We previously reported that an increase in cellular Mg2+ content can suppress defects in 70S ribosome formation and growth rate caused by the absence of ribosomal protein L34. In the present study, we demonstrated that, even in mutants lacking individual ribosomal proteins other than L34 (L1, L23, L36, and S6), an increase in the cellular Mg2+ content could restore 70S ribosome formation. Moreover, the defect in sporulation caused by the absence of L1 was also suppressed by an increase in the cellular Mg2+ content. These findings indicate that at least part of the function of these ribosomal proteins can be complemented by Mg2+, which is essential for all living cells.
KEYWORDS: ribosome, ribosomal protein, magnesium, Bacillus subtilis, ribosomes
ABSTRACT
Individually, the ribosomal proteins L1, L23, L36, and S6 are not essential for cell proliferation of Bacillus subtilis, but the absence of any one of these ribosomal proteins causes a defect in the formation of the 70S ribosomes and a reduced growth rate. In mutant strains individually lacking these ribosomal proteins, the cellular Mg2+ content was significantly reduced. The deletion of YhdP, an exporter of Mg2+, and overexpression of MgtE, the main importer of Mg2+, increased the cellular Mg2+ content and restored the formation of 70S ribosomes in these mutants. The increase in the cellular Mg2+ content improved the growth rate and the cellular translational activity of the ΔrplA (L1) and the ΔrplW (L23) mutants but did not restore those of the ΔrpmJ (L36) and the ΔrpsF (S6) mutants. The lack of L1 caused a decrease in the production of Spo0A, the master regulator of sporulation, resulting in a decreased sporulation frequency. However, deletion of yhdP and overexpression of mgtE increased the production of Spo0A and partially restored the sporulation frequency in the ΔrplA (L1) mutant. These results indicate that Mg2+ can partly complement the function of several ribosomal proteins, probably by stabilizing the conformation of the ribosome.
IMPORTANCE We previously reported that an increase in cellular Mg2+ content can suppress defects in 70S ribosome formation and growth rate caused by the absence of ribosomal protein L34. In the present study, we demonstrated that, even in mutants lacking individual ribosomal proteins other than L34 (L1, L23, L36, and S6), an increase in the cellular Mg2+ content could restore 70S ribosome formation. Moreover, the defect in sporulation caused by the absence of L1 was also suppressed by an increase in the cellular Mg2+ content. These findings indicate that at least part of the function of these ribosomal proteins can be complemented by Mg2+, which is essential for all living cells.
INTRODUCTION
The bacterial 70S ribosome is a complex macromolecule that is composed of small (30S) subunit and large (50S) subunits. The small subunit is comprised of the 16S rRNA and more than 20 proteins, whereas the large subunit is comprised of the 23S and 5S rRNAs and more than 30 proteins (1, 2). Protein synthesis by the ribosome requires the coordinated action of these subunits. The small subunit associates with the mRNA and the anticodon stem-loop of the bound tRNA, and it engages in ensuring the fidelity of translation by checking for correct pairing between the codon and anticodon (3–7). The large subunit associates with the acceptor arms of the tRNA and catalyzes the formation of a peptide bond between the amino acid attached to the tRNA in the A site and the nascent peptide chain bound to the tRNA in the P site (8, 9). The ribosomal proteins that constitute these subunits play an important role(s) in translation. For instance, ribosomal protein L1, which is localized to the stalk region near the E site (10, 11), plays a critical role in the translocation of the newly deacylated tRNA from the P to the E sites (12). Ribosomal protein L2 plays important roles in binding of the tRNA to the A and P sites, peptidyltransferase activity, and formation of the peptide bond (13–17). Therefore, the mature conformation of the 70S ribosomes is required for efficient translation activity.
Although the ribosomal proteins are important in the translation process, as well as in the association of the ribosomal subunits (13, 18, 19), several genes encoding ribosomal proteins can be deleted. In Escherichia coli, 22 of the 54 genes for ribosomal proteins are not individually essential for cell proliferation (20, 21). Similarly, in Bacillus subtilis, 22 of the 57 genes for ribosomal proteins can be individually deleted (22). The rpmH gene, encoding ribosomal protein L34, which is a component of the large subunit, is one of the nonessential genes. Mutants lacking L34 have a severe defect in the formation of the 70S ribosome and a reduced growth rate (22). However, we found that the defect in the formation of 70S ribosomes and the reduction in the growth rate could be suppressed by an increase in the Mg2+ content in the cell (23).
Magnesium ions are the most abundant divalent cations in living cells (24, 25), and are important for the maintenance of ribosome structure. Mg2+ is required for both stabilization of the secondary structure of rRNA and binding of the ribosomal proteins to the rRNA (26–28). The in vitro association of the 30S and 50S ribosomal subunits to form intact 70S ribosomes depends strongly on the concentration of Mg2+ (29–31). Therefore, we believe that Mg2+ can partly complement the L34 function by stabilizing both the conformation of the 50S subunit and the intersubunit bridges.
The goal of the present study was to elucidate whether Mg2+ can also complement mutant strains lacking ribosomal proteins other than L34. We examined the effect of increasing the Mg2+ content in mutant strains individually lacking ribosomal proteins L1, L23, L36, and S6 on the formation of 70S ribosomes, the growth rate, the cellular translational activity, and on sporulation.
RESULTS
Reduction in the cellular Mg2+ content caused by lack of ribosomal proteins was restored by disruption of yhdP and overexpression of mgtE.
