Despite being one of the most common zoonoses worldwide, there is currently no human vaccine to combat brucellosis. Therefore, a better understanding of the pathogenesis and biology of Brucella spp., the causative agent of brucellosis, is essential for the discovery of novel therapeutics against these highly infectious bacteria. In this study, we further characterize the virulence-associated transcriptional regulator VtlR in Brucella abortus. Our findings not only shed light on our current understanding of a virulence related genetic system in Brucella spp. but also increase our knowledge of small proteins in the field of bacteriology.
KEYWORDS: Brucella, small protein, VtlR regulon
ABSTRACT
Elucidating the function of proteins <50 amino acids in length is no small task. Nevertheless, small proteins can play vital roles in the lifestyle of bacteria and influence the virulence of pathogens; thus, the investigation of the small proteome is warranted. Recently, our group identified the Brucella abortus protein VtlR as a transcriptional activator of four genes, one of which is the well-studied small regulatory RNA AbcR2, while the other three genes encode hypothetical small proteins, two of which are highly conserved among the order Rhizobiales. This study provides evidence that all three genes encode authentic small proteins and that all three are highly expressed under oxidative stress, low-pH, and stationary-phase growth conditions. Fractionation of the cells revealed that the proteins are localized to the membranes of B. abortus. We demonstrate that the small proteins under the transcriptional control of VtlR are not accountable for attenuation observed with the B. abortus vtlR deletion strain. However, there is an association between VtlR-regulated genes and growth inhibition in the presence of the sugar l-fucose. Subsequent transcriptomic analyses revealed that B. abortus initiates the transcription of a locus encoding a putative sugar transport and utilization system when the bacteria are cultured in the presence of l-fucose. Altogether, our observations characterize the role of the VtlR-controlled small proteins BAB1_0914, BAB2_0512, and BAB2_0574 in the biology of B. abortus, particularly in the capacity of the bacteria to utilize l-fucose.
IMPORTANCE Despite being one of the most common zoonoses worldwide, there is currently no human vaccine to combat brucellosis. Therefore, a better understanding of the pathogenesis and biology of Brucella spp., the causative agent of brucellosis, is essential for the discovery of novel therapeutics against these highly infectious bacteria. In this study, we further characterize the virulence-associated transcriptional regulator VtlR in Brucella abortus. Our findings not only shed light on our current understanding of a virulence related genetic system in Brucella spp. but also increase our knowledge of small proteins in the field of bacteriology.
INTRODUCTION
The term “small protein” is an unrestricting term encompassing any protein that is less than 50 amino acids in length without posttranslational processing (1). Generally considered too small to contribute to the physiology and biology of bacteria, small proteins are often left unannotated, and without optimized protocols to investigate small proteins, the study of small proteins has been neglected (2). This is not completely unwarranted due to the large number of open reading frames, ranging from 2 to 99 codons in prokaryotic and eukaryotic genomes. It was calculated that the number of open reading frames between 2 and 99 codons exceeded 250,000 in Saccharomyces cerevisiae (3). Several studies have been published exploring the ability to identify small proteins in both eukaryotes and bacteria (3, 4). This presents the problem of distinguishing between genes that encode functional small proteins and those that are meaningless. However, in the last decade, several small proteins have been well characterized in bacteria, and small proteins have been characterized for their various roles in sporulation, cell division, oxidative stress response, and as regulators of other proteins (5–8).
To date, only a few studies have described the role of small proteins found in Brucella spp., which are Gram-negative intracellular bacterial pathogens that are members of the Alphaproteobacteria class of bacteria. Each species preferentially infects a specific host; however, several Brucella strains adapted the ability to infect other hosts, including humans (9). Infection leads to spontaneous abortions and infertility in cows and swine, while causing a debilitating relapsing fever in humans (10). Upon infection, Brucella spp. traffic through human macrophages and dendritic cells to form an intracellular replicative niche (11). Regarding small proteins in Brucella spp., one of the most robust studies was performed when Sun et al. characterized whether 30 putative small proteins in Brucella abortus were important for the colonization of J774 macrophage-like cells or infection of a mouse model. Their findings indicated that four of the identified small proteins were important for either the infection of J774 macrophage-like cells or mice (6). The authors went on to characterize one of these small proteins, CydX, as a contributor to the function of cytochrome bd oxidase, but did not elucidate the mechanistic roles of the other small proteins. While microbiologists have historically overlooked small proteins, this study is a prime example of the potentially large value they bring to microbial physiology.
Recently, our group described the role of VtlR, a LysR-type transcriptional regulator (LTTR), in the virulence of B. abortus 2308 (12). We showed that VtlR is required for the ability of B. abortus to (i) survive and replicate in naive peritoneal macrophages and (ii) colonize the spleens of BALB/c mice. Microarray and Northern blot analyses revealed that VtlR transcriptionally activates four genes, abcR2, bab1_0914 (bab_rs20300), bab2_0512 (bab_rs28790), and bab2_0574 (bab_rs29075) (12). The small regulatory RNA (sRNA) AbcR2 has a sibling sRNA, AbcR1, that has regulatory functions redundant of those of AbcR2, and a deletion of one abcR gene does not cause a decrease in the ability of B. abortus to colonize the spleens of C57BL/6 mice (13). Since VtlR only activates the transcription of abcR2, and not abcR1, we hypothesized that a lack of expression of bab1_0914, bab2_0512, and/or bab2_0574 in a ΔvtlR mutant strain of B. abortus was accountable for the attenuation of the B. abortus ΔvtlR mutant strain in macrophages and mice. The data in the present study do not support this hypothesis, since isogenic deletions or the deletion of all three genes encoding small proteins do not lead to the attenuation of B. abortus in a BALB/c mouse model of infection. Despite this, we were able to further characterize the localization and expression profiles of the three small proteins. Moreover, we also observed an interesting phenotype involving the ability of B. abortus to utilize l-fucose during growth and sensitivity to this sugar when vtlR is deleted from the Brucella chromosome. Altogether, this study describes three novel small proteins in B. abortus and furthermore characterizes the link between these small proteins and the utilization of l-fucose by Brucella species.
RESULTS
BAB1_0914 and BAB2_0512 are well conserved among the order Rhizobiales and are highly similar in amino acid sequence.
bab1_0914 and bab2_0512 encode small hypothetical proteins of unknown function, and these putative proteins are predicted to be 48 amino acids in length (Fig. 1). bab1_0914 is flanked by bab1_0915, which encodes a hypothetical protein, and by the methyltransferase-encoding gene bab1_0913. bab2_0512 is flanked by bab2_0513 that encodes the glycine cleavage system protein T (GcvT) and bab2_0511 that encodes an oxidoreductase. Of note, the amino acid sequences of BAB1_0914 and BAB2_0512 are highly similar (Fig. 1D). In fact, BAB1_0914 and BAB2_0512 share over 75% amino acid identity and over 85% similarity. Another gene of interest whose expression is linked to VtlR in B. abortus is bab2_0574, which encodes a small hypothetical protein of 45 amino acids in length (Fig. 1C). bab2_0574 is located on chromosome II and is surrounded by bab2_0573 that encodes a transporter and bab2_0575 that encodes a hypothetical protein. While BAB2_0574 is well conserved across Brucella strains, bab1_0914 and bab2_0512 are well conserved throughout many members of the order Rhizobiales in the class Alphaproteobacteria. Moreover, it appears that bab1_0914 and bab2_0512 are homologous to a single gene found in other bacteria in the Rhizobiales. These include sMc02051 in Sinorhizobium meliloti 1021, atu1667 in Agrobacterium tumefaciens C58, and rleg2_1502 in Rhizobium leguminosarum bv. trifolii WSM2304.
FIG 1.
