Interestingly, the binding site interactions between the CLD of HlyB and HlyA's C terminus are essential for HlyA secretion (30). Thus, bacteriocin and RTX toxin ABC transporters secrete very different substrates, bind at opposite ends of their respective substrates to facilitate secretion, and can be distinguished on the basis of their primary amino acid sequences. Surprisingly, although the C39 and CLD are structurally conserved, Lecher et al. demonstrated the ligand-binding regions are on opposite sides of these domains (30).
It has been proposed that HlyB-CLD binding at the C terminus of HlyA may discourage premature folding of HlyA in the cytoplasm. Although many T1SS ABC transporters contain the N-terminal CLD or C39 domain, these domains are absent in the ABC transporters of various small T1SS proteases and lipases (4), and it is currently unclear if this is a general strategy among CLD-containing T1SS ABC transporters.
BACTERIAL TRANSGLUTAMINASE-LIKE CYSTEINE PROTEINASES ARE CONSERVED T1SS ACCESSORY PROTEINS
Several classes of T1SS substrates require accessory proteins for posttranslational activation/modification or proper localization. For example, HlyA and other T1SS RTX toxins are synthesized in the immature protoxin form. In the case of HlyA, an acyl carrier protein (ACP)-dependent protein acyltransferase called HlyC transfers fatty acyl moieties to Lys564 and Lys690 residues of pro-HlyA, maturing this α-hemolysin of E. coli (31). This posttranslational acylation activates the toxin but does not impact secretion. Surface retention of the adhesin SiiE, which mediates host cell contact for the intracellular pathogen Salmonella enterica, illustrates another example of a role for accessory proteins. SiiE is encoded by the Salmonella pathogenicity island (SPI) 4 alongside its cognate T1SS machinery and the inner membrane accessory proteins, SiiAB. The SiiAB accessory proteins, which resemble the MotAB proton channel proteins of the flagellar motor, are required for modulating SiiE surface retention and release (32). The deletion of the siiA or siiB genes nearly abolishes S. enterica adherence to and invasion of polarized Madin-Darby canine kidney (MDCK) cells (33). Interestingly, that study (33) also demonstrated SiiB interacts with the T1SS ABC transporter of the SiiE adhesin, called SiiF, indicating that accessory proteins may interact with the secretion machinery.
Ginalski et al. (34) described a novel family of bacterial transglutaminase-like cysteine proteinases (BTLCPs) with invariant Cys-His-Asp catalytic triads (previously belonging to DUF920 and COG3672) that are often linked with T1SS machinery and large RTX-containing proteins. While the function of these accessory BTLCPs was unclear at the time, their structural predictions led the group to posit that these proteins may posttranslationally modify target proteins via transamidase, acetylase, or hydrolase activity (34). Genetic and biochemical studies of a BTLCP of Pseudomonas fluorescens Pf0-1, called LapG, demonstrated that LapG is a periplasmic calcium-dependent cysteine protease responsible for the posttranslational cleavage of the giant T1SS RTX adhesin LapA (Fig. 1C). Cleavage occurs in the periplasm at an N-terminal dialanine motif, leading to the removal of LapA from the cell surface and thus decreasing biofilm formation (35–37). Further studies indicated that LapG activity is controlled by the inner membrane-bound c-di-GMP receptor, LapD (38). LapD contains cytoplasmic catalytically inactive phosphodiesterase and diguanylate cyclase domains that are unable to synthesize or degrade c-di-GMP, as well as a periplasmic output domain (LapDoutput). The phosphodiesterase domain of LapD was shown to bind c-di-GMP (39). Newell et al. developed an inside-out signaling model whereby LapD binds cytoplasmic c-di-GMP, leading to conformational changes in the LapDoutput domain that promote LapG binding (35, 37, 39). This LapD-LapG interaction, and the sequestration of LapG from its substrate, enables LapA to remain at the cell surface where it is able to promote biofilm formation. When cellular levels of c-di-GMP are depleted, LapG is liberated from LapD; LapG is then able to cleave LapA and inhibit biofilm formation (37, 39–41). Thus, LapG and LapD act as a regulatory hub to modulate the cell surface localization of a large adhesin in response to the levels of a cytoplasmic second messenger (42).
Genes for LapG homologs and the associated LapD receptor are found together in the genomes of over 1,300 bacterial species spanning 120 genera in the Proteobacteria (43), suggesting that this adhesin-localization strategy is quite common among this group of organisms. In support of this idea, there is a fair degree of cross-functionality between these proteins despite their low sequence identity. For example, the CdgS9 protein, the LapD homolog from L. pneumophila, can bind both its cognate BTLCP, lpg0827, and LapG from P. fluorescens Pf0-1 (44). Also, the L. pneumophila LapG homolog can cleave the N terminus of LapA from P. fluorescens Pf0-1 (44). Mechanistic insights into LapG regulation by LapD were revealed by the structure of LapG from P. fluorescens in complex with the LapD periplasmic output domain from L. pneumophila (CdgS9) (45). In support of previous biochemical and genetic evidences, a surface-exposed tryptophan residue (W126 in CdgS9, W125 in P. fluorescens LapD) found in the conserved GWXQ loop of the LapD output domain is crucial for the LapDoutput-LapG interaction (37, 44). The conserved tryptophan in CdgS9 inserts into a hydrophobic pocket in LapG, mediating the interactions between these two proteins. Finally, the expression of either the LapG or LapD homolog from Bordetella bronchiseptica partially complements the biofilm phenotype of the corresponding P. fluorescens mutant strain (46). Thus, BTLCPs from different organisms appear to be functionally conserved.