The defect in the formation of 70S ribosomes caused by the absence of L34 could be suppressed by increasing the cellular Mg2+ content (23). To investigate the generality of the partial complementation of ribosomal protein function by Mg2+, disruptions of yhdP and the multicopy plasmid pDGmgtE, which can induce the overexpression of mgtE, were introduced into mutants lacking individual ribosomal proteins L1, L23, L36, and S6. Because we previously characterized the phenotypes of 19 mutants individually lacking ribosomal proteins and found that 70S ribosome formation by mutants individually lacking L1, L23, L36, and S6 was decreased (22), we chose these four mutants as the targets for analysis in this study.
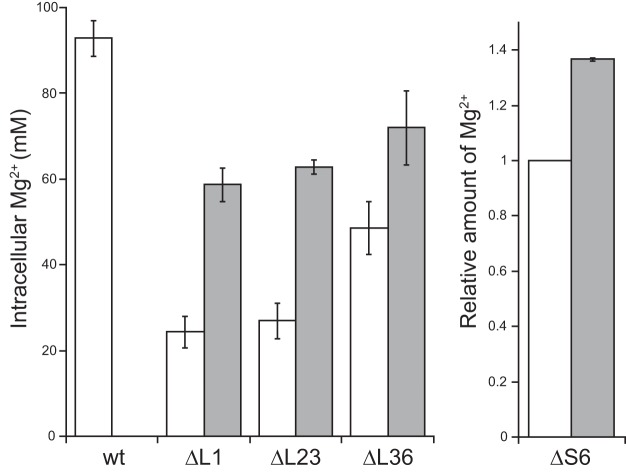
MgtE is the main importer of Mg2+ (32), whereas YhdP is probably an exporter of Mg2+ in B. subtilis (23). We previously reported that the absence of L34 (RpmH) caused a decrease in the Mg2+ content in the cell, probably due to a reduced number of 70S ribosomes, and that the Mg2+ content in the ΔrpmH mutant was restored by disruption of yhdP and overexpression of mgtE (23). Similarly, the Mg2+ contents in the ΔrplA (L1), ΔrplW (L23) and ΔrpmJ (L36) mutants were also significantly reduced (Fig. 1). However, the Mg2+ content in these three mutants was restored, albeit incompletely, by disruption of yhdP and overexpression of mgtE (Fig. 1). In these experiments, the cellular Mg2+ concentration was calculated by dividing the amount of Mg2+ per cell by the cell volume. The cell volume of each mutant was estimated from the cell size, which was measured by microscopic analysis, as described in Materials and Methods. However, the cell size of the ΔrpsF (S6) mutant could not be defined because the cellular morphology of the ΔrpsF mutant was aberrantly filamentous (see Fig. S1 in the supplemental material). Thus, in the ΔrpsF mutant, the relative Mg2+ amount per cell when the Mg2+ amount of a cell in the parental strain was defined as 1 is shown in Fig. 1. Although a comparison of the Mg2+ content in the ΔrpsF mutants with that in the wild type was difficult, the Mg2+ content in the ΔrpsF mutants was certainly increased by disruption of yhdP and overexpression of mgtE.
FIG 1.
Reduction in the Mg2+ content in mutant strains lacking individual ribosomal proteins and partial restoration by disruption of yhdP and overexpression of mgtE. The Mg2+ content per cell in exponential phase, which was measured as described in Materials and Methods, is shown. In the case of the ΔrpsF (S6) mutant, the relative amount of Mg2+ per cell is shown (see the text for details). White bars indicate the wild type and each mutant lacking individual ribosomal proteins. Gray bars indicate the results when yhdP was disrupted and mgtE was overexpressed. The means from three independent experiments are shown. Error bars indicate standard deviations.
It should be noted that the Mg2+ concentrations that were measured in this study are total Mg2+ concentrations, including Mg2+ ions chelated in proteins and nucleic acids, and not only the free Mg2+ concentrations. Because the cells were completely disrupted by sonication and proteins were denatured by acid treatment, the Mg2+ ions that were chelated in ribosomes, other enzymes, and nucleic acids were also detected by this method. The Mg2+ ions that are contained in a cell are usually chelated by proteins and nucleic acids, and thus only 5% of the Mg2+ ions in a cell are present as free metal ions (33–35).
The effect of increasing the cellular Mg2+ content of mutants lacking individual ribosomal proteins on the formation of 70S ribosomes, the growth rate, and the cellular translational activity.
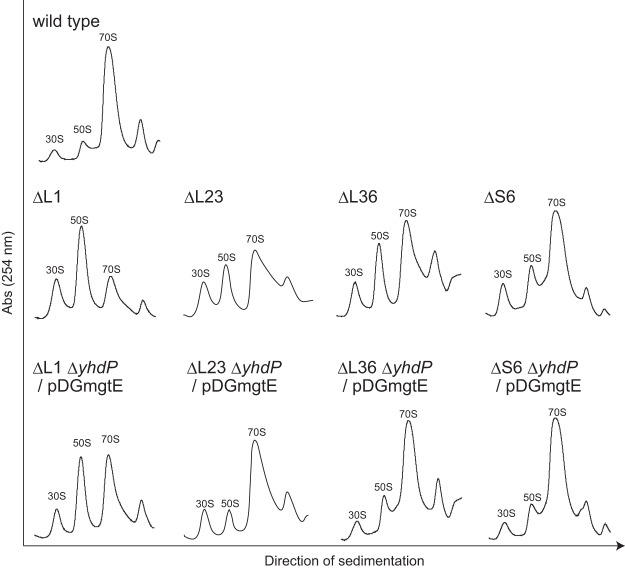
As shown in Fig. 2, the lack of individual ribosomal proteins (L1, L23, L36, and S6) caused defects in the formation of 70S ribosomes that are consistent with our previous data for several mutants lacking individual ribosomal proteins (22). The defect in 70S ribosome formation observed in these mutants was suppressed to various degrees by disruption of yhdP and overexpression of mgtE (Fig. 2). In all of the mutants investigated here, the amount of 70S ribosomes relative to the amount of dissociated subunits was restored by increasing the cellular Mg2+ content. These results indicate that Mg2+ can suppress the defect in the formation of 70S ribosomes and/or in the reduction in the stability of the 70S complex caused by the absence of several individual ribosomal proteins.