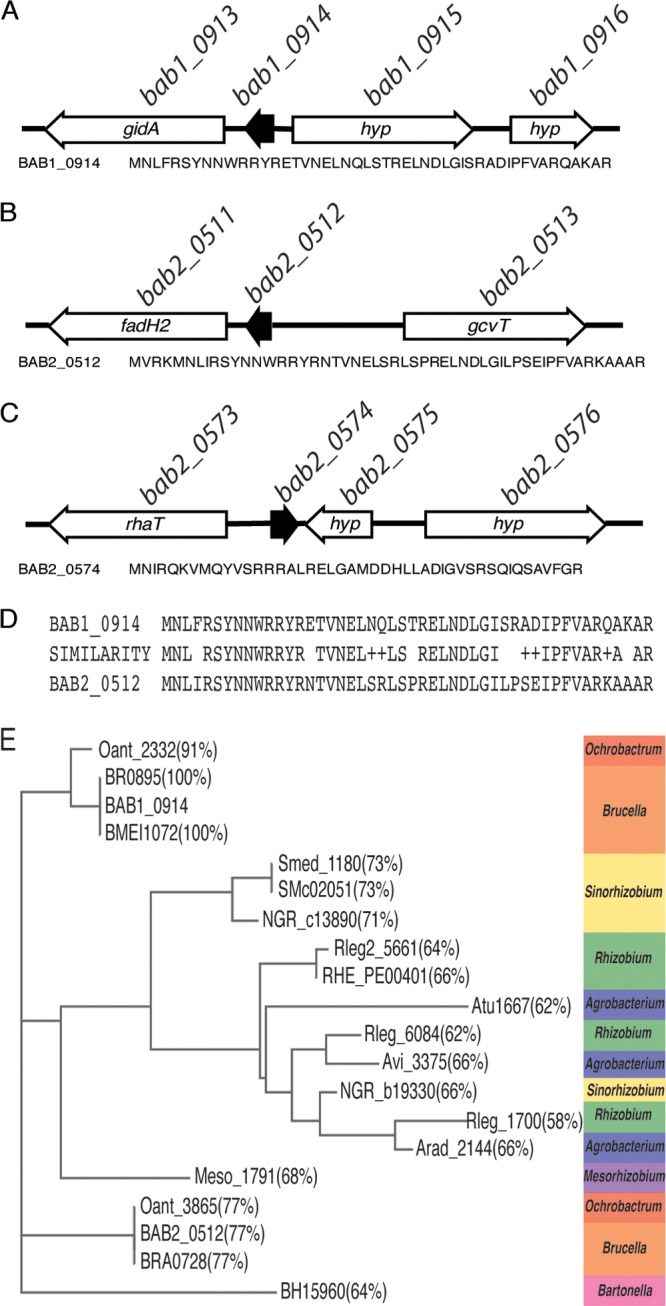
Organization of bab1_0914, bab2_0512, and bab2_0574 in Brucella abortus 2308 and amino acid sequence similarity among BAB1_0914 and BAB2_0512. (A to C) Genetic contexts and encoding amino acid sequences of bab1_0914 (bab_rs20300) (A), bab2_0512 (bab_rs28790) (B), and bab2_0574 (bab_rs29075) (C). (D) Amino acid sequence alignment of BAB1_0914 and BAB2_0512 created by NCBI Align Sequences Protein Blast. Identical amino acids are depicted by abbreviated amino acid symbols, and similar amino acids are depicted by +. (E) PHYLIP ML phylogenetic tree of BAB1_0914 and BAB2_0512 homologs created using the “Determine sequence relationships” tool at https://dnasubway.cyverse.org/. Percentages represent amino acid identity to BAB1_0914.
Utilizing BAB1_0914 as the template amino acid sequence and the online tool DNA Subway, a PHYLIP maximum likelihood (ML) phylogenetic tree was constructed showing the homologs of BAB1_0914 throughout the Rhizobiales (Fig. 1E). This analysis illustrated that genes encoding BAB1_0914-type small proteins are highly abundant in bacteria of the Rhizobiales, an order of bacteria that are extremely divergent in terms of their environmental niches. Related to this analysis, it should be noted that a homolog of BAB2_0512 is not listed for Brucella melitensis, because the gene encoding this protein is not annotated in the genome; however, an unannotated region of DNA with 100% nucleotide identity to bab2_0512 can be found in the B. melitensis genome downstream of bmeII0557.
bab1_0914, bab2_0512, and bab2_0574 encode small proteins and are expressed under oxidative and acidic stress.
The first question we sought to answer was whether or not bab1_0914, bab2_0512, and bab2_0574 encode authentic peptides. For this, a 3×FLAG tag was incorporated onto the C termini of BAB1_0914, BAB2_0512, and BAB2_0574 at each chromosomal locus, and we demonstrated the production of each protein (Fig. 2A). According to the annotated B. melitensis biovar Abortus 2308 genome in NCBI, BAB1_0914 and BAB2_0512 are both predicted to be 48 amino acids in length, and BAB2_0574 is predicted to be 45 amino acids in length. With the addition of the 22-amino-acid 3×FLAG tag, each protein is predicted to be 8 to 10 kDa. This is consistent with the bands observed in the Western blot analyses depicted in Fig. 2A.
FIG 2.

bab1_0914, bab2_0512, and bab2_0574 are activated during stationary phase of growth, low pH, and in the presence of H2O2. (A) Western blot analyses for protein expression of B. abortus 2308, B. abortus 2308 bab1_0914-3×FLAG, B. abortus 2308 bab2_0512-3×FLAG, and B. abortus 2308 bab2_0574-3×FLAG. Probes include anti-FLAG and anti-Omp89. (B) Northern blot analyses for RNA expression of AbcR1, AbcR2, BAB1_0914, BAB2_0512, and BAB2_0574 during stationary phase and exponential phase of growth in brucella broth, GMM, and E medium, and in the presence of H2O2 (5 μM), pH 4.5, and deoxycholate (0.5%) in brucella broth. (C to E) Western blot analyses for protein expression of BAB1_0914-3×FLAG, BAB2_0512-3×FLAG, BAB2_0574-3×FLAG, respectively, during stationary phase and exponential phase of growth in brucella broth, GMM, and E medium, and in the presence of H2O2 (5 μM), pH 4.5, and deoxycholate (0.5%) in brucella broth. Probes include anti-FLAG and anti-Omp89.
To begin to understand if bab1_0914, bab2_0512, and bab2_0574 are transcribed constitutively or under specific conditions, Northern blot analysis was conducted on RNA isolated from cultures of B. abortus 2308 grown under a variety of conditions. B. abortus 2308 was grown to mid-log phase in brucella broth, washed with phosphate-buffered saline (PBS), and then used to inoculate either brucella broth, Gerhardt's minimal medium (GMM), E medium (low nutrients and low pH), or brucella broth with the addition of hydrogen peroxide (5 μM), citric acid to lower the pH to 4.5, or deoxycholate (0.5% final concentration). A culture of B. abortus 2308 was also grown to stationary phase in brucella broth. Cultures were incubated for 30 min, and then RNA was extracted. Northern blot analysis was performed to determine the expression of AbcR1, AbcR2, BAB1_0914, BAB2_0512, BAB2_0574, and 5S rRNA as a control (Fig. 2B). This experiment revealed that AbcR2 was expressed at higher levels in stationary phase of growth and in the presence of H2O2, low pH, and in E medium than under the other conditions. BAB1_0914, BAB2_0512, and BAB2_0574 showed expression patterns similar to that of AbcR2, with the exception of E medium. BAB1_0914 and BAB2_0512 were not expressed at a detectable level in the presence of E medium. It is unknown why bab2_0512 and bab2_0574 produce two bands in the Northern blot. It is possible that one of the bands represents a processed or truncated form of the transcript; however, it should be noted that a deletion of bab2_0512 or bab2_0574 abolished the presence of both bands in a Northern blot, indicating that the two RNA bands are encoded by a single gene (data not shown).
The experimental design described above was repeated with the B. abortus strains harboring 3×FLAG versions of BAB1_0914, BAB2_0512, and BAB2_0574, with the intent of assessing protein production under the tested conditions. Using Western blot analysis, we observed expression levels of protein similar to that of RNA in the BAB1_0914-3×FLAG-tagged strain, with higher expression in stationary-phase cells and those exposed to lower pH and oxidative stress (Fig. 2C). Alternatively, the BAB2_0512 protein exhibited robust production in both exponential and stationary phases of growth, as well as in response to hydrogen peroxide stress, growth in E medium, and in the presence of deoxycholate (Fig. 2D). This observed discrepancy between RNA and protein levels of BAB2_0512 is interesting, and it is possible that a posttranscriptional regulatory mechanism exists to coordinate the levels of BAB2_0512 protein under certain environmental conditions. The increased production of BAB2_0574 was observed in cells grown to stationary phase and when the bacteria were exposed to hydrogen peroxide, and BAB2_0574 production was slightly induced when the bacteria were cultured in nutrient-limiting GMM and in medium with increased acidity (i.e., at pH 4.5) (Fig. 2E). Altogether, these experiments demonstrate that bab1_0914, bab2_0512, and bab2_0574 encode authentic proteins in B. abortus 2308 and, moreover, that the production of these small proteins is driven by conditions that are biological relevant to the brucellae.
Localization of BAB1_0914, BAB2_0512, and BAB2_0574 in the B. abortus cell.
Fractionation of B. abortus was developed from Thein et al., and Western blot analysis was applied to localize the three small proteins to the membrane fractions of B. abortus (Fig. 3) (14). Control Western blot analyses to support the successful fractionation method performed included anti-SodC antibody for the periplasmic fraction, anti-GroEL antibody for the cytoplasmic fraction, anti-Omp89 antibody for the membrane fraction. Anti-FLAG antibody was used to detect each 3×FLAG-tagged small protein.
FIG 3.