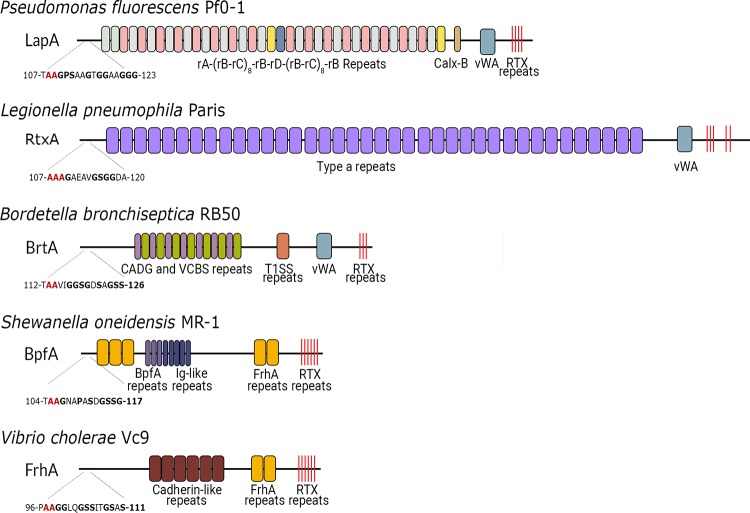
The LapD/LapG accessory proteins have been implicated in or shown to control aspects of biofilm formation in multiple bacterial species, including B. bronchiseptica (46), Shewanella spp. (47), Pectobacterium atrosepticum (48), Desulfovibrio vulgaris Hildenborough (49), P. putida (50), and Pseudomonas aeruginosa (51, 52). Interestingly, the LapG substrate in P. aeruginosa is not a T1SS RTX adhesin but rather a two-partner secreted (TPS) adhesin, CdrA. CdrA is involved in reinforcing the biofilm matrix and contributes to cell-cell interactions by binding mannose moieties of the Psl polysaccharide, a critical component of the biofilm extracellular matrix in P. aeruginosa (53). Thus, although our bioinformatic analysis suggests several species have coopted the LapGD accessory proteins to process TPS adhesins, the vast majority of LapG targets are T1SS RTX adhesins such as LapA (Fig. 2, top).
FIG 2.
BTLCP-linked adhesins are diverse. Schematic representation of adhesive repeat and domain architectures from select BTLCP-linked adhesins: LapA (Pfl01_0133), RtxA (lpp0699), BrtA (BB1186), BpfA (SO4317), and FrhA (VC1620). Below each adhesin, the putative dialanine BTLCP cleavage site is bolded in red and the glycine-rich linker is bolded in black, with the corresponding amino acid positions on either side of the sequence.
BTLCP-LINKED ADHESINS ARE DIVERSE
BTLCP substrates are some of the largest and most diverse proteins encoded by bacterial genomes (Fig. 2), likely highlighting their importance for binding a wide variety of substrata. These RTX adhesins contain elaborate repeat sequences that occur in seemingly countless combinations, making them difficult to detect via amino acid sequence-based alignments. However, two shared features among these adhesins are the presence of multiple C-terminal RTX motifs and N-terminal dialanine cleavage sites. LapA of P. fluorescens Pf0-1 contains 37 100-amino acid (aa) repeats (D273 to S3982) followed by a Calx-β domain (S4019 to D4094), a von Willebrand factor type A domain ([vWA] N4736 to S4963), and 6 RTX motifs (G5020 to W5144). These features are packaged between an N-terminal retention module (M1 to S125) and a C-terminal secretion signal (N5151 to S5218) (43, 54). The retention module also contains the dialanine LapG cleavage site (A108 to A109). From an evolutionary standpoint, such protein architecture and domain modularity could be exploited to rapidly adapt to each suitable environment.
In L. pneumophila, the presence of the BTLCP-linked adhesin RtxA is associated with enhanced virulence (55). The size and repeat composition of RtxA vary between L. pneumophila strains, from 7,910 aa in the Lens strain to 3,033 aa in the Philadelphia strain; however, the N-terminal retention module and C-terminal region show a high degree of similarity to each other and resemble the corresponding regions in LapA (11). The repeats found in RtxA homologs have been characterized as types a, b1, b2, c1, c2, and d. The c2 repeats are similar to the SiiE Ig-like repeats and are found in the RtxAs harbored by Corby and Alcoy strains of L. pneumophila. All RtxA proteins characterized thus far contain N-terminal dialanine LapG cleavage sites (11) as well as C-terminal vWA domains; the vWA domain is found in proteins involved in cell-cell interactions and protein multimerization (56). A P. fluorescens Pf0-1 LapA mutant lacking the vWA revealed that this domain is critical for biofilm formation on borosilicate glass, a model hydrophilic surface (54). Because RtxA is involved in host cell invasion, the repeat number and domain arrangement likely impact strain virulence, as an rtxA mutant shows a marked decrease in macrophage adherence and invasion (12, 57). Interestingly, the LapA-like T1SS adhesins are restricted to L. pneumophila strains that show increased intracellular growth within the human host compared to that of other Legionella species (58).
A bioinformatics analysis indicates the BTLCP-linked RTX adhesin BrtA of the respiratory pathogen B. bronchiseptica shows similar N- and C-terminal hallmarks of LapA and RtxA (Fig. 2). BrtA contributes to biofilm formation on glass (hydrophilic) and plastic (hydrophobic) surfaces (59). In strain RB50, BrtA is a 3,346-aa adhesin with 8 CADG-VCBS repeats. Like RtxA, BrtA variants all contain vWA domains, highly similar N and C termini, and variable internal repeat numbers, leading to stark differences in repeat compositions and overall protein sizes. LapG cleaves BrtA at the conserved dialanine cleavage motif within the N-terminal retention module (46), demonstrating the conservation of LapA's cell retention strategy despite the low overall sequence similarity among LapA, BrtA, and RtxA. It is currently unclear if the repeat number or composition contributes to B. bronchiseptica pathogenicity. Ambrosis et al. (46) demonstrated that a B. bronchiseptica RB50 lapG mutant showed a slight but significant increase in lung bacterial burden 14 days postinfection, despite BrtA not being essential for initial colonization. These data suggest that while BrtA is not required for colonization, appropriate regulation of this cell surface adhesin may play a role in the maintenance of infection.