FIG 2.
Defect in 70S ribosome formation in the absence of each ribosomal protein and its suppression by the disruption of yhdP and overexpression of mgtE. Crude cell extracts were sedimented through a 10 to 40% sucrose gradient as described in Materials and Methods. The 30S, 50S, and 70S peaks are indicated in each individual profile. The term “/pDGmgtE” indicates the overexpression of mgtE in the mutant cells.
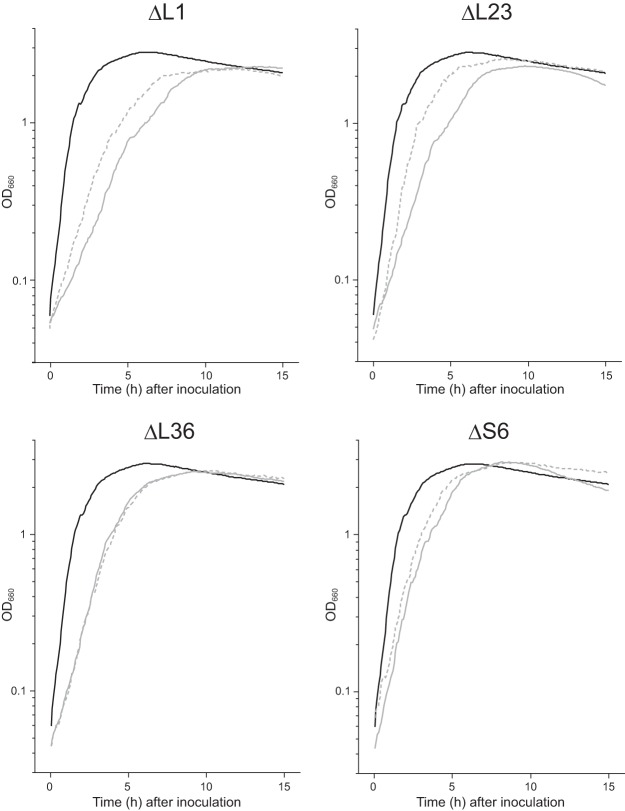
We next investigated the effect of the cellular Mg2+ content on the growth rate of the mutants. We have reported that the slow growth observed for the ΔrpmH (L34) mutant was suppressed by an increase in the Mg2+ content, probably due to the restoration of the amount of 70S ribosomes (23). A reduction in the growth rate was observed for the mutants lacking individual ribosomal proteins, which agrees with our previous results for several mutants lacking individual ribosomal proteins (22). As expected, in the ΔrplA (L1) and ΔrplW (L23) mutants, the growth rate was partially restored by disruption of yhdP and overexpression of mgtE (Fig. 3; Table 1). When only mgtE was overexpressed in the ΔrplA (L1) mutant, its effect on the growth rate was minimal (23). The combination of overexpression of mgtE and disruption of yhdP, however, increased the growth rate of the ΔrplA (L1) mutant. In contrast, the growth rates of the ΔrpmJ (L36) and ΔrpsF (S6) mutants were not significantly increased when the cellular Mg2+ content was increased (Fig. 3; Table 1).
FIG 3.
Effects of the increase in cellular Mg2+ content on the growth rate of the mutant strains lacking individual ribosomal proteins. Cells were grown in LB at 37°C, and the optical density at 660 nm was measured. Growth curves of the wild type and each mutant lacking individual ribosomal proteins are shown using black and gray solid lines, respectively. Growth curves of each mutant in which yhdP was disrupted and mgtE was overexpressed are shown by dotted lines.
TABLE 1.
Doubling times of mutants lacking ribosomal proteins
| Strain description | Doubling time ± SD (min)a |
|
|---|---|---|
| Parental | ΔyhdP/pDGmgtE | |
| wt | 21.4 ± 1.4 | |
| ΔrplA (L1) | 67.7 ± 1.7 | 54.6 ± 0.49 |
| ΔrplW (L23) | 56.7 ± 1.9 | 37.0 ± 2.4 |
| ΔrpmJ (L36) | 43.3 ± 1.3 | 41.2 ± 0.54 |
| ΔrpsF (S6) | 38.2 ± 1.1 | 36.4 ± 0.49 |
Means from three independent experiments are shown.
To investigate whether the partial restoration of the growth rate of the ΔrplA (L1) and ΔrplW (L23) mutants by the increase in the cellular Mg2+ content was dependent on an increase in cellular translational activity, we measured the translational activity in cells lacking individual ribosomal proteins. As expected, the cellular translational activity of the ΔrplA (L1) and ΔrplW (L23) mutants was severely reduced (Fig. 4). In the ΔrplA (L1) and ΔrplW (L23) mutants, an increase in the cellular Mg2+ content partially restored the cellular translational activity, whereas in the ΔrpmJ (L36) and ΔrpsF (S6) mutants, the effect of increased cellular Mg2+ content on the translational activity was minimal. These results are correlated with the effect that the increased cellular Mg2+ content had on the growth rate. Therefore, the partial restoration of the cellular translational activity by the increased cellular Mg2+ content improved the growth rate of the ΔrplA (L1) and ΔrplW (L23) mutants.