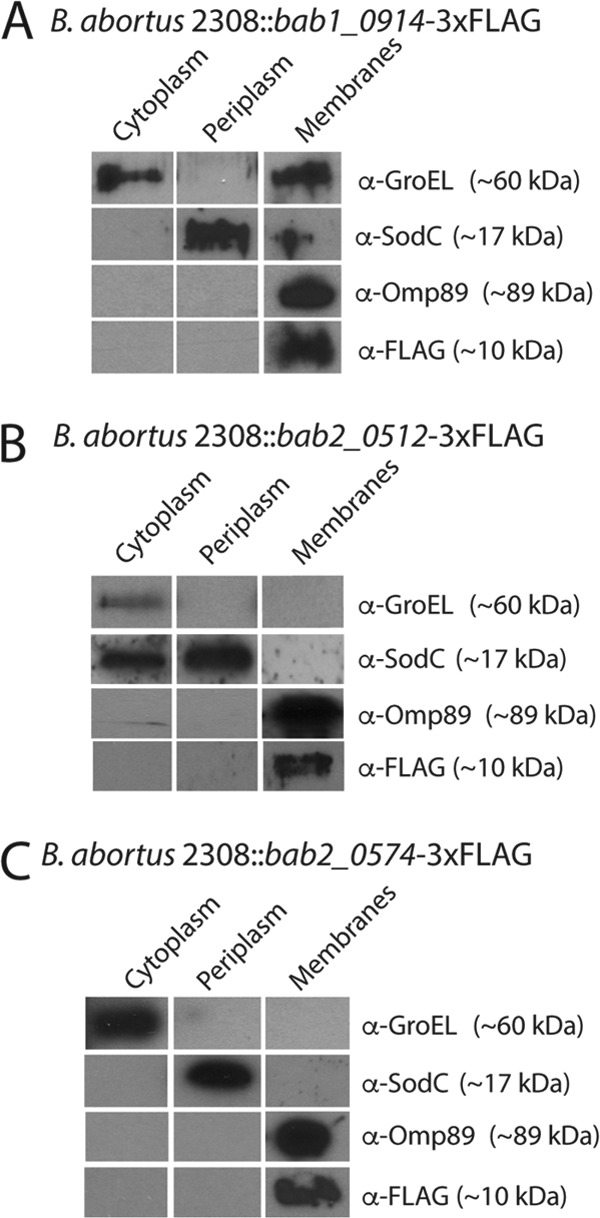
Localization of BAB1_0914, BAB2_0512, and BAB2_0574. Western blot analysis of Brucella strains grown to stationary phase, followed by fractionation methods described in Materials and Methods for B. abortus 2308 bab1_0914-3×FLAG (A), B. abortus 2308 bab2_0512-3×FLAG (B), and B. abortus 2308 bab2_0574-3×FLAG (C). Antibodies utilized include anti-FLAG, anti-Omp89, anti-SodC, and anti-GroEL.
The fractionation contained little contamination between the cytoplasmic, periplasmic, and membrane fractions in all three samples. The membrane fraction isolated from BAB1_0914-3×FLAG was contaminated with cytoplasmic proteins, as indicated by the presence of GroEL in the membrane fraction, and the cytoplasmic fraction from BAB2_0512-3×FLAG was contaminated with periplasmic proteins, as indicated by SodC in the cytoplasmic fraction. Despite this, no fractions other than the membrane fraction contained the protein Omp89, and thus, the appearance of FLAG-tagged protein in the membrane fraction alone reveals that BAB1_0914, BAB2_0512, and BAB2_0574 are localized to the membrane alone (Fig. 3).
These data are supported by the large number of charged and hydrophobic residues in the amino acid sequences of BAB1_0914, BAB2_0512, and BAB2_0574 (Fig. 1), and protein structure homology modeling via the free online server SWISS-MODEL revealed that the predicted structures for BAB1_0914, BAB2_0512, and BAB2_0574 all contain alpha-helices (see Fig. S1 in the supplemental material) (15–19). It should be noted that homology modeling provided BAB1_0914 and BAB2_0512 with the same predicted structure due to the highly similar amino acid sequences of the two small proteins.
Phenotypic analysis of the VtlR regulon.
Biolog Phenotype MicroArray plates for microbial cells were utilized to identify phenotypic differences between wild-type B. abortus 2308 and a deletion of the entire VtlR regulon (i.e., a strain with deletions of abcR2, bab1_0914, bab2_0512, and bab2_0574) to tease out any potential functions for any of the small proteins and/or AbcR2. For this, the parental B. abortus 2308 and the B. abortus 2308 ΔabcR2-Δbab1_0914 Δbab2_0512 Δbab2_0574 mutant strain (i.e., Δquad mutant) strains were used to inoculate 20 Biolog Phenotype MicroArray plates and grown statically at 37°C for 84 h. Metabolic activity, measured by cell respiration due to the reduction of a tetrazolium dye (clear to purple), was measured via the optical density at 590 nm (OD590) every 12 to 24 h (see Data Set S1 in the supplemental material). In a comparison with B. abortus 2308, the quadruple-deletion strain contained two major differences in growth. The B. abortus Δquad mutant strain was unable to utilize l-fucose as the sole carbon source for growth in minimal medium in plate 1, well B4 and showed minimal growth over the 84-h incubation time (Fig. 4A), while the parental strain was able to utilize l-fucose as a carbon source and grew to an OD of over 0.8. However, the Δquad mutant was able to grow in minimal medium (pH 4.5) with the addition of hydroxylysine in plate 10, well D8, while the growth of the parental strain was inhibited (Fig. 4B). The experiment was repeated by inoculating plate 1 and plate 10 of the Biolog system with the Δbab1_0914, Δbab2_0512, Δbab2_0574, and ΔabcR2 mutant strains, as well as a bab1_0914 bab2_0512 double-deletion strain and a bab1_0914 bab2_0512 bab2_0574 triple-deletion strain (Δtriple mutant) (Data Set S1). With regard to plate 10, the quadruple-deletion strain was able to grow in the presence of pH 4.5 minimal medium plus hydroxylysine. Conversely, the double- and triple-deletion strains had intermediate growth, and B. abortus 2308 and the isogenic bab1_0914 and bab2_0512 deletion mutants exhibited little to no growth in the presence of pH 4.5 minimal medium plus hydroxylysine. In plate 1, only B. abortus 2308 and the isogenic bab2_0574 deletion mutant were able to utilize fucose as the sole carbon source in minimal media. We sought to further examine these phenotypes outside the Biolog system.
FIG 4.
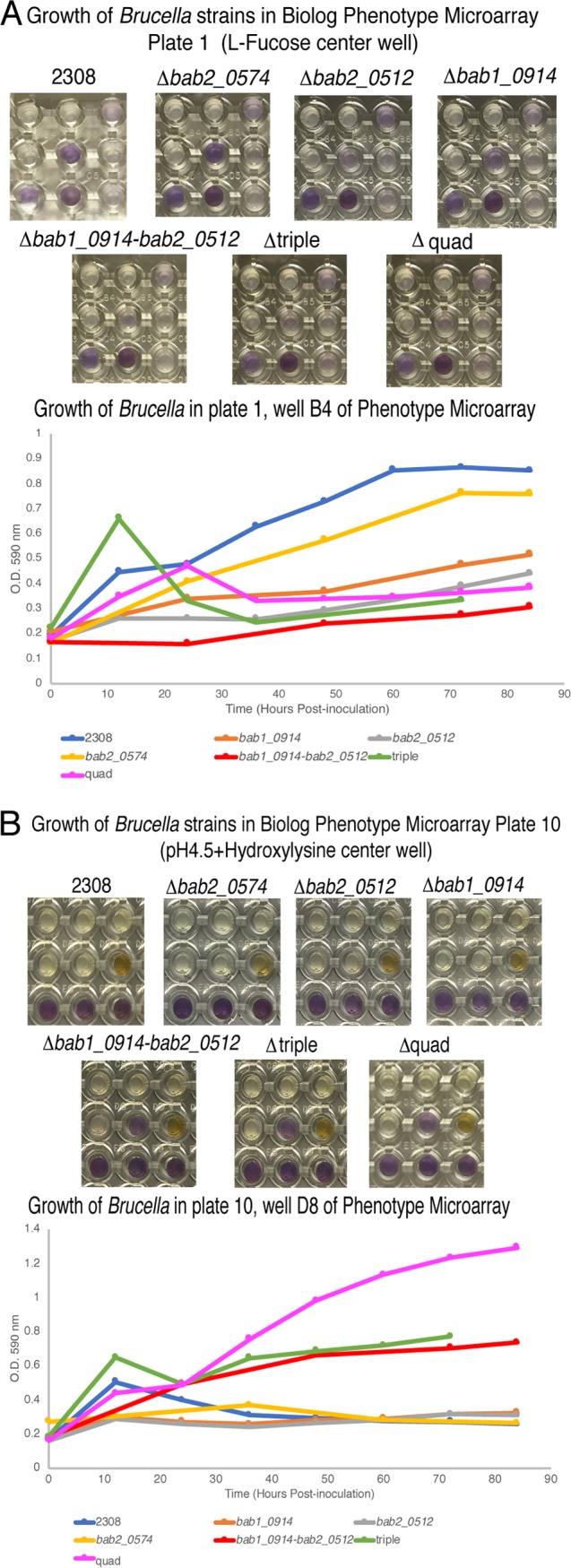
Characterization of B. abortus 2308 and VtlR regulon mutants using Biolog Phenotype MicroArray plates. (A) Growth of Brucella strains in plate 1, well B4, in minimal medium plus 24 mM l-fucose, over 84 h of growth. Shown are pictures of plate 1, well B4, (center well) after 84 h of growth. The graph shows the OD590 of plate 1, well B4, over 84 h of incubation. (B) Growth of Brucella strains in plate 10, well D8, in minimal medium, pH 4.5, plus hydroxylysine over 84 h of growth. Shown are pictures of plate 10, well D8 (center well), after 84 h of growth. The graph shows the OD590 of plate 10, well D8 over 84 h of incubation.
Effect of l-fucose on B. abortus growth.