BTLCP-LINKED ADHESINS ARE TETHERED AT THE CELL SURFACE THROUGH THEIR T1SS OUTER MEMBRANE PORES
A bioinformatics analysis of the N termini of several BTLCP-linked adhesins revealed short polyglycine regions shortly following the dialanine cleavage sites (Fig. 2). A mutational analysis showed that a LapA N-terminal truncation mutant lacking residues D31 to A95 was unable to associate with the cell surface, secreted directly into the supernatant, and was inoperative for biofilm formation (43). Thus, the N-terminal region of LapA encompassing the first 125 amino acids, including the polyglycine linker, was termed LapA's “retention module” (43). A LapA chimera containing the retention module from a predicted BTLCP-linked adhesin harbored by Vibrio cholerae, VC0395_0388, complements adhesin localization and LapG surface regulation (43), suggesting this large group of low-identity BTLCP-linked adhesins is anchored to the cell surface through a common mechanism.
LapA and other BTLCP-linked adhesins contain dialanine motifs that are the site of cleavage by the BTLCP family periplasmic protease. This cleavage event is required to release LapA from the cell surface, suggesting these adhesins are accessible from the periplasm when anchored at the cell surface (54). Importantly, LapA and other BTLCP-linked RTX adhesins lack any obvious pore-forming motifs. Together, these data hinted that these adhesins are not secreted directly into the extracellular environment, in canonical T1SS fashion, to subsequently reassociate with the outer membrane. Consistent with the idea that LapA is anchored in the TolC-like outer membrane component of the T1SS, secretion competition experiments demonstrated that cell surface-associated LapA blocks the secretion of a model secretion peptide containing only LapA's secretion signal (43), suggesting LapA occupies a portion of its cognate transport machinery when anchored to the cell surface (Fig. 1C). Secretion of the small peptide is restored upon stimulation of LapG proteolysis to remove LapA from the cell surface when the small peptide is expressed in a lapA mutant that is unable to associate with the cell surface or when expressed as a transcriptional fusion with the T1SS OM TolC-like protein, LapE. A comparable secretion competition strategy was previously employed to demonstrate that surface-associated HMW1 of Haemophilus influenzae is tethered to the cell surface through its cognate two-partner secretion (TPS) OM pore, HMW1B (60). In this class of adhesins, which includes CdrA of P. aeruginosa, a bulky disulfide C-terminal hook anchors HMW1 through its HMW1B secretion partner rather than anchoring with an N-terminal retention module via a T1SS. However, in both systems, a large bulky domain acts to block the secretion of the substrate until this domain is cleaved by a BTLCP family periplasmic protease.
The nuclear magnetic resonance (NMR) structure of a portion of the putative retention module from MpIBP of Marinomonas primoryensis, an adhesin with an N-terminal domain similar to LapA, revealed this region adopts a novel β-sandwich fold with a triangular shaped structure (10). Because this folded structure (24 Å by 28 Å by 26 Å) is too large to pass through TolC (20-Å internal diameter), the authors reasoned this structure could prevent the extracellular release of the adhesin. The folded domain is followed by a proteolysis-sensitive region that also includes the LapG cleavage site (61). A circular dichroism (CD)-based analysis indicated that of the structural domains tested from MpIBP, only the region corresponding to the N-terminal retention module did not require Ca2+ for folding. Additionally, thermal unfolding, solution X-ray scattering, and limited proteolysis analyses of LapA's N terminus (43) suggest that the retention module contains a well-folded far N-terminal region that prevents the extracellular secretion of the adhesin and a poorly folded C-terminal region that is sensitive to limited proteolysis by subtilisin and proteinase K and which could serve to thread through the OM LapE pore (Fig. 1C, step 3). This model is consistent with that proposed for MpIBP (61).
The structure of the N-terminal folded retention domain from MpIBP also revealed a hydrophobic surface patch that is conserved with that of LapA (61). Its functional relevance is not entirely clear at the moment, but Guo et al. speculate it could aid in stabilizing the anchoring of the adhesin. The retention domain is preceded by a random coil motif that in MpIBP contains two phenylalanine residues, hypothesized to interact directly with the outer membrane (61). Assuming both hydrophobic features mediate membrane binding, it is equally plausible that their interaction is with the inner membrane, bringing the LapG-processing site closer to the LapD-LapG complex, which may increase the efficiency of protease targeting. Notably, unlike the conserved hydrophobic surface patch on the folded retention domain, the phenylalanine residues in the disordered N terminus are not conserved in LapA, indicating that they are not part of a consensus mechanism.
Altogether, these complementary structural and mechanistic studies present strong evidence that a large group of BTLCP-linked adhesins utilize a novel cell surface localization mechanism for T1SS substrates and represent an important advance in our understanding of T1SS functional diversity. Rather than following the canonical one-step translocation strategy of T1SS, the release of these adhesins involves a two-step secretion mechanism whereby a large adhesin is first loaded into the outer membrane as a secretion intermediate through interactions with its cognate T1SS OM pore and a periplasmic N-terminal retention module (Fig. 1C, steps 1 to 3). Proteolysis at the dialanine cleavage motif within the retention module releases the adhesin from the cell surface in a separate well-regulated step (Fig. 1C, steps 4 and 5).
T1SS MACHINERY LINKED WITH BTLCPs IS A DISTINCT SUBFAMILY OF THIS SECRETION SYSTEM
AggA of Shewanella spp. is a TolC-like outer membrane protein shown to transport the RTX adhesin BpfA (62) (Fig. 2), which is critical for biofilm formation and pellicle formation (63), a floating biofilm-like lifestyle. The genes for BpfA, AggA, and the remaining T1SS machinery are next to the genes for LapD and LapG accessory proteins in this organism, which coordinate to modulate biofilm formation by Shewanella (47). Interestingly, AggA is upregulated in a hyperaggregating mutant of Shewanella oneidensis MR-1 isolated from a rifampin-challenged culture. This hyperaggregating mutant, termed S. oneidensis COAG (64), indicates that increased production of AggA is associated with increased microbial community formation. A bioinformatics analysis suggests that AggA belongs to a subclass of TolC-like proteins involved with secreting large T1SS adhesins (65). Included in this group are LapE of P. fluorescens Pf0-1 and other LapE-like proteins linked with the LapD and LapG accessory proteins, suggesting the outer membrane component of T1SS adhesin transporters is distinct from those of general TolC, RND, and major facilitator outer membrane pores. Consistent with the bioinformatics evidence, biophysical studies suggest that AggA has features distinct from TolC. Single-channel conductance recordings of the closed state show that AggA has a wider periplasmic channel than TolC, indicating this group of OM pores may be more relaxed when rested. However, it is currently unclear how these molecular features promote the secretion and/or anchoring of large adhesins such as BpfA.