FIG 4.
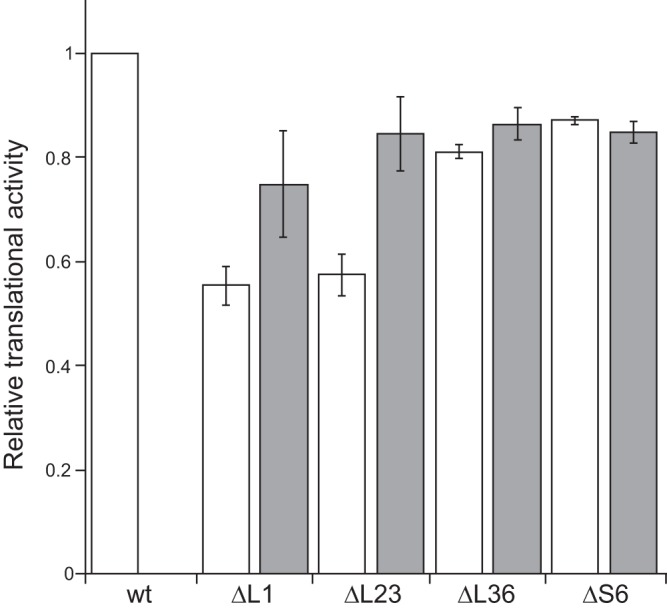
Effects of the increase in cellular Mg2+ content on the cellular translational activity of the mutant strains lacking individual ribosomal proteins. The relative translational activity in mutant cells compared to that of the wild type is shown. White bars indicate the wild type and each mutant lacking individual ribosomal proteins. Gray bars indicate the results when yhdP was disrupted and mgtE was overexpressed. The means from three independent experiments are shown. Error bars indicate standard deviations.
The increase in the cellular Mg2+ content suppresses the defect in sporulation caused by the absence of ribosomal protein L1.
We previously found that the absence of ribosomal protein L1 causes a defect in sporulation (22). It should be noted that this phenotype was not caused solely by the decreased growth rate, because the sporulation frequency of the ΔrpmH (L34) mutant, which also showed a severe growth defect similar to that of the ΔrplA (L1) mutant, was almost the same as that of the wild type (22). We therefore investigated whether the sporulation defect of the ΔrplA (L1) mutant could be suppressed by Mg2+. Consistent with our previous data, the ΔrplA (L1) mutant was severely defective in forming heat-resistant spores (the sporulation frequency was less than 0.01%), whereas the other mutants formed spores with frequencies of >70% (Table 2). However, the sporulation frequency of the ΔrplA (L1) mutant was partially restored by disruption of yhdP and overexpression of mgtE (Table 2). In addition, the growth rate of the ΔrplA (L1) mutant in sporulation medium was also restored by disruption of yhdP and overexpression of mgtE (Fig. 5A). These results indicate that increasing the cellular Mg2+ content can partially suppress not only the growth defect but also the sporulation defect in the ΔrplA (L1) mutant.
TABLE 2.
Restoration of sporulation frequency of the ΔL1 mutant by Mg2+
| Strain description | CFU · ml−1a |
Frequency (%) ± SDa | |
|---|---|---|---|
| Total | Spores | ||
| wt | 6.4 × 108 | 5.8 × 108 | 90 ± 7.9 |
| ΔL23 | 4.9 × 108 | 3.5 × 108 | 71 ± 5.8 |
| ΔL36 | 6.4 × 108 | 4.6 × 108 | 73 ± 13 |
| ΔS6 | 3.5 × 108 | 2.7 × 108 | 77 ± 12 |
| ΔL1 | 1.6 × 108 | 2.3 × 102 | (1.7 ±1.2) × 10−4 |
| ΔL1 ΔyhdP/pDGmgtE | 3.1 × 108 | 7.5 × 107 | 25 ± 4.7 |
Means from three independent experiments.
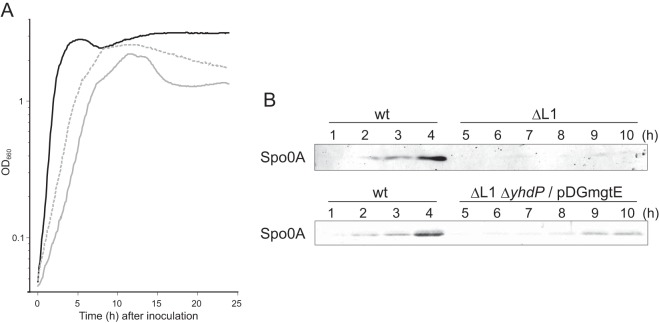
FIG 5.
Reduction in the growth rate and in the production of Spo0A (caused by lack of L1) and their suppression by the disruption of yhdP and overexpression of mgtE. (A) Cells were grown in sporulation medium (2×SG) at 37°C, and the optical density at 660 nm was measured. Growth curves of the wild type and the ΔrplA (L1) mutant are shown using black and gray solid lines, respectively. The growth curve of the L1 mutant in which yhdP was disrupted and mgtE was overexpressed is shown by a dotted line. (B) Cells were grown in sporulation medium at 37°C and were collected at the indicated times. Crude cell extracts were subjected to Western blot analysis using antisera against Spo0A. The term “/pDGmgtE” indicates the overexpression of mgtE in the mutant cells.