While we were unsuccessful in reproducing the growth characteristics of B. abortus in the presence of hydroxylysine that we observed in the Biolog plates, we were able to independently verify the ability of B. abortus to utilize fucose during growth outside the Biolog system. First, we tested the effect of supplementing minimal medium with 24 mM l-fucose on B. abortus growth. GMM with or without 24 mM l-fucose was inoculated with ∼5 × 104 CFU/ml and incubated for 96 h. Samples were collected every 24 h and serially diluted to calculate the CFU brucellae per milliliter. The data revealed that after 96 h of growth, B. abortus 2308 grew to a larger cell density in the presence of 24 mM l-fucose (Fig. 5A). The following experiment tested the ability of VtlR regulon deletion strains to utilize l-fucose when grown in GMM. The data revealed that after 96 h of growth, the B. abortus Δquad mutant strain grew to a lower cell density than the parental strain B. abortus 2308 (Fig. 5B). B. abortus 2308 Δbab2_0574 showed no significant difference compared to B. abortus 2308. Surprisingly, the growth of B. abortus 2308 Δbab1_0914 was not significantly decreased, but B. abortus 2308 Δbab2_0512 cell density was significantly lower than that of B. abortus 2308 after 96 h of incubation. As a proof of principle, we tested the effect of l-fucose on B. abortus 2308 ΔvtlR. Not only was cell density significantly less in B. abortus 2308 ΔvtlR cultures, but growth was inhibited by the presence of l-fucose (Fig. 5C). Indeed, the vtlR deletion strain exhibits significantly decreased bacterial numbers compared to the parental strain 2308 at all time points during the growth experiment (data not shown), indicating that the vtlR deletion strain is unable to utilize fucose. Rhamnose and fucose are degraded similarly in Escherichia coli, and both lead to the secretion of the by-product propanediol (20). Thus, we tested the sensitivity of the ΔvtlR mutant strain to these compounds, as well as glucose to test whether there is a general sensitivity to sugars (Fig. S1). Our data demonstrate that ΔvtlR mutant growth was not inhibited by rhamnose, propanediol, or glucose when incubated in GMM, suggesting that the phenotype observed is reserved solely for l-fucose.
FIG 5.
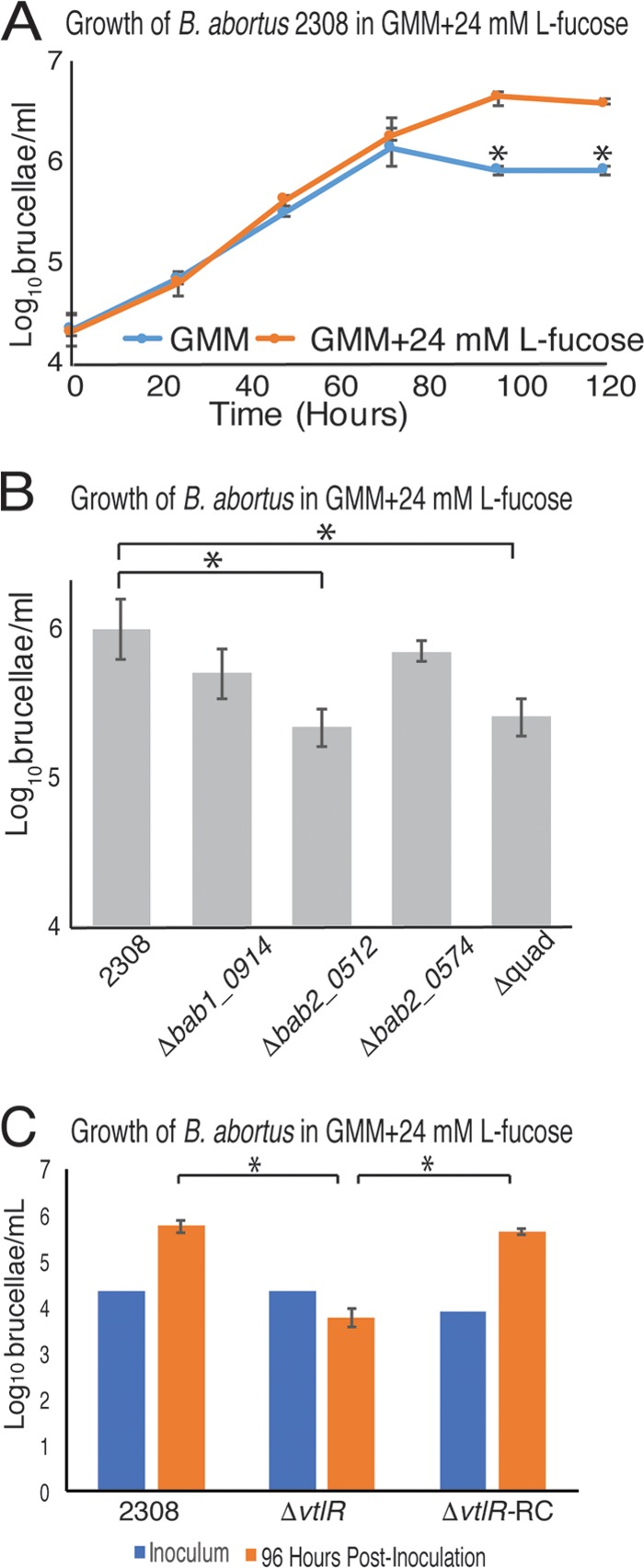
l-Fucose contribution to B. abortus growth and growth inhibition in VtlR regulon mutants. Cultures of Gerhardt's minimal medium (GMM) supplemented with and without the addition of 24 mM l-fucose were inoculated with Brucella strains at an initial concentration of 5 × 104 CFU/ml and incubated at 37°C. (A) B. abortus 2308 grows to a higher cell density in GMM supplemented with 24 mM l-fucose than GMM without 24 mM l-fucose. (B) In the presence of l-fucose, B. abortus 2308 Δbab2_0512 and B. abortus 2308 ΔabcR2 Δbab1_0914 Δbab2_0512 Δbab2_0574 (Δquad) growth is significantly lower than that of B. abortus 2308. (C) B. abortus 2308 ΔvtlR is sensitive to the presence of 24 mM l-fucose in GMM. Statistical significance (*) was determined using one-way analysis of variance (ANOVA; P < 0.05).
Identification of genes induced in the presence of l-fucose.
Due to our observations that B. abortus grows to a higher density when exposed to l-fucose (Fig. 5A), we hypothesized that B. abortus imports l-fucose and expression of a fucose import system and genes related to fucose import and utilization would fluctuate in the presence of l-fucose. Therefore, we set out to define the transcriptional response of B. abortus to l-fucose in order to identify potential l-fucose transport and/or utilization systems, and RNA sequencing (RNA-seq) analysis was employed to detect differences in the gene expression in cultures of B. abortus growth in the presence or absence of l-fucose. RNA was isolated from B. abortus 2308 incubated in GMM with and without the addition of 100 μM l-fucose, and RNA-seq analysis revealed very few genes that displayed significantly different levels of expression between the conditions tested (Data Set S2). Indeed, only 12 genes exhibited >4-fold (i.e., log2 ≥ 2) increased expression in cultures with the addition of 100 μM l-fucose compared to cultures without the addition of l-fucose (Table 1). All 12 genes belong to a single genetic locus on chromosome I of B. abortus 2308, and the genes in this locus encode a putative IclR-family transcriptional regulator (BAB1_0237), a putative ABC-type sugar transport system (BAB1_0238 to BAB1_0241), and proteins potentially involved in sugar metabolism (BAB1_0242 to BAB1_0248).
TABLE 1.
Differential gene expression of B. abortus 2308 in GMM plus 100 μM l-fucose versus GMMa
| BAB designation | BAB_RS designation | Protein description | Log2 fold change for GMM + 100 μM l-fucose vs GMM |
|---|---|---|---|
| BAB1_0238 | BAB_RS17035 | Sugar ABC transporter substrate-binding protein | 4.4 |
| BAB1_0239 | BAB_RS17040 | Sugar ABC transporter permease | 4.3 |
| BAB1_0241 | BAB_RS17050 | Sugar ABC transporter ATPase | 4.2 |
| BAB1_0240 | BAB_RS17045 | Maltose ABC transporter permease | 4.1 |
| BAB1_0242 | BAB_RS17055 | Mandelate racemase | 4.1 |
| BAB1_0244 | BAB_RS17065 | Oxidoreductase | 4.0 |
| BAB1_0246 | BAB_RS17075 | Oxidoreductase | 4.0 |
| BAB1_0247 | BAB_RS17080 | 2-Hydroxyhepta-2,4-diene-1,7-dioate isomerase | 3.9 |
| BAB1_0248 | BAB_RS17085 | Fuconate dehydratase | 3.8 |
| BAB1_0243 | BAB_RS17060 | l-Rhamnose mutarotase | 3.7 |
| BAB1_0237 | BAB_RS17030 | IclR family transcriptional regulator | 3.2 |
| BAB1_0236 | BAB_RS17025 | Amidohydrolase | 2.2 |
RNA-seq analysis was performed using total cellular RNA from Brucella strains grown in minimal medium to late exponential phase, and those genes whose expression was shown to be more than 4-fold altered in minimal medium with 100 μM l-fucose compared to minimal media without 100 μM l-fucose are shown in the list.