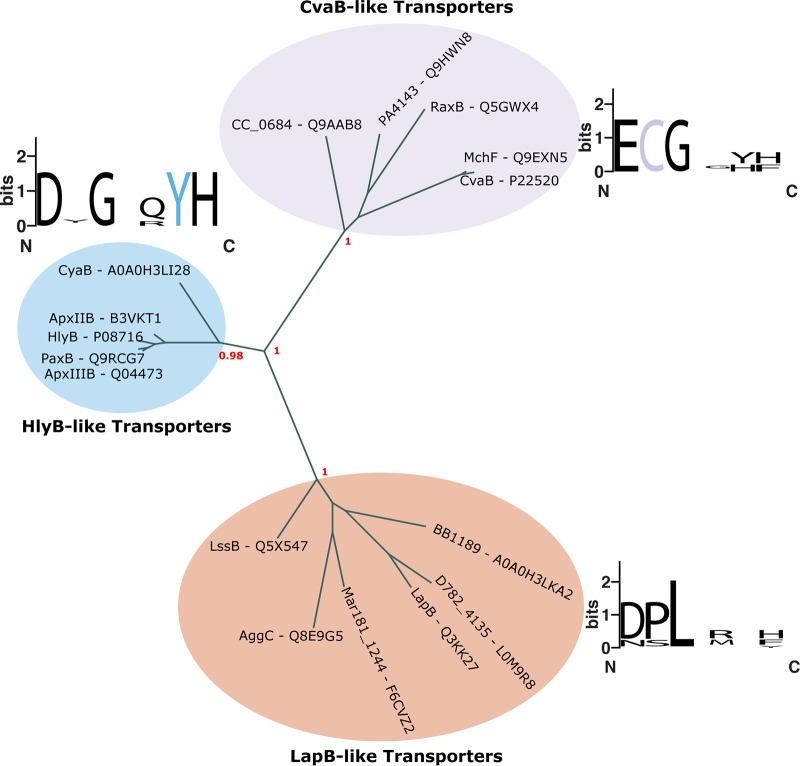
A phylogenetic analysis of the inner membrane ABC transporters linked with the LapD and LapG accessory proteins also suggests that these transporters represent a novel subclass of T1SS adhesin ABC transporters. Like HlyB, LapB and other RTX adhesin transporters contain previously characterized N-terminal CLDs. However, the amino acid signature found in the LapB CLD is distinct from the HlyB CLD, suggesting RTX toxin transporters can be differentiated from RTX adhesin transporters linked with LapD and LapG accessory proteins (Fig. 3). Indeed, the ABC transporters of the RTX toxin CyaA and RTX adhesin BrtA harbored by B. bronchiseptica cluster with HlyB and LapE transporters, respectively, suggesting their functional differences are reflected by distinct sequences.
FIG 3.
The phylogenic relationship between BTLCP-linked and classical ABC transporters. Phylogenetic analysis of bacteriocin, RTX toxin, and BTLCP-linked ABC transporters involved in T1SS. The web tool http://www.phylogeny.fr/ was used to construct the phylogenic tree with default settings. Bootstrap values are indicated in red and bolded. WebLogo results of residues at regions critical for bacteriocin processing (purple, C) and RTX binding (blue, Y) are shown. The corresponding region for the BTLCP-linked ABC transporters, lacking these residues, is also shown.
Importantly, the CLDs from BTLCP-linked ABC transporters lack the conserved residues critical for HlyB CLD binding of the unfolded HlyA C-terminal RTX motifs. It is currently unknown if LapB binds the C-terminal RTX motifs of LapA via a distinct mechanism or binds elsewhere in LapA. Given that a LapA mutant lacking RTX motifs efficiently localizes to the cell surface and promotes biofilm formation (54), it seems unlikely that the RTX motifs have a critical role in LapA secretion. Of note, the C39 and CLD bind glycine-rich regions in their respective substrates. LapA's N terminus contains an additional glycine-rich region, which could be targeted by the LapB CLD to discourage premature cytoplasmic folding of the gigantic adhesin. To date, no chaperone for LapA or any other RTX-containing protein has been characterized.
COOPTING THE Lap SYSTEM FOR SYNTHETIC BIOLOGY
Although bacteria have been studied in recent years for their roles in human disease, the recent upsurge in microbiome research has led many to attempt to harness bacterial metabolic and protective capabilities to promote human health. Similarly, microbial engineering has addressed a variety of industrial needs, including the production of microbial metabolites. Such processes might often benefit from strains that are effectively retained during fermentation processes. One ambition of synthetic biology is to program bacterial cell surfaces for downstream purposes, such as probiotic development (66), biofilm engineering (67, 68), bioremediation of toxic metals (69, 70), and biomining (71). Given the role of various Gram-negative bacteria in these processes, which naturally express at least one T1SS variant, it is enticing to consider the Lap and related systems as a viable platform for these engineering purposes. As mentioned above, the diversity of the internal regions of these RTX adhesins is far ranging, and multiple groups have shown the classical T1SS can translocate a variety of mammalian, Gram-positive, and cytoplasmic proteins directly to the extracellular environment when fused to a C-terminal T1SS secretion signal. Such fusion proteins include alkaline phosphatase (72), streptokinase harbored by beta-hemolytic streptococci (73), β-lactamase (74), maltose binding protein (75), the intracellular mammalian intestinal fatty acid binding protein IFABP (76), PhoA (77), and subtilisin E of Bacillus subtilis (78). The amenability of the T1SS to secrete diverse substrates suggests the Lap system could be coopted to display large multifunctional cargo proteins at the cell surface. Importantly, the release of the surface-displayed cargo can be tied to internal c-di-GMP levels (or other signals), enabling directed and reversible biofilm engineering. Furthermore, c-di-GMP-producing and -degrading enzymes are linked to nearly countless natural and engineered stimuli, including near-infrared light (79), oxygen (62), metal availability (80), phosphorylation (81), and blue light (82, 83), indicating LapG activity, which is controlled by a c-di-GMP receptor, can be tailored to a host of cues for engineering purposes. For example, the ability to regulate the display of these adhesins could aid in controlling gut colonization of engineered probiotic strains or recruiting bacteria with unique metabolic capabilities to a surface for engineering microbial consortia.