The restoration of spore formation by the ΔrplA (L1) mutant prompted us to identify which stage of sporulation was affected by the absence of L1 and restoration by Mg2+. At the initiation stage of B. subtilis sporulation, cells divide asymmetrically, and chromosomal DNA is concentrated in the forespore (36). In fact, asymmetric septation and concentration of chromosomal DNA were detected in the wild-type cells 5 h after inoculation into sporulation medium (see Fig. S2 in the supplemental material). In contrast, in the ΔrplA (L1) cells, asymmetric septation was not observed, even 24 h after inoculation (Fig. S2). However, asymmetric septation and concentration of chromosomal DNA were detected in the ΔrplA (L1) cells 15 h after inoculation when yhdP was disrupted and mgtE was overexpressed (Fig. S2). We next examined the level of Spo0A in the ΔrplA (L1) mutant. Phosphorylation of Spo0A, the master transcriptional regulator of sporulation, governs the decision to initiate sporulation (37–39). In the wild type and mutants other than the ΔrplA (L1) mutant, the level of Spo0A increased 4 h and 6 to 8 h after inoculation into sporulation medium, respectively (Fig. 5B; see also Fig. S3 in the supplemental material). In contrast, Spo0A was barely detectible in ΔrplA (L1) cells even 10 h after inoculation (Fig. 5B). However, the disruption of yhdP and overexpression of mgtE in the ΔrplA (L1) cells increased the amount of Spo0A by 9 h after inoculation, although the level of Spo0A remained lower than that in the wild type (Fig. 5B). These results indicate that the defect in the initiation stage of sporulation caused by the absence of L1 can be at least partially suppressed by an increase in the Mg2+ content in the cell.
DISCUSSION
The cellular Mg2+ contents of the mutant strains individually lacking L1, L23, or L36 were reduced compared to that of the wild type, while that of the ΔrpsF (S6) mutant was difficult to calculate because of its filamentous cellular morphology (Fig. 1; also see Fig. S1 in the supplemental material). The reduction in the amount of Mg2+ was probably caused by the smaller amount of 70S ribosomes, which harbor more than 170 Mg2+ ions per complex (40), and by a decrease in the amount of protein and RNA other than ribosomes that can chelate Mg2+. In fact, we previously showed that the reduction in the cellular Mg2+ content correlated with the decrease in the amount of 70S ribosomes (23). On the other hand, the disruption of yhdP and overexpression of mgtE increased cellular Mg2+ content and restored the formation of 70S ribosomes in the mutants tested here that lacked individual ribosomal proteins (Fig. 2). Although the absence of L34 causes ribosomal protein L16 to dissociate from the 50S subunit, the increase in the Mg2+ content restores the binding of L16 to the 50S subunit, indicating that Mg2+ can stabilize the conformation of 50S subunits lacking L34 (23). Likewise, stabilization of the conformation of each subunit as well as bridges between the subunits by Mg2+ probably restored 70S formation and/or stability of 70S complexes in the mutants lacking individual ribosomal proteins tested here (L1, L23, L36, and S6).
The increase in 70S ribosome formation restored cellular translational activity and growth rates of the ΔrplA (L1) and ΔrplW (L23) mutants (Fig. 4, Fig. 3, and Table 1). However, the restoration of cellular translational activity and growth rates of these mutants was only partial. Possible reasons for these partial restorations are (i) an incomplete restoration of the normal amount of 70S ribosomes and (ii) functions of the ribosomal proteins other than in stabilizing the 70S ribosomes could not be complemented by Mg2+. Ribosomal protein L1 plays a critical role in the translocation of the newly deacylated tRNA from the P to the E site (12), while ribosomal protein L23, which is located at the polypeptide exit channel of the large subunit, tethers trigger factor to the ribosome (41). Trigger factor, which is the first molecular chaperone interacting with newly synthesized polypeptides by the ribosome, promotes protein folding (42–44). The functions of these ribosomal proteins are probably essential for efficient growth. In contrast to mutants lacking L1 or L23, the cellular translational activity and growth rates of the ΔrpmJ (L36) and ΔrpsF (S6) mutants were not significantly increased when the Mg2+ content was increased, although 70S ribosome formation was restored to near wild-type levels (Fig. 2 and 3). Although the detailed functions of L36 and S6 in protein synthesis are unknown, their role(s) in translation or other function(s) does not appear to be complemented by Mg2+. In addition, the filamentous morphology of cells caused by the absence of S6 was not suppressed by increasing cellular Mg2+ content (Fig. S2).
The increase in cellular Mg2+ content suppressed not only the defect of 70S ribosome formation but also the sporulation defect of the ΔrplA (L1) mutant (Table 2). Although the level of Spo0A, the master regulator of sporulation, was drastically reduced in the ΔrplA (L1) mutant, Spo0A levels were partially restored when cellular Mg2+ content was increased (Fig. 5B) and resulted in the restoration of sporulation frequency of the ΔrplA (L1) mutant. The phosphorylated form of Spo0A activates expression of sporulation genes, as well as its own gene, via a positive feedback loop (45, 46). Therefore, possible reasons why the level of Spo0A decreased in the ΔrplA (L1) mutant include inhibition of phosphorylation of Spo0A, which in turn causes reduction in the expression level of Spo0A, and/or simply a decrease in the stability of Spo0A. The increase in the cellular Mg2+ content in the ΔrplA (L1) mutant may suppress these defects by increasing the amount of 70S ribosomes and/or by increasing the cellular translational activity. However, the ΔrplW (L23) mutant, whose amount of 70S ribosomes and cellular translational activity were decreased similarly to those in the ΔrplA (L1) mutant, formed spores with an almost normal frequency (Table 2), implying a complex mechanism for the sporulation defect caused by the absence of L1.