To test whether genes in this locus are involved in fucose metabolism or virulence, an isogenic deletion of bab1_0238, the putative periplasmic binding protein gene in the identified putative fucose transporter (Table 1), was constructed, and the deletion strain was evaluated for the capacity to grow in minimal medium with the addition of l-fucose and for the ability to survive and replicate within macrophages. A deletion of bab1_0238 resulted in no difference in bacterial growth in the presence of l-fucose or difference in the ability to survive and replicate in macrophages compared to the parental strain B. abortus 2308 (Fig. S3).
bab1_0914, bab2_0512, or bab2_0574 does not contribute to the ability of the brucellae to colonize BALB/c mice.
It was previously shown that bab1_0914, bab2_0512, and bab2_0574 do not contribute to the ability of B. abortus to survive and replicate in a macrophage model of infection (12). However, a lack of attenuation by a mutant strain of B. abortus in a macrophage model does not always correlate with a difference in the ability of the strain to infect a mouse model. Therefore, multiple strains of B. abortus 2308 were constructed with different combinations of mutations in the genes associated with the VtlR regulon to determine which, if any, genes in the described VtlR regulon are responsible for the decrease in spleen colonization previously observed in the vtlR mutant in the mouse model. Isogenic mutants of each small hypothetical protein were constructed, a strain containing a mutation of all three hypothetical small proteins (Δbab1_0914 Δbab2_0512 Δbab2_0574) and a quadruple mutant (Δbab1_0914 Δbab2_0512 Δbab2_0574 ΔabcR2) were also constructed. Female BALB/c mice were infected intraperitoneally with approximately 5 × 104 CFU/ml of either B. abortus 2308 or the mutant strains. After 8 weeks of infection, spleens were removed, homogenized, and serially diluted to determine the CFU per spleen, and it was determined that there is no difference in the ability of B. abortus 2308 and any of the deletion strains tested to colonize the spleens of BALB/c mice (Fig. 6).
FIG 6.
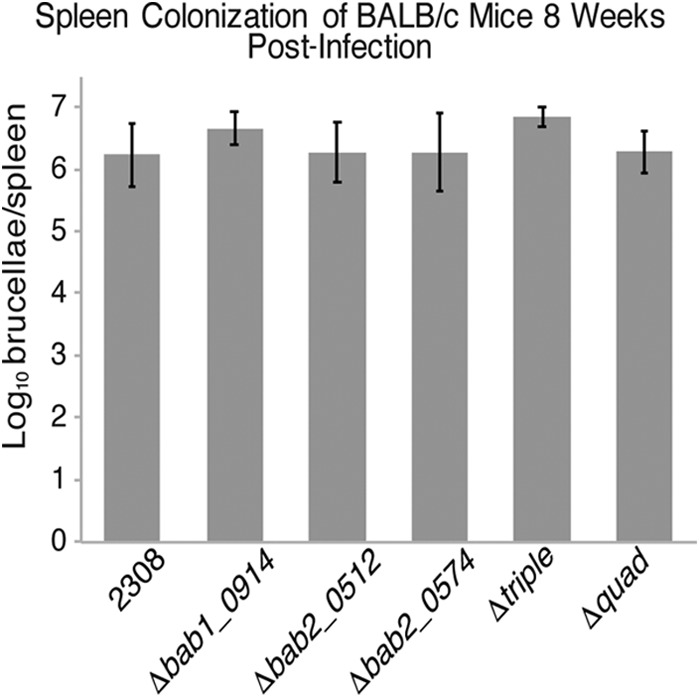
bab1_0914, bab2_0512, and bab2_0574 do not contribute to the ability of B. abortus 2308 to colonize the spleens of BALB/c mice. BALB/c mice were infected intraperitoneally with 105 CFU B. abortus 2308, B. abortus 2308 Δbab1_0914, B. abortus 2308 Δbab2_0512, B. abortus 2308 Δbab2_0574, B. abortus 2308 Δbab1_0914 Δbab2_0512 Δbab2_0574 (Δtriple), or B. abortus 2308 ΔabcR2 Δbab1_0914 Δbab2_0512 Δbab2_0574 (Δquad). Mice were sacrificed at 8 weeks postinfection, and the number of brucellae colonizing the spleens was determined. The data are presented as the average brucellae ± the standard deviation from the 5 mice colonized with a specific Brucella strain.
DISCUSSION
In the present study, we characterize three hypothetical small protein-encoding genes activated by VtlR in B. abortus 2308, and importantly, none of these hypothetical small proteins nor their homologous proteins found in other bacteria have been characterized previously. Our previous study describing VtlR in B. abortus 2308 revealed that isogenic mutations of the small hypothetical proteins, as well as a quadruple mutant of the entire VtlR regulon, survive and replicate in naive peritoneal macrophages at a degree similar to that of parental strain B. abortus 2308. In the present study, we sought to further characterize the role that the small hypothetical proteins activated by VtlR contribute to B. abortus biology and/or virulence in vivo. We demonstrated that similar to the previously performed infection of naive macrophages, B. abortus strains containing deletions of the VtlR regulon do not significantly differ in their ability to colonize the spleens of BALB/c mice compared to B. abortus 2308 and do not account for the observed attenuation of the ΔvtlR mutant strain in vivo (Fig. 6). Despite this, we further characterized the three small proteins and proved their translation, defined their expression under biologically relevant conditions, localized them within B. abortus, and propose a functional role for bab1_0914 and bab2_0512 in the utilization of l-fucose by B. abortus.
Our virulence data lead us to believe that VtlR has a separate in vivo regulon from our previously published in vitro regulon. The original VtlR regulon was characterized via microarray analysis, and this transcriptional analysis was performed on RNA from brucellae cultured in nutrient-rich broth. In that in vitro analysis, all three genes encoding the small hypothetical proteins showed decreased expression in a ΔvtlR mutant strain. We formed the hypothesis that these small hypothetical proteins constituted the VtlR regulon, and therefore, deletions of these small hypothetical proteins would have phenotypes in vivo similar to that of an isogenic deletion of vtlR. Conversely, the deletion of individual or combinations of the genes in the observed VtlR regulon showed no difference in the ability to successfully colonize and survive in naive peritoneal macrophages or a BALB/c mouse model compared to the parental strain (Fig. 6) (12). This was the first indication that VtlR may have an alternative transcriptional regulon in vivo. The second indication of multiple VtlR regulons was observed by measuring the transcriptional expression of BAB1_0914, BAB2_0512, and BAB2_0574 under different biological conditions. The transcription of all three genes changed under the different conditions tested. bab1_0914, bab2_0512, and bab2_0574 were highly expressed during the stationary phase of growth, under oxidative stress and acidic conditions. There are multiple hypotheses to account for the change in expression of bab1_0914, bab2_0512, and bab2_0574 under different culture conditions. One hypothesis is that regulators other than VtlR could regulate the expression of bab1_0914, bab2_0512, and bab2_0574 in B. abortus. Another hypothesis is that the activity of VtlR changes under different growth conditions. Thus, under changing environmental conditions, such as those encountered during intramacrophagic trafficking, the expression of VtlR transcriptional targets may also change. To date, we have not empirically tested this hypothesis, and future experiments are under way to identify and characterize the in vivo gene targets of VtlR transcriptional regulation.
Our results also further our understanding of the AbcR small RNAs in B. abortus, as it was previously unknown whether AbcR1 and AbcR2 were differentially expressed in response to various growth and stress conditions. Our data show that under the conditions tested, AbcR1 is consistently produced, while AbcR2 is activated in response to specific conditions, including stationary growth, oxidative stress, acidic growth medium, and E medium (Fig. 2). AbcR1 and AbcR2 have been shown to be redundant regulators of target mRNAs in B. abortus (12), yet our expression data indicate that AbcR1 is constitutively expressed under the conditions tested. This does not seem beneficial to the cell, because if AbcR1 is always present, all of the AbcR target mRNAs would be constitutively regulated as well. Thus, AbcR1 is most likely not constitutively expressed within Brucella species, and our group is currently working to identify a transcriptional regulator of AbcR1 in B. abortus. Moreover, while all of the AbcR target mRNAs have not been individually studied in Brucella species, homologs of AbcR1 and AbcR2 target mRNAs are necessary for the pathogenesis or symbiosis of other closely related bacteria (12, 21), and ongoing work in our laboratory is continuing to define the links between the control of abcR expression, AbcR-mediated regulation of mRNA levels, and Brucella virulence.
Up to this point, we have discussed how our findings have changed our understanding of the activity of the transcriptional regulator VtlR and new discoveries of AbcR sRNA expression in Brucella species, but perhaps the most intriguing data observed from this study are the potential interaction between l-fucose and Brucella species. It has been shown that many strains of B. melitensis and B. abortus can utilize l-fucose, but in general, very little is known about l-fucose metabolism in Brucella spp. (22). The ability of a bacterium to sense and potentially utilize l-fucose has recently been documented as an important virulence determinant, as is has been demonstrated that some enteric bacteria can use l-fucose to aid in their ability to persist in the gut. For example, Campylobacter jejuni can transport l-fucose, and strains containing a mutation in this transport system are less able to successfully colonize the host (23). Similar to in vitro growth analyses in C. jejuni, we demonstrated that B. abortus 2308 can grow to a higher density in minimal medium supplemented with l-fucose (Fig. 5). Enterohemorrhagic E. coli (EHEC) can both metabolize host-derived l-fucose and sense l-fucose using a two-component regulatory system, which regulates virulence-associated genes (24, 25). Additionally, Klebsiella pneumoniae incorporates fucose into its capsule during infection of a mouse model (26). While these studies highlight the ability of different bacteria to use fucose to their benefit, little information is available about bacterial sensitivity to fucose, as we observed in our experiments with B. abortus mutants (Fig. 4 and 5). Regarding fucose availability, Brucella infections commonly occur via the oral route, and thus, it is likely that Brucella spp. would encounter l-fucose in the gut during infection.