CONCLUSIONS AND FUTURE DIRECTIONS
The ability to interact with and respond to environmental factors is integral to bacterial survival. The T1SS was first characterized as secreting pore-forming toxins, such as HlyA, involved with virulence. The role of this secretion system has expanded over the last several decades to include large biofilm-promoting adhesins, MARTX, proteases, protective S-layer proteins, bacterial lipases, small bacteriocins involved in bacterial warfare, and iron-scavenging proteins (9, 22). A central tenet of this secretion strategy is that substrate translocation into the extracellular environment occurs in a single step, bypassing the periplasmic space. The identification of type 1-secreted adhesins that are often encoded near the gene for a periplasmic BTLCP revealed a novel subgroup of T1SSs that anchor their substrates at the cell surface through a cognate TolC-like OM pore. In contrast to the classical T1SS, a second-step—periplasmic proteolysis by the BTLCP—is required to release the substrate into the extracellular environment. In the absence of proteolysis, the substrate remains threaded through its TolC-like pore with the periplasmic retention module blocking the extracellular release of the adhesin. The ABC transporter and OM TolC-like pore of this two-step mechanism form a distinct phylogenic group compared to their classical T1SS counterparts, suggesting that these translocation components have evolved to secrete and retain this large group of RTX adhesins with a periplasmic intermediate. Interestingly, BTLCP-linked T1SS may be derived from a hybrid of the bacteriocin and RTX toxin secretion systems, with the calcium-dependent bacteriocin-activating C39 proteolysis event being allocated to a calcium-dependent periplasmic BTLCP that releases the RTX adhesin from its cognate OM pore. Instead of the bacteriocin N-terminal secretion signal, these RTX adhesins use a C-terminal signal similar to that used for the RTX toxins. Interestingly, the LapA-like proteins, like bacteriocins, are cleaved at their N termini to release these large adhesins from the cell, albeit by a different class of cysteine proteases.
Despite several decades of study, there are still many unanswered questions concerning the in RTX adhesin transporters such as LapB. One gap in our current knowledge is how such a large protein such as LapA remains unfolded in the cytoplasm in the apparent absence of a chaperone. RTX motifs have been shown to discourage premature folding within the cytoplasm, but it is unclear how these motifs accomplish this task when attached to the end of an up to 5,000-aa protein. Future biochemical, biophysical, and microscopy analyses to probe the LapA-LapE complex, as well as the role of the CLD of LapB in LapA folding and secretion, will significantly contribute to our understanding of this novel subgroup of T1SSs. It will be exciting to see future studies of this novel retention strategy and the role it plays in promoting biofilm formation in both commensal and pathogenic proteobacteria. If the wealth of classical T1SS literature amassed over the last several decades is any indicator, there is plenty left to discover about this two-step secretion strategy, and there will likely be many surprises along the way.
ACKNOWLEDGMENT
This work was supported by the NIH via grant R01GM123609 (to H.S. and G.A.O.).
Biographies

T. Jarrod Smith is a graduate student in the laboratory of George O'Toole at the Geisel School of Medicine at Dartmouth in Hanover, NH. His thesis work focuses on large biofilm-promoting adhesins in Gram-negative bacteria and their mechanisms of retention and surface regulation.

Holger Sondermann is a professor of molecular medicine at the College of Veterinary Medicine, Cornell University, Ithaca, NY. He received his Ph.D. in 2001 from the University of Cologne, Germany, for studies he conducted with Franz-Ulrich Hartl at the Max Planck Institute of Biochemistry, Martinsried, Germany. From 2001 to 2005, he was a postdoctoral fellow with John Kuriyan, first at the Rockefeller University and then at the University of California, Berkeley. In 2005, Dr. Sondermann joined the faculty at Cornell, where he started his work on bacterial c-di-GMP signaling and biofilm formation.

George A. O'Toole is a professor of microbiology and immunology at the Geisel School of Medicine at Dartmouth in Hanover, NH. He received his Ph.D. with Jorge Escalante-Semerena at the University of Wisconsin—Madison in 1994 and was a postdoctoral fellow with Roberto Kolter at Harvard Medical School. He started his faculty position at Dartmouth in 1999. From the beginning, Dr. O'Toole's lab has studied the mechanism of bacterial biofilm formation by Pseudomonas fluorescens. The Sondermann and O'Toole labs have been collaborating for over a decade.