In the present study, we demonstrated that the defect in the formation of 70S ribosomes as well as in sporulation caused by the absence of individual ribosomal proteins can be suppressed by increasing the cellular Mg2+ content. Mg2+ plays a crucial role not only in the ribosome but also in numerous biological processes and cellular functions, such as the activation and catalytic reactions of hundreds of enzymes, utilization of ATP, and maintenance of genomic stability (47, 48). Clarifying the relationship between the ribosome and Mg2+, both of which are essential to living cells, is important for understanding cellular function. It has been suggested that the sizes of ribosomal proteins have increased during evolution to complement the function of the rRNA, which originally acted as a ribozyme (49, 50). From another point of view, increasing of the sizes of ribosomal proteins and/or binding of ribosomal proteins to the ribosome during evolution can be considered to complement the Mg2+ function in the ribosome, because in the ribosome, the relative abundance of Mg2+ is decreased, whereas that of ribosomal proteins is increased (50, 51). Further investigation to reveal the mechanism of complementation of the ribosomal protein function by Mg2+ may provide important information about the evolution of the ribosome.
MATERIALS AND METHODS
Media and culture conditions.
LB medium (52), LB agar, and 2× Schaeffer's sporulation medium supplemented with 0.1% glucose (2×SG) (53) were used. The culture conditions and media for preparing competent cells have been described previously (54). When required, 5 μg · ml−1 chloramphenicol, 5 μg · ml−1 kanamycin, and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) were added to the media. Growth curves of B. subtilis cells were generated by automatically measuring the optical density at 660 nm (OD660) value of each culture every 5 min using a TVS062CA incubator (Advantec).
Bacterial strains.
All of the B. subtilis strains used in this study are isogenic with B. subtilis strain 168 trpC2. The ΔrplA::cat, ΔrplW::cat, ΔrpmJ::cat, and ΔrpsF::cat strains, which were constructed by replacing the open reading frame of each gene with a promoterless cat gene lacking an intrinsic terminator sequence, were described previously (22). Chromosomal DNA extracted from the ΔrplA::cat, ΔrplW::cat, ΔrpmJ::cat, and ΔrpsF::cat strains was used to transform the strain harboring ΔyhdP::erm and the plasmid pDGmgtE, which carries the mgtE gene under the control of an IPTG-inducible Pspac promoter (23), and the transformants were selected on the basis of their chloramphenicol-resistant phenotype.
Measurement of the cellular Mg2+ content.
The cellular Mg2+ content was measured as described previously (23). Briefly, B. subtilis cells were grown in LB medium to exponential phase and harvested. Simultaneously, viable cells were counted by plating the culture on LB agar plates. The cells were resuspended in lysis buffer and disrupted by sonication, and then the pH of the crude extract was adjusted to approximately 3.0 with hydrochloric acid to denature the proteins. The amount of Mg2+ in the cell lysate was measured with a Metallo assay kit for magnesium (Metallogenics). The Mg2+ content per cell was calculated by dividing the amount of Mg2+ in the crude extract by the number of viable cells. The concentration of Mg2+ was calculated by assuming that a B. subtilis cell is a cylinder. To measure the cell size (radius and length), microscopic images were analyzed by MicrobeJ, an ImageJ plug-in (55). The mean size of >30 cells was used for the calculation.
Sucrose density gradient sedimentation analysis.
B. subtilis cells were grown in LB medium at 37°C with shaking to exponential phase (OD600, ∼0.4) and harvested. Sucrose density gradient sedimentation analysis was performed as described previously (22). Briefly, the cells were disrupted by passage through a French pressure cell, and cell debris was removed by centrifugation. Aliquots of extract were layered onto 10 to 40% sucrose density gradients, which were subjected to centrifugation at 4°C for 17.5 h at 65,000 × g (Hitachi P40ST rotor). Samples were collected with a Piston Gradient Fractionator (BioComP), and absorbance profiles were monitored at 254 nm using a Bio-Mini UV monitor (ATTO, Japan). When normalizing the applied volume by the total absorbance at 260 nm, 10 A260 units of crude extract per tube were used.
Measurement of the cellular translational activity.
B. subtilis cells were grown in LB medium at 37°C with shaking to exponential phase (OD600, ∼0.4) and harvested. The cellular translational activity was measured according to the manufacturer's instructions (Protein Synthesis assay kit; Cayman Chemical). Briefly, cells that were harvested from 100 μl of culture were incubated at 37°C with shaking for 30 min in 100 μl of O-propargyl-puromycin (OPP) working solution, which contained a 2-fold higher concentration of OPP than that recommended by the manufacturer. After incubation in the assay fixative and washing, cells were stained with 5-fluorescein amidite-azide (5-FAM-azide). The number of cells was normalized by the OD600 value, and fluorescence was detected using a Typhoon FLA 9500 (GE Healthcare). The fluorescence intensities were calculated by ImageJ (56).
Sporulation assay.
B. subtilis cells were grown in 2×SG medium for 24 h at 37°C with shaking. Heat-resistant spores were counted by heating the cells at 80°C for 10 min, plating them on LB agar plates, and then incubating the plates at 37°C for 24 h.