Our data indicate that B. abortus strains containing mutations in vtlR, bab1_0914, and bab2_0512 exhibit increased growth inhibition to the presence of l-fucose in the growth medium, but what accounts for this l-fucose toxicity? While fucose metabolism has not been studied in Brucella spp., it has been shown in E. coli and Clostridium phytofermentans that fucose and rhamnose are digested similarly to produce lactaldehyde, which is a precursor of lactic acid and 1,2-propanediol (20, 27). Our data demonstrate that the vtlR deletion strain is not sensitive to the presence of rhamnose or 1,2-propanediol during growth (see Fig. S2 in the supplemental material), and thus, it is possible that an accumulation of lactaldehyde in the ΔvtlR mutant strain leads to the toxicity observed when the strain is grown in the presence of l-fucose. However, this hypothesis is based on the assumption that the metabolism of fucose in B. abortus is similar to fucose metabolism in E. coli and C. phytofermentans. As such, a more detailed analysis of fucose metabolism in Brucella species is needed to elucidate the mechanism of l-fucose-mediated toxicity observed for the B. abortus ΔvtlR mutant strain. While at least one small protein, SgrT, has been shown to be involved in sugar transport in E. coli, there are few examples of the role of small proteins with sugar utilization in general (28, 29). The presented study warrants further research to examine the role of small proteins in fucose utilization and metabolism in other organisms.
Regarding the response of B. abortus to l-fucose, RNA sequencing analysis revealed a putative fucose-activated sugar transport and utilization system (Table 1), and we hypothesize that this system is responsible for the transport and utilization of fucose by B. abortus. Of the genes exhibiting elevated expression in response to l-fucose, 10 genes (i.e., bab1_0238 to bab1_0248) encode a putative sugar ABC transport system and proteins potentially involved in the metabolism of sugars (e.g., oxidoreductases, isomerases, and dehydratases). Our data revealed that an isogenic deletion of bab1_0238 from the B. abortus genome did not result in any significant differences in the capability to grow in minimal medium with the addition of l-fucose or the ability to survive and replicate in a macrophage model of infection compared to the parental strain B. abortus 2308 (Fig. S3). Nonetheless, it is possible that the presence of orphan periplasmic binding proteins encoded in the Brucella genome functions in place of BAB1_0238, and this may account for the lack of an observable fucose-linked phenotype in the bab1_0238 deletion strain. Moreover, it is possible that another ABC transport system can transport l-fucose in the absence of the BAB1_0238 to BAB1_0248 system (30, 31). Overall, additional studies are needed to fully characterize the function of the putative fucose transport and utilization system, as well as the role of fucose in the biology and virulence of B. abortus.
Two additional genes, bab1_0236 and bab1_0237, are divergently oriented from the putative fucose ABC transporter and utilization genes, and bab1_0237 encodes a putative transcriptional regulatory protein from the IclR family, while bab1_0236 encodes a putative amidohydrolase. A highly similar genomic locus in Sinorhizobium meliloti 1021 is induced in cultures by the addition of l- and d-fucose (32). With regard to Brucella spp., Lamontagne et al. showed that within 24 h of infection of murine RAW 264.7 macrophages with B. abortus, the expression levels of bab1_0238 and bab1_0246 were both significantly decreased, along with several other sugar transport systems (33). This led the authors to conclude that sugars imported by these systems may be in low abundance intracellularly (33). Indeed, we hypothesize that fucose may play a more important role in the gut of an infected host rather than during intracellular trafficking. It is important to note that while many studies have focused on the interactions between fucose and microbes in the gut, fucose can be found throughout the mammalian body, including the reproductive tract and the brain (34–37). Thus, the role of fucose in the virulence of Brucella spp. could be important during different stages of Brucella pathogenesis, different routes of infection, or the infection of different hosts.
To summarize, while many small proteins do not have enzymatic functions, several have been shown to act as facilitators of enzymatic function for larger proteins (1). Because of this, observable phenotypes in strains with mutated or deleted small proteins can be subtle or nonexistent. The deletion of bab1_0914 and bab2_0512 in B. abortus revealed a minor sensitivity to the sugar l-fucose, which led us to discover a robust phenotype in a deletion of the transcriptional regulator VtlR. As previously stated, it is hypothesized that VtlR may be operating in response to an environmental stimulus, which could include the sugar l-fucose during infection (12). Overall, this study serves as an example of the benefits of studying small proteins, which are continually overlooked as insignificant but may play major roles in bacterial physiology and virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Brucella abortus 2308 and derivative strains were routinely grown on Schaedler blood agar (SBA), composed of Schaedler agar (Acumedia, Burton, MI) containing 5% defibrinated bovine blood (Quad Five, Ryegate, MT), or in brucella broth (BD, Sparks, MD). For cloning, Escherichia coli strain DH5α was grown on tryptic soy agar (MP Biomedicals, Solon, OH) or in Luria-Bertani (LB) broth. When appropriate, growth media were supplemented with kanamycin (45 μg/ml) or l-fucose (24 mM). All work with live Brucella strains was performed in a biosafety level 3 (BSL3) facility.
Construction of Brucella abortus mutants and 3×FLAG tag strains by complementation.
Strains containing nonpolar unmarked isogenic deletions of bab1_0914, bab2_0512, and bab2_0574 as well as a quadruple-deletion strain of abcR2, bab1_0914, bab2_0512, and bab2_0574 in Brucella abortus 2308 were previously constructed and utilized in the publication by Sheehan et al. (12) and are utilized in this publication. A bab1_0914 bab2_0512 double-deletion strain, bab1_0914, bab2_0512, and bab2_0574 triple-deletion strain, and an isogenic deletion strain of bab1_0238 were also constructed using the strategy previously stated (12). Briefly, a 1-kb fragment upstream of the gene of interest plus the addition of the first codon of the gene of interest was amplified using Up-For and Up-Rev primers, and a 1-kb fragment downstream of the gene of interest plus the addition of the last codon of the gene of interest was amplified using Dn-For and Dn-Rev primers and genomic B. abortus 2308 DNA. The upstream fragment was digested with BamHI, and the downstream fragment was digested with PstI. Both fragments were phosphorylated using polynucleotide kinase in the presence of ATP. The upstream and downstream fragments were then ligated to each other and then into a BamHI/PstI digest pNPTS138 vector (M. R. K. Alley, unpublished data) using T4 DNA ligase (Monserate Biotechnology Group, San Diego, CA). The primers utilized for the construction of the deletion constructs were used in our previous study and can be found in Table 2. Plasmids used can be found in Table 3. The deletion constructs were introduced into B. abortus 2308, and merodiploid transformants were selected on SBA plus kanamycin. Merodiploid transformants were then incubated at 37°C for 7 h and then plated onto SBA plus 10% sucrose. Colonies that were sucrose resistant and kanamycin sensitive were screened via PCR for a loss of the genes of interest. The double- and triple-mutant strains were constructed by sequential mutagenesis of B. abortus with the deletion constructs to obtain the correct combination of genetic deletions. The bab1_0914 bab2_0512 double mutant was named JB003, and the bab1_0914 bab2_0512 bab2_0574 triple mutant was named JB004.
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| 1_0914-Up-For | TAGGATCCCCAGTATCAGCCCTATGCCAATC |
| 1_0914-Dn-Rev | TACTGCAGAGCGTTTCCGGTCCGCGCTC |
| 1_0914-Up-3×-FLAG-Rev | ATCTTTATAATCACCGTCATGGTCTTTGTAGTCGCGCGCTTTGGCCTGACG |
| 1_0914-Dn-3×-FLAG-For | CATGACATCGACTACAAGGATGACGATGACAAGTAATCACAAGTTTCTCCG |
| 2_0512-Up-For | TAGGATCCCTCGATGTCGGCATCGGCATTG |
| 2_0512-Dn-Rev | TACTGCAGGGACGTGCGCAATGTCGTGAT |
| 2_0512-Up-3×-FLAG-Rev | ATCTTTATAATCACCGTCATGGTCTTTGTAGTCGCGGGCAGCGGCCTTGCG |
| 2_0512-Dn-3×-FLAG-For | CATGACATCGACTACAAGGATGACGATGACAAGTGATAGCGCCGCCTCCAA |
| 2_0574-Dn-Rev | TACTGCAGTCCAACGCAAAACCGCTACG |
| 2_0574-Up-For | TAGGATCCGATCATCTGGCTATATTGCGGTG |
| 2_0574-Up-3×-FLAG-Rev | ATCTTTATAATCACCGTCATGGTCTTTGTAGTCTCGACCGAAGACGGCGCT |
| 2_0574-Dn-3×-FLAG-For | CATGACATCGACTACAAGGATGACGATGACAAGTAACAGTTATGGCGCGGC |
| 1_0238-Up-For | ATGGATCCCACGCCTACATCCGCGCCCAC |
| 1_0238-Up-Rev | TTTGAAATCCTGCATTCGTCTCC |
| 1_0238-Dn-For | GCCGGCGACCTTTGACC |
| 1_0238-Dn-Rev | ATCTGCAGACTACCACTCCGCTCCGATCTGC |
Underlined sequences depict a restriction endonuclease recognition site. Bold letters depicts 3×FLAG sequence.