REFERENCES
- 1.Welch R. 1991. Pore-forming cytolysins of Gram-negative bacteria. Mol Microbiol 5:512–528. doi: 10.1111/j.1365-2958.1991.tb00723.x. [DOI] [PubMed] [Google Scholar]
- 2.Andersen C, Hughes C, Koronakis V. 2000. Chunnel vision: export and efflux through bacterial channel-tunnels. EMBO Rep 1:313–318. doi: 10.1093/embo-reports/kvd075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackman N, Nicaud JM, Gray L, Holland IB. 1985. Identification of polypeptides required for the export of haemolysin 2001 from E. coli. Mol Gen Genet 201:529–536. doi: 10.1007/BF00331351. [DOI] [PubMed] [Google Scholar]
- 4.Kanonenberg K, Schwarz CKW, Schmitt L. 2013. Type I secretion systems - a story of appendices. Res Microbiol 164:596–604. doi: 10.1016/j.resmic.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Koronakis V, Koronakis E, Hughes C. 1989. Isolation and analysis of the C-terminal signal directing export of Escherichia coli hemolysin protein across both bacterial membranes. EMBO J 8:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangola P, Rosen BP. 1987. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem 262:12570–12574. [PubMed] [Google Scholar]
- 7.Chenal A, Guijarro JI, Raynal B, Delepierre M, Ladant D. 2009. RTX calcium binding motifs are intrinsically disordered in the absence of calcium implication for protein secretion. J Biol Chem 284:1781–1789. doi: 10.1074/jbc.M807312200. [DOI] [PubMed] [Google Scholar]
- 8.Bumba L, Masin J, Macek P, Wald T, Motlova L, Bibova I, Klimova N, Bednarova L, Veverka V, Kachala M, Svergun DI, Barinka C, Sebo P. 2016. Calcium-driven folding of RTX domain β-rolls ratchets translocation of RTX proteins through type I secretion ducts. Mol Cell 62:47–62. doi: 10.1016/j.molcel.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Linhartová I, Bumba L, Mašn J, Basler M, Osička R, Kamanová J, Procházková K, Adkins I, Hejnová-Holubová J, Sadílková L, Morová J, Šebo P. 2010. RTX proteins: a highly diverse family secreted bya common mechanism. FEMS Microbiol Rev 34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo S, Stevens CA, Vance TDR, Olijve LLC, Graham LA, Campbell RL, Yazdi SR, Escobedo C, Bar-Dolev M, Yashunsky V, Braslavsky I, Langelaan DN, Smith SP, Allingham JS, Voets IK, Davies PL. 2017. Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice. Sci Adv 3:e1701440. doi: 10.1126/sciadv.1701440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Auria G, Jiménez N, Peris-Bondia F, Pelaz C, Latorre A, Moya A. 2008. Virulence factor Rtx in Legionella pneumophila, evidence suggesting it is a modular multifunctional protein. BMC Genomics 9:14. doi: 10.1186/1471-2164-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo SLG, Bermudez LE, El-Etr SH, Duhamel GE, Cirillo JD. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect Immun 69:508–517. doi: 10.1128/IAI.69.1.508-517.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yousef-Coronado F, Travieso ML, Espinosa-Urgel M. 2008. Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida. FEMS Microbiol Lett 288:118–124. doi: 10.1111/j.1574-6968.2008.01339.x. [DOI] [PubMed] [Google Scholar]
- 14.Wagner W, Vogel M, Goebel W. 1983. Transport of haemolysin across the outer membrane of Escherichia coli requires two functions. J Bacteriol 154:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wandersman C, Delepelaire P. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci U S A 87:4776–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thanabalu T, Koronakis E, Hughes C, Koronakis V. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J 17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray L, Mackman N, Nicaud JM, Holland IB. 1986. The carboxy-terminal region of haemolysin 2001 is required for secretion of the toxin from Escherichia coli. Mol Gen Genet 205:127–133. doi: 10.1007/BF02428042. [DOI] [PubMed] [Google Scholar]
- 18.Mackman N, Baker K, Gray L, Haigh R, Nicaud JM, Holland IB. 1987. Release of a chimeric protein into the medium from Escherichia coli using the C-terminal secretion signal of haemolysin. EMBO J 6:2835–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarchau T, Chakraborty T, Garcia F, Goebel W. 1994. Selection for transport competence of C-terminal polypeptides derived from Escherichia coli hemolysin: the shortest peptide capable of autonomous HIyB/HIyD-dependent secretion comprises the C-terminal 62 amino acids of HlyA. Mol Gen Genet 245:53–60. doi: 10.1007/BF00279750. [DOI] [PubMed] [Google Scholar]
- 20.Hantke K, Winkler K, Schultz JE. 2011. Escherichia coli exports cyclic AMP via TolC. J Bacteriol 193:1086–1089. doi: 10.1128/JB.01399-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Gilson L, Mahanty HK, Kolter R. 1990. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J 9:3875–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satchell KJF. 2011. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol 65:71–90. doi: 10.1146/annurev-micro-090110-102943. [DOI] [PubMed] [Google Scholar]
- 23.Higgins CF, Hiles ID, Salmond GPC, Gill DR, Downie JA, Evans IJ, Holland IB, Gray L, Buckel SD, Bell AW, Hermodson MA. 1986. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323:448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- 24.ter Beek J, Guskov A, Slotboom DJ. 2014. Structural diversity of ABC transporters. J Gen Physiol 143:419–435. doi: 10.1085/jgp.201411164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenders MHH, Weidtkamp-Peters S, Kleinschrodt D, Jaeger K-E, Smits SHJ, Schmitt L. 2015. Directionality of substrate translocation of the hemolysin A type I secretion system. Sci Rep 5:12470. doi: 10.1038/srep12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 27.Pei X-Y, Hinchliffe P, Symmons MF, Koronakis E, Benz R, Hughes C, Koronakis V. 2011. Structures of sequential open states in a symmetrical opening transition of the TolC exit duct. Proc Natl Acad Sci U S A 108:2112–2117. doi: 10.1073/pnas.1012588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zgurskaya HI, Nikaido H. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci U S A 96:7190–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaur SJ, Rahman MS, Ammerman NC, Beier-sexton M, Ceraul SM, Gillespie JJ. 2012. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J Bacteriol 194:4920–4932. doi: 10.1128/JB.00793-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lecher J, Schwarz CKW, Stoldt M, Smits SHJ, Willbold D, Schmitt L. 2012. An RTX transporter tethers its unfolded substrate during secretion via a unique N-terminal domain. Structure 20:1778–1787. doi: 10.1016/j.str.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig a, Garcia F, Bauer S, Jarchau T, Benz R, Hoppe J, Ludwig A, Garcia F, Bauer S, Jarchau T, Hoppe R, Goebel W, Benz R. 1996. Analysis of the in vivo activation of hemolysin (HlyA) from Escherichia coli. Microbiology 178:5422–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barlag B, Hensel M. 2015. The giant adhesin SiiE of Salmonella enterica. Molecules 20:1134–1150. doi: 10.3390/molecules20011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wille T, Wagner C, Mittelstädt W, Blank K, Sommer E, Malengo G, Döhler D, Lange A, Sourjik V, Hensel M, Gerlach RG. 2014. SiiA and Siib are novel type I secretion system subunits controlling SPI4-mediated adhesion of Salmonella enterica. Cell Microbiol 16:161–178. doi: 10.1111/cmi.12222. [DOI] [PubMed] [Google Scholar]
- 34.Ginalski K, Kinch L, Rychlewski L, Grishin N. 2004. BTLCP proteins: a novel family of bacterial transglutaminase-like cysteine proteinases. Trends Biochem Sci 29:392–395. doi: 10.1016/j.tibs.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyd CD, Chatterjee D, Sondermann H, O'Toole GA. 2012. LapG, required for modulating biofilm formation by Pseudomonas fluorescens Pf0-1, is a calcium-dependent protease. J Bacteriol 194:4406–4414. doi: 10.1128/JB.00642-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navarro MVAS, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol 9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monds RD, Newell PD, Gross RH, O'Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 41.Cooley RB, O'Donnell JP, Sondermann H. 2016. Coincidence detection and bi-directional transmembrane signaling control a bacterial second messenger receptor. Elife 5:e21848. doi: 10.7554/eLife.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dahlstrom KM, O'Toole GA. 2017. A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu Rev Microbiol 71:179–195. doi: 10.1146/annurev-micro-090816-093325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith TJ, Font ME, Kelly CM, Sondermann H, O'Toole GA. 2018. An N-terminal retention module anchors the giant adhesin LapA of Pseudomonas fluorescens at the cell surface: a novel sub-family of type I secretion systems. J Bacteriol doi: 10.1128/JB.00734-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee D, Boyd CD, O'Toole GA, Sondermann H. 2012. Structural characterization of a conserved, calcium-dependent periplasmic protease from Legionella pneumophila. J Bacteriol 194:4415–4425. doi: 10.1128/JB.00640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatterjee D, Cooley RB, Boyd CD, Mehl RA, O'Toole GA, Sondermann H. 2014. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. Elife 3:e03650. doi: 10.7554/eLife.03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambrosis N, Boyd CD, O'Toole GA, Fernández J, Sisti F. 2016. Homologs of the LapD-LapG c-di-GMP effector system control biofilm formation by Bordetella bronchiseptica. PLoS One 11:e0158752. doi: 10.1371/journal.pone.0158752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou G, Yuan J, Gao H. 2015. Regulation of biofilm formation by BpfA, BpfD, and BpfG in Shewanella oneidensis. Front Microbiol 6:790. doi: 10.3389/fmicb.2015.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez-Mendoza D, Coulthurst SJ, Humphris S, Campbell E, Welch M, Toth IK, Salmond GPC. 2011. A multi-repeat adhesin of the phytopathogen, Pectobacterium atrosepticum, is secreted by a type I pathway and is subject to complex regulation involving a non-canonical diguanylate cyclase. Mol Microbiol 82:719–733. doi: 10.1111/j.1365-2958.2011.07849.x. [DOI] [PubMed] [Google Scholar]
- 49.De León KB, Zane GM, Trotter VV, Krantz GP, Arkin AP, Butland GP, Walian PJ, Fields MW, Wall JD. 2017. Unintended laboratory-driven evolution reveals genetic requirements for biofilm formation by Desulfovibrio vulgaris Hildenborough. mBio 8:e01696-17. doi: 10.1128/mBio.01696-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 51.Cooley RB, Smith TJ, Leung W, Tierney V, Borlee BR, O'Toole GA, Sondermann H. 2016. Cyclic di-GMP-regulated periplasmic proteolysis of a Pseudomonas aeruginosa type Vb secretion system substrate. J Bacteriol 198:66–76. doi: 10.1128/JB.00369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rybtke M, Berthelsen J, Yang L, Høiby N, Givskov M, Tolker-Nielsen T. 2015. The LapG protein plays a role in Pseudomonas aeruginosa biofilm formation by controlling the presence of the CdrA adhesin on the cell surface. Microbiologyopen 4:917–930. doi: 10.1002/mbo3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyd CD, Smith TJ, El-Kirat-Chatel S, Newell PD, Dufrêne YF, O'Toole GA. 2014. Structural features of the Pseudomonas fluorescens biofilm adhesin LapA required for LapG-dependent cleavage, biofilm formation, and cell surface localization. J Bacteriol 196:2775–2788. doi: 10.1128/JB.01629-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cirillo SLG, Lum J, Cirillo JD. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345–1359. doi: 10.1099/00221287-146-6-1345. [DOI] [PubMed] [Google Scholar]
- 56.Ruggeri ZM, Ware J. 1993. von Willebrand factor. FASEB J 7:308–316. doi: 10.1096/fasebj.7.2.8440408. [DOI] [PubMed] [Google Scholar]
- 57.Cirillo SLG, Yan L, Samrakandi MM, Cirillo JD. 2002. Role of the Legionella pneumophila rtxA gene in amoebae. Microbiology 148:1667–1677. doi: 10.1099/00221287-148-6-1667. [DOI] [PubMed] [Google Scholar]