Microscopic imaging.
B. subtilis cells were grown in 2×SG medium at 37°C with shaking. At the indicated times, 500 μl of the culture was removed and subjected to centrifugation at 12,000 × g for 1 min. The cell pellet was resuspended in 40 μl of culture supernatant and then FM 4-64 (Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI) (Wako Pure Chemical Industries) were added to final concentrations of 10 μg/ml and 5 μg/ml, respectively. The cell suspension was mounted on a microscope slide and coated with poly-l-lysine to fix the cells, and differential interference contrast and fluorescence images were obtained with a confocal fluorescence microscope (LSM800; Carl Zeiss).
Western blot analysis.
Western blot analysis was performed according to a previously described method (57). Aliquots (15 μg of protein) of crude cell extracts were loaded onto a sodium dodecyl sulfate polyacrylamide gel (12%) and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore Co., Japan). This membrane was then used in the Western blot assay, using antisera (1:10,000 dilution) against Spo0A (58).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by Grants-in-Aid for Scientific Research (C) (grants 26450101 and 15K07013 to G.A. and Y.K.-Y., respectively), Grants-in-Aid for Young Scientists (B) (grants 17K15253 and 23770157 to G.A. and Y.K.-Y., respectively), and a Strategic Research Foundation Grant-aided Project for Private Universities (grant S1201003 to F.K. and Y.K.-Y.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00212-18.
REFERENCES
- 1.Kurland CG. 1972. Structure and function of the bacterial ribosome. Annu Rev Biochem 41:377–408. doi: 10.1146/annurev.bi.41.070172.002113. [DOI] [PubMed] [Google Scholar]
- 2.Nomura M. 1970. Bacterial ribosome. Bacteriol Rev 34:228–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenner L, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol 17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 4.Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 5.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102:615–623. doi: 10.1016/S0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 6.Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 7.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. 2001. The path of messenger RNA through the ribosome. Cell 106:233–241. doi: 10.1016/S0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 8.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 9.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 10.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal RK, Lata RK, Frank J. 1999. Conformational variability in Escherichia coli 70S ribosome as revealed by 3D cryo-electron microscopy. Int J Biochem Cell Biol 31:243–254. doi: 10.1016/S1357-2725(98)00149-6. [DOI] [PubMed] [Google Scholar]
- 12.Fei J, Kosuri P, MacDougall DD, Gonzalez RL. 2008. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell 30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Diedrich G, Spahn CM, Stelzl U, Schafer MA, Wooten T, Bochkariov DE, Cooperman BS, Traut RR, Nierhaus KH. 2000. Ribosomal protein L2 is involved in the association of the ribosomal subunits, tRNA binding to A and P sites and peptidyl transfer. EMBO J 19:5241–5250. doi: 10.1093/emboj/19.19.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khaitovich P, Mankin AS, Green R, Lancaster L, Noller HF. 1999. Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc Natl Acad Sci U S A 96:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulze H, Nierhaus KH. 1982. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. EMBO J 1:609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlein M, Weglöhner W, Urlaub H, Wittmann-Liebold B. 1998. Functional implications of ribosomal protein L2 in protein biosynthesis as shown by in vivo replacement studies. Biochem J 331:423–430. doi: 10.1042/bj3310423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willumeit R, Forthmann S, Beckmann J, Diedrich G, Ratering R, Stuhrmann HB, Nierhaus KH. 2001. Localization of the protein L2 in the 50S subunit and the 70S E. coli ribosome. J Mol Biol 305:167–177. doi: 10.1006/jmbi.2000.4289. [DOI] [PubMed] [Google Scholar]
- 18.Teraoka H, Nierhaus KH. 1978. Protein L16 induces a conformational change when incorporated into a L16-deficient core derived from Escherichia coli ribosomes. FEBS Lett 88:223–226. doi: 10.1016/0014-5793(78)80179-3. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki S, Tanigawa O, Akanuma G, Nanamiya H, Kawamura F, Tagami K, Nomura N, Kawabata T, Sekine Y. 2014. Enhanced expression of Bacillus subtilis yaaA can restore both the growth and sporulation defects caused by mutation of rplB, encoding ribosomal protein L2. Microbiology 160:1040–1053. doi: 10.1099/mic.0.076463-0. [DOI] [PubMed] [Google Scholar]
- 20.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG. 2011. Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol 413:751–761. doi: 10.1016/j.jmb.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akanuma G, Nanamiya H, Natori Y, Yano K, Suzuki S, Omata S, Ishizuka M, Sekine Y, Kawamura F. 2012. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J Bacteriol 194:6282–6291. doi: 10.1128/JB.01544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akanuma G, Kobayashi A, Suzuki S, Kawamura F, Shiwa Y, Watanabe S, Yoshikawa H, Hanai R, Ishizuka M. 2014. Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. J Bacteriol 196:3820–3830. doi: 10.1128/JB.01896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maguire ME, Cowan JA. 2002. Magnesium chemistry and biochemistry. Biometals 15:203–210. doi: 10.1023/A:1016058229972. [DOI] [PubMed] [Google Scholar]
- 25.Wacker WE. 1969. The biochemistry of magnesium. Ann N Y Acad Sci 162:717–726. doi: 10.1111/j.1749-6632.1969.tb13003.x. [DOI] [PubMed] [Google Scholar]