TABLE 3.
Plasmids used in this study
| Plasmid name | Description | Reference or source |
|---|---|---|
| pNPTS138 | Cloning vector; contains sacB gene; Kanra | M. R. K. Alley, unpublished data |
| pJB006 | Intact bab2_0512 gene plus coding sequence for 3×FLAG tag at C terminus and 1 kb of each flanking region in pNPTS138 | This study |
| pJB007 | Intact bab2_0574 gene plus coding sequence for 3×FLAG tag at C terminus and 1 kb of each flanking region in pNPTS138 | This study |
| pJB008 | Intact bab1_0914 gene plus coding sequence for 3×FLAG tag at C terminus and 1 kb of each flanking region in pNPTS138 | This study |
| pΔbab1_0238 | In-frame deletion of bab1_0238 plus 1 kb of each flanking region in pNPTS138 | This study |
Kanr, kanamycin resistance.
The addition of a C-terminal 3×FLAG tag (5′-GACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGACTACAAGGATGACGATGACAAG-3′) to the small hypothetical protein nucleotide sequence was carried out by reconstruction of the deleted loci on the B. abortus chromosome. This process is similar to the one described above; however, new primers were constructed to replace Up-Rev and Down-For, each containing half of the 3×FLAG tag. The amplified upstream and downstream regions encompassing the genes of interest and the addition of the 3×FLAG tag were digested with BamHI and PstI, respectively. The fragments were phosphorylated and ligated together and then ligated into the pNPTS138 plasmid, and transformation and selection for the reconstructed tagged strains were carried out as described above.
RNA expression and protein production under different growth conditions.
For expression experiments under different growth conditions, B. abortus 2308, B. abortus 2308 bab1_0914-3×FLAG, or B. abortus 2308 bab2_0512-3×FLAG was used to inoculate brucella broth at a concentration of 109 CFU/ml or 106 CFU/ml for cells grown to stationary phase and exponential phase, respectively, and grown for approximately 30 h to the appropriate OD. Exponential-phase cells at an OD of 0.15 were collected, washed with PBS, and used to inoculate the appropriate media at a final concentration of 109 CFU/ml. Brucella broth, brucella broth at pH 4.5, brucella broth supplemented with 0.5% deoxycholate, brucella broth supplemented with 5 μM H2O2, GMM, and E medium were utilized. After 30 min of incubation at 37°C with shaking, cultures were diluted 1:1 with ethanol-acetone, and RNA or protein was extracted as previously described (38).
Fractionation of Brucella abortus 2308.
Fractionation of B. abortus 2308 was performed as previously described by Thein et al., with revisions optimized for B. abortus 2308 (14). One hundred milliliters of brucella broth (BD) was inoculated with an appropriate strain of B. abortus and grown to late-exponential phase at 37°C. One hundred milliliters of 1:1 ethanol-acetone was added to the culture, and death of Brucella spp. was established for 10 days postaddition of ethanol-acetone prior to the removal of the samples from the BSL3 facility. Cells were pelleted at 10,000 × g for 10 min, and the supernatant was discarded. The pellet was resuspended in 10 ml of solution 1 (0.2 M Tris-HCl [pH 8], 1 M sucrose, 1 mM EDTA, lysozyme [1 mg/ml]). The suspension was vortexed and incubated at room temperature for 5 min. Forty milliliters of distilled water (dH2O) was added to the suspension and placed on ice for 10 min. The suspension was centrifuged at 200,000 × g for 45 min at 4°C. The supernatant was collected as the periplasmic fraction. The pellet was resuspended in 7.5 ml of ice-cold solution 2 (10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 0.2 mM dithiothreitol [DTT] supplemented with 50 μl DNase [1 mg/ml]). Cells were ruptured via 2 passes through a French press at 106 Pa. Unbroken cells were spun down by centrifugation at ∼2,000 × g for 10 min. The pellet was discarded. The supernatant was centrifuged at 300,000 × g for 3 h at 4°C. The supernatant was collected as the cytoplasmic fraction. Pellet containing the crude membranes was separated by a sucrose gradient. The pellet was resuspended in a 1-ml solution (10 mM Tris [pH 7.5], 15% sucrose, 5 mM EDTA, 0.2 mM DTT). Samples were stored at −20°C until analyzed via Western blotting.
Western blot analyses.
Protein samples were prepared with the addition of 4× Laemmli buffer and boiled for 5 min prior to being loaded into a 10 to 20% polyacrylamide gel. BAB1_0914-3×FLAG, BAB2_0512-3×FLAG, and BAB2_0574-3×FLAG were detected using mouse monoclonal anti-FLAG antibody at a 1:1,000 dilution (Thermo Fisher Scientific). SodC was detected using rabbit polyclonal anti-SodC at a dilution of 1:1,000. Omp89 was detected with the addition of serum directly isolated from rabbit serum. GroEL was detected using mouse monoclonal anti-GroEL at a 1:1,000 dilution (Origene). Horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit antibody was used as secondary antibody when appropriate, and protein detection was carried out via the femtoLUCENT Luminol solution (G Biosciences).
Growth in Biolog Phenotype MicroArray plates.
Phenotype MicroArray plates (Biolog, Inc., Hayward, CA) were utilized to determine phenotypic differences between different B. abortus strains. Strains were grown on SBA plates to produce a lawn of bacteria. Bacteria were collected and suspended in IF-0a GN/GP Base (Biolog). The protocol “PM procedures for GN fastidious bacteria” provided by Biolog was followed, and Biolog Phenotype MicroArray plates 1 to 20 were inoculated at a final concentration of 109 CFU/ml. Plates were grown statically at 37°C for 84 h, and the OD590 was measured every 12 to 24 h.
RNA sequencing analysis of l-fucose-treated cultures.
Brucella broth was inoculated with B. abortus 2308 and incubated at 37°C with shaking for ∼24 h until the cultured obtained had an OD600 of 0.15. Cells were then washed with PBS, and cells were used to inoculate either GMM or GMM with the addition of 100 μM l-fucose at a concentration of 109 CFU/ml. Cultures were incubated for 20 min at 37°C with shaking. Following incubation, an equal volume of 1:1 ethanol-acetone was added to each culture, and cultures were frozen at −80°C until RNA isolation. This was performed in triplicate for each condition. RNA was isolated from each culture and DNase treated prior to submission for RNA-seq analysis.
Stranded RNA library construction for prokaryotic RNA-seq.
One microgram of total RNA with an RNA integrity number (RIN) of ≥8.0 was depleted of rRNA using Illumina's Ribo-Zero rRNA removal kit (Gram-positive and Gram-negative bacteria) (P/N MRZB12424; Illumina, CA). The depleted RNA is fragmented and converted to first-strand cDNA using reverse transcriptase and random primers using Illumina's TruSeq stranded mRNA high-throughput (HT) sample prep kit (catalog no. RS-122-2103; Illumina). This is followed by second-strand synthesis using polymerase I and RNase H, and dinucleotide triphosphates (dNTPs) that contain dUTP instead of dTTP. The cDNA fragments then go through end repair, the addition of a single “A” base, and then ligation of adapters and indexed individually. The products are then purified and the second strand digested with N-glycosylase, thus resulting in a stranded template. The template molecules with the adapters are enriched by 10 cycles of PCR to create the final cDNA library. The library generated was validated using Agilent 2100 Bioanalyzer and quantitated using the Quant-iT double-stranded DNA (dsDNA) HS kit (Invitrogen) and quantitative PCR (qPCR). Sixteen individually indexed cDNA libraries were pooled and sequenced on an Illumina NextSeq platform.
Illumina NextSeq sequencing.
The libraries were clustered and sequenced using the NextSeq 500/550 high-output kit V2 (150 cycles) (P/N FC-404-2002) to 2 × 75 cycles to generate paired-end reads. The Illumina NextSeq control software version 2.1.0.32 with Real Time Analysis (RTA) version 2.4.11.0 was used to provide the management and execution of the NextSeq 500 kit and to generate BCL files. The BCL files were converted to FASTQ files and demultiplexed using bcl2fastq conversion software version 2.20. The FASTQ files are provided to the investigator for further analysis.
RNA-Seq data processing and analysis.
The Brucella abortus strain 2308 gene and genome sequences, as well as corresponding annotations from NCBI (https://www.ncbi.nlm.nih.gov/), were used as the references. Raw reads were quality controlled and filtered with FastqMcf (39), resulting in an average of 1,971 Mbp (1,730 to 2,305 Mbp) of nucleotides (Fig. S4). The remaining reads were mapped to the gene reference using BWA (40), with default parameters. Differential expression of genes was calculated using the edgeR package (41) in the R software (http://www.r-project.org/), with Benjamini-Hochberg-adjusted P values of 0.05 considered to be significant.
Virulence of Brucella strains in experimentally infected mice and macrophages.