- 58.Qin T, Zhou H, Ren H, Liu W. 2017. Distribution of secretion systems in the genus Legionella and its correlation with pathogenicity. Front Microbiol 8:388. doi: 10.3389/fmicb.2017.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishikawa S, Shinzawa N, Nakamura K, Ishigaki K, Abe H, Horiguchi Y. 2016. The Bvg-repressed gene brtA, encoding biofilm-associated surface adhesin, is expressed during host infection by Bordetella bronchiseptica. Microbiol Immunol 60:93–105. doi: 10.1111/1348-0421.12356. [DOI] [PubMed] [Google Scholar]
- 60.Buscher AZ, Grass S, Heuser J, Roth R, St Geme JW. 2006. Surface anchoring of a bacterial adhesin secreted by the two-partner secretion pathway. Mol Microbiol 61:470–483. doi: 10.1111/j.1365-2958.2006.05236.x. [DOI] [PubMed] [Google Scholar]
- 61.Guo S, Langelaan DN, Phippen SW, Smith SP, Voets IK, Davies PL. 2018. Conserved structural features anchor biofilm-associated RTX–adhesins to the outer membrane of bacteria. FEBS J 285:1812–1826. [DOI] [PubMed] [Google Scholar]
- 62.Wu C, Cheng Y-Y, Yin H, Song X-N, Li W-W, Zhou X-X, Zhao L-P, Tian L-J, Han J-C, Yu H-Q. 2013. Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci Rep 3:1945. doi: 10.1038/srep01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang Y, Gao H, Chen J, Dong Y, Wu L, He Z, Liu X, Qiu G, Zhou J. 2010. Pellicle formation in Shewanella oneidensis. BMC Microbiol 10:291. doi: 10.1186/1471-2180-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Vriendt K, Theunissen S, Carpentier W, De Smet L, Devreese B, Van Beeumen J. 2005. Proteomics of Shewanella oneidensis MR-1 biofilm reveals differentially expressed proteins, including AggA and RibB. Proteomics 5:1308–1316. doi: 10.1002/pmic.200400989. [DOI] [PubMed] [Google Scholar]
- 65.Theunissen S, Vergauwen B, De Smet L, Van Beeumen J, Van Gelder P, Savvides SN. 2009. The agglutination protein AggA from Shewanella oneidensis MR-1 is a TolC-like protein and forms active channels in vitro. Biochem Biophys Res Commun 386:380–385. doi: 10.1016/j.bbrc.2009.06.044. [DOI] [PubMed] [Google Scholar]
- 66.Mathipa MG, Thantsha MS. 2017. Probiotic engineering: towards development of robust probiotic strains with enhanced functional properties and for targeted control of enteric pathogens. Gut Pathog 9:28. doi: 10.1186/s13099-017-0178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen F, Wegner SV. 2017. Blue light switchable bacterial adhesion as a key step toward the design of biofilms. ACS Synth Biol 6:2170–2174. doi: 10.1021/acssynbio.7b00197. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen PQ, Botyanszki Z, Tay PKR, Joshi NS. 2014. Programmable biofilm-based materials from engineered curli nanofibres. Nat Commun 5:4945. doi: 10.1038/ncomms5945. [DOI] [PubMed] [Google Scholar]
- 69.Tay PKR, Nguyen PQ, Joshi NS. 2017. A synthetic circuit for mercury bioremediation using self-assembling functional amyloids. ACS Synth Biol 6:1841–1850. doi: 10.1021/acssynbio.7b00137. [DOI] [PubMed] [Google Scholar]
- 70.Eskandari V, Yakhchali B, Sadeghi M, Karkhane AA, Ahmadi-Danesh H. 2015. Efficient cadmium bioaccumulation by displayed hybrid CS3 pili: effect of heavy metal binding motif insertion site on adsorption capacity and selectivity. Appl Biochem Biotechnol 177:1729–1741. doi: 10.1007/s12010-015-1849-y. [DOI] [PubMed] [Google Scholar]
- 71.Brune KD, Bayer TS. 2012. Engineering microbial consortia to enhance biomining and bioremediation. Front Microbiol 3:203. doi: 10.3389/fmicb.2012.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Erb K, Vogel M, Wagner W, Goebel W. 1987. Alkaline phosphatase which lacks its own signal sequence becomes enzymatically active when fused to N-terminal sequences of Escherichia coli haemolysin (HlyA). Mol Gen Genet 208:88–93. doi: 10.1007/BF00330427. [DOI] [PubMed] [Google Scholar]
- 73.Kern I, Ceglowski P. 1995. Secretion of streptokinase fusion proteins from Escherichia coli cells through the hemolysin transporter. Gene 163:53–57. doi: 10.1016/0378-1119(95)00395-M. [DOI] [PubMed] [Google Scholar]
- 74.Chervaux C, Sauvonnet N, Le Clainche A, Kenny B, Hunt AL, Broome-Smith JK, Holland IB. 1995. Secretion of active β-lactamase to the medium mediated by the Escherichia coli haemolysin transport pathway. Mol Gen Genet 249:237–245. doi: 10.1007/BF00290371. [DOI] [PubMed] [Google Scholar]
- 75.Bakkes PJ, Jenewein S, Smits SHJ, Holland IB, Schmitt L. 2010. The rate of folding dictates substrate secretion by the Escherichia coli hemolysin type 1 secretion system. J Biol Chem 285:40573–40580. doi: 10.1074/jbc.M110.173658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwarz CKW, Landsberg CD, Lenders MHH, Smits SHJ, Schmitt L. 2012. Using an E. coli type 1 secretion system to secrete the mammalian, intracellular protein IFABP in its active form. J Biotechnol 159:155–161. doi: 10.1016/j.jbiotec.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 77.Hess J, Gentschev I, Goebel W, Jarchau T. 1990. Analysis of the haemolysin secretion system by PhoA-HlyA fusion proteins. Mol Gen Genet 224:201–208. doi: 10.1007/BF00271553. [DOI] [PubMed] [Google Scholar]
- 78.Sugamata Y, Shiba T. 2005. Improved secretory production of recombinant proteins by random mutagenesis of HlyB, an alpha-hemolysin transporter from Escherichia coli improved secretory production of recombinant proteins by random mutagenesis of HlyB, an alpha-hemolysin transporter. Appl Environ Microbiol 71:656–662. doi: 10.1128/AEM.71.2.656-662.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ryu M, Gomelsky M. 2014. Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth Biol 3:802–810. doi: 10.1021/sb400182x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gomelsky M. 2013. A zinc lock on GGDEF domain dimerization inhibits E. coli biofilms. Structure 21:1067–1068. doi: 10.1016/j.str.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 81.Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem 282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- 82.Tschowri N, Busse S, Hengge R. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev 23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Savakis P, De Causmaecker S, Angerer V, Ruppert U, Anders K, Essen LO, Wilde A. 2012. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol Microbiol 85:239–251. [DOI] [PubMed] [Google Scholar]