- 26.Drygin D, Zimmermann RA. 2000. Magnesium ions mediate contacts between phosphoryl oxygens at positions 2122 and 2176 of the 23S rRNA and ribosomal protein L1. RNA 6:1714–1726. doi: 10.1017/S1355838200001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein DJ, Moore PB, Steitz TA. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrov AS, Bernier CR, Hsiao C, Okafor CD, Tannenbaum E, Stern J, Gaucher E, Schneider D, Hud NV, Harvey SC, Williams LD. 2012. RNA-magnesium-protein interactions in large ribosomal subunit. J Phys Chem B 116:8113–8120. doi: 10.1021/jp304723w. [DOI] [PubMed] [Google Scholar]
- 29.Blaha G, Burkhardt N, Nierhaus KH. 2002. Formation of 70S ribosomes: large activation energy is required for the adaptation of exclusively the small ribosomal subunit. Biophys Chem 96:153–161. doi: 10.1016/S0301-4622(02)00021-2. [DOI] [PubMed] [Google Scholar]
- 30.Liiv A, O'Connor M. 2006. Mutations in the intersubunit bridge regions of 23 S rRNA. J Biol Chem 281:29850–29862. doi: 10.1074/jbc.M603013200. [DOI] [PubMed] [Google Scholar]
- 31.Tissieres A, Watson JD, Schlessinger D, Hollingworth BR. 1959. Ribonucleoprotein particles from Escherichia coli. J Mol Biol 1:221–233. doi: 10.1016/S0022-2836(59)80029-2. [DOI] [Google Scholar]
- 32.Wakeman CA, Goodson JR, Zacharia VM, Winkler WC. 2014. Assessment of the requirements for magnesium transporters in Bacillus subtilis. J Bacteriol 196:1206–1214. doi: 10.1128/JB.01238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moncany ML, Kellenberger E. 1981. High magnesium content of Escherichia coli B. Experientia 37:846–847. [DOI] [PubMed] [Google Scholar]
- 34.Alatossava T, Jutte H, Kuhn A, Kellenberger E. 1985. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol 162:413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. 2004. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett 237:49–55. doi: 10.1111/j.1574-6968.2004.tb09677.x. [DOI] [PubMed] [Google Scholar]
- 36.Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoch JA. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol 47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 38.Stephenson K, Hoch JA. 2002. Evolution of signalling in the sporulation phosphorelay. Mol Microbiol 46:297–304. doi: 10.1046/j.1365-2958.2002.03186.x. [DOI] [PubMed] [Google Scholar]
- 39.Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. 2003. The Spo0A regulon of Bacillus subtilis. Mol Microbiol 50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 40.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 41.Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. 2002. L23 protein functions as a chaperone docking site on the ribosome. Nature 419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- 42.Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, Deuerling E. 2008. Molecular mechanism and structure of trigger factor bound to the translating ribosome. EMBO J 27:1622–1632. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartl FU, Hayer-Hartl M. 2009. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 44.Hoffmann A, Bukau B, Kramer G. 2010. Structure and function of the molecular chaperone Trigger Factor. Biochim Biophys Acta 1803:650–661. doi: 10.1016/j.bbamcr.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 45.Hoch JA. 1991. spo0 genes, the phosphorelay, and the initiation of sporulation, p 747–755. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and other Gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC. [Google Scholar]
- 46.Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552. doi: 10.1016/0092-8674(91)90238-T. [DOI] [PubMed] [Google Scholar]
- 47.Cowan JA. 2002. Structural and catalytic chemistry of magnesium dependent enzymes. Biometals 15:225–235. doi: 10.1023/A:1016022730880. [DOI] [PubMed] [Google Scholar]
- 48.Hartwig A. 2001. Role of magnesium in genomic stability. Mutat Res 475:113–121. doi: 10.1016/S0027-5107(01)00074-4. [DOI] [PubMed] [Google Scholar]
- 49.Bokov K, Steinberg SV. 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457:977–980. doi: 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- 50.Wachowius F, Attwater J, Holliger P. 2017. Nucleic acids: function and potential for abiogenesis. Q Rev Biophys 50:e4. doi: 10.1017/S0033583517000038. [DOI] [PubMed] [Google Scholar]
- 51.Hsiao C, Mohan S, Kalahar BK, Williams LD. 2009. Peeling the onion: ribosomes are ancient molecular fossils. Molecular Biology and Evolution 26:2415–2425. doi: 10.1093/molbev/msp163. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 53.Leighton TJ, Doi RH. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem 246:3189–3195. [PubMed] [Google Scholar]
- 54.Ashikaga S, Nanamiya H, Ohashi Y, Kawamura F. 2000. Natural genetic competence in Bacillus subtilis natto OK2. J Bacteriol 182:2411–2415. doi: 10.1128/JB.182.9.2411-2415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang C, Brown PJ, Ducret A, Brun YV. 2014. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature 506:489–493. doi: 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nanamiya H, Shiomi E, Ogura M, Tanaka T, Asai K, Kawamura F. 2003. Involvement of ClpX protein in the post-transcriptional regulation of a competence specific transcription factor, ComK protein, of Bacillus subtilis. J Biochem 133:295–302. doi: 10.1093/jb/mvg040. [DOI] [PubMed] [Google Scholar]
- 58.Nanamiya H, Ohashi Y, Asai K, Moriya S, Ogasawara N, Fujita M, Sadaie Y, Kawamura F. 1998. ClpC regulates the fate of a sporulation initiation sigma factor, σH protein, in Bacillus subtilis at elevated temperatures. Mol Microbiol 29:505–513. doi: 10.1046/j.1365-2958.1998.00943.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.