The infection of primary peritoneal murine macrophages was described previously by Sheehan et al. (12). Macrophages were harvested from mice and seeded into 96-well plates in Dulbecco's modified Eagle's medium with 5% fetal bovine serum. The following day, macrophages were infected with opsonized brucellae at a multiplicity of infection of 100:1. After 2 h, the infected macrophages were treated with gentamicin (50 μg · ml−1) for 1 h. The macrophages were then lysed with 0.1% deoxycholate in PBS, and serial dilutions were plated on Schaedler blood agar (SBA). For the 24- and 48-h time points, macrophages were washed with PBS following gentamicin treatment, and fresh cell culture medium containing gentamicin (20 μg · ml−1) was added to the monolayer. At the indicated time points, macrophages were lysed, and serial dilutions were plated on SBA in triplicates.
The infection of mice by Brucella strains was performed as described previously by Sheehan et al. (12). Female BALB/c mice (5 per Brucella strain per experiment) were infected intraperitoneally with ∼105 CFU of each Brucella strain in sterile PBS. The mice were sacrificed at 8 weeks postinfection, and serial dilutions of spleen homogenates were plated on SBA to determine the CFU per spleen of each strain.
Accession number(s).
The NCBI Sequence Read Archive (SRA) accession number for the RNA-seq data is SRP141183.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jean-Jacques Letesson for the generous contribution of the anti-Omp89 antibody.
This study was supported by a grant from the National Institute of Allergy and Infectious Diseases to C.C.C. (grant AI117648).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00127-18.
REFERENCES
- 1.Storz G, Wolf YI, Ramamurthi KS. 2014. Small proteins can no longer be ignored. Annu Rev Biochem 83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramamurthi KS, Storz G. 2014. The small protein floodgates are opening; now the functional analysis begins. BMC Biol 12:96. doi: 10.1186/s12915-014-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basrai MA, Hieter P, Boeke JD. 1997. Small open reading frames: beautiful needles in the haystack. Genome Res 7:768–771. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- 4.Friedman RC, Kalkhof S, Doppelt-Azeroual O, Mueller SA, Chovancova M, von Bergen M, Schwikowski B. 2017. Common and phylogenetically widespread coding for peptides by bacterial small RNAs. BMC Genomics 18:553. doi: 10.1186/s12864-017-3932-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting S, Anderson M, Lysenko E, Page A, Tomoyasu T, Tatematsu K, Tatsuta T, Kroos L, Ogura T. 1997. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J Bacteriol 179:5534–5542. doi: 10.1128/jb.179.17.5534-5542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun YH, de Jong MF, den Hartigh AB, Roux CM, Rolan HG, Tsolis RM. 2012. The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front Cell Infect Microbiol 2:47. doi: 10.3389/fcimb.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisson-Filho AW, Discola KF, Castellen P, Blasios V, Martins A, Sforca ML, Garcia W, Zeri AC, Erickson HP, Dessen A, Gueiros-Filho FJ. 2015. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc Natl Acad Sci U S A 112:E2130–E2138. doi: 10.1073/pnas.1414242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lippa AM, Goulian M. 2009. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet 5:e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atluri VL, Xavier MN, de Jong MF, den Hartigh AB, Tsolis RM. 2011. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol 65:523–541. doi: 10.1146/annurev-micro-090110-102905. [DOI] [PubMed] [Google Scholar]
- 10.de Figueiredo P, Ficht TA, Rice-Ficht A, Rossetti CA, Adams LG. 2015. Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am J Pathol 185:1505–1517. doi: 10.1016/j.ajpath.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Bargen K, Gorvel JP, Salcedo SP. 2012. Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol Rev 36:533–562. doi: 10.1111/j.1574-6976.2012.00334.x. [DOI] [PubMed] [Google Scholar]
- 12.Sheehan LM, Budnick JA, Blanchard C, Dunman PM, Caswell CC. 2015. A LysR-family transcriptional regulator required for virulence in Brucella abortus is highly conserved among the alpha-proteobacteria. Mol Microbiol 98:318–328. doi: 10.1111/mmi.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, Sayood K, Dunman PM, Roop RM II. 2012. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol 85:345–360. doi: 10.1111/j.1365-2958.2012.08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thein M, Sauer G, Paramasivam N, Grin I, Linke D. 2010. Efficient subfractionation of Gram-negative bacteria for proteomics studies. J Proteome Res 9:6135–6147. doi: 10.1021/pr1002438. [DOI] [PubMed] [Google Scholar]
- 15.Benkert P, Biasini M, Schwede T. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T. 2017. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep 7:10480. doi: 10.1038/s41598-017-09654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, Schwede T. 2017. The SWISS-MODEL Repository–new features and functionality. Nucleic Acids Res 45:D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guex N, Peitsch MC, Schwede T. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 20.Baldoma L, Aguilar J. 1988. Metabolism of l-fucose and l-rhamnose in Escherichia coli: aerobic-anaerobic regulation of l-lactaldehyde dissimilation. J Bacteriol 170:416–421. doi: 10.1128/jb.170.1.416-421.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker A, Overlöper A, Schlüter J-P, Reinkensmeier J, Robledo M, Giegerich R, Narberhaus F, Evguenieva-Hackenberg E. 2014. Riboregulation in plant-associated α-proteobacteria. RNA Biol 11:550–562. doi: 10.4161/rna.29625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Dahouk S, Scholz HC, Tomaso H, Bahn P, Gollner C, Karges W, Appel B, Hensel A, Neubauer H, Nockler K. 2010. Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system. BMC Microbiol 10:269. doi: 10.1186/1471-2180-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahl M, Friis LM, Nothaft H, Liu X, Li J, Szymanski CM, Stintzi A. 2011. l-Fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc Natl Acad Sci U S A 108:7194–7199. doi: 10.1073/pnas.1014125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keeney KM, Finlay BB. 2013. Microbiology: EHEC downregulates virulence in response to intestinal fucose. Curr Biol 23:R108–R110. doi: 10.1016/j.cub.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu JH, Wu AM, Tsai CG, Chang XY, Tsai SF, Wu TS. 2008. Contribution of fucose-containing capsules in Klebsiella pneumoniae to bacterial virulence in mice. Exp Biol Med (Maywood) 233:64–70. doi: 10.3181/0706-RM-170. [DOI] [PubMed] [Google Scholar]
- 27.Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. 2013. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8:e54337. doi: 10.1371/journal.pone.0054337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd CR, Park S, Fei J, Vanderpool CK. 2017. The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system. J Bacteriol 199:e00869-16. doi: 10.1128/JB.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raina M, Storz G. 2017. SgrT, a small protein that packs a sweet punch. J Bacteriol 199:e00130-17. doi: 10.1128/JB.00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas GH. 2010. Homes for the orphans: utilization of multiple substrate-binding proteins by ABC transporters. Mol Microbiol 75:6–9. doi: 10.1111/j.1365-2958.2009.06961.x. [DOI] [PubMed] [Google Scholar]
- 31.Durmort C, Brown JS. 2015. Chapter 10–Streptococcus pneumoniae lipoproteins and ABC transporters, p 181–206, Streptococcus Pneumoniae. Academic Press, Amsterdam, The Netherlands. [Google Scholar]
- 32.Mauchline TH, Fowler JE, East AK, Sartor AL, Zaheer R, Hosie AH, Poole PS, Finan TM. 2006. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc Natl Acad Sci U S A 103:17933–17938. doi: 10.1073/pnas.0606673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamontagne J, Forest A, Marazzo E, Denis F, Butler H, Michaud JF, Boucher L, Pedro I, Villeneuve A, Sitnikov D, Trudel K, Nassif N, Boudjelti D, Tomaki F, Chaves-Olarte E, Guzman-Verri C, Brunet S, Cote-Martin A, Hunter J, Moreno E, Paramithiotis E. 2009. Intracellular adaptation of Brucella abortus. J Proteome Res 8:1594–1609. doi: 10.1021/pr800978p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staudacher E, Altmann F, Wilson IB, Marz L. 1999. Fucose in N-glycans: from plant to man. Biochim Biophys Acta 1473:216–236. doi: 10.1016/S0304-4165(99)00181-6. [DOI] [PubMed] [Google Scholar]
- 35.Ma B, Simala-Grant JL, Taylor DE. 2006. Fucosylation in prokaryotes and eukaryotes. Glycobiology 16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 36.Domino SE, Zhang L, Gillespie PJ, Saunders TL, Lowe JB. 2001. Deficiency of reproductive tract alpha(1,2)fucosylated glycans and normal fertility in mice with targeted deletions of the FUT1 or FUT2 alpha(1,2)fucosyltransferase locus. Mol Cell Biol 21:8336–8345. doi: 10.1128/MCB.21.24.8336-8345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mountford C, Quadrelli S, Lin A, Ramadan S. 2015. Six fucose-alpha(1-2) sugars and alpha-fucose assigned in the human brain using in vivo two-dimensional MRS. NMR Biomed 28:291–296. doi: 10.1002/nbm.3239. [DOI] [PubMed] [Google Scholar]
- 38.Caswell CC, Gaines JM, Roop RM Jr.. 2012. The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J Bacteriol 194:3–14. doi: 10.1128/JB.05623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aronesty E. 2013. Comparison of sequencing utility programs. Open Bioinformatics J 7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 40.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


