Abstract
Background
Autism spectrum disorder (ASD) has been associated with brain inflammation as indicated by the activation of microglia, but the triggers are not known. Extracellular vesicles (EVs) are secreted from many cells in the blood and other biological fluids and carry molecules that could influence the function of target cells. EVs have been recently implicated in several diseases, but their presence or function in ASD has not been studied.
Methods
EVs were isolated from the serum of children with ASD (n = 20, 16 males and 4 females, 4–12 years old) and unrelated age and sex-matched normotypic controls (n = 8, 6 males and 2 females, 4–12 years old) using the exoEasy Qiagen kit. EVs were characterized by determining the CD9 and CD81 membrane-associated markers with Western blot analysis, while their morphology and size were assessed by transmission electron microscopy (TEM). Human microglia SV40 were cultured for 24 h and then stimulated with EVs (1 or 5 μg/mL), quantitated as total EV-associated protein, for 24 or 48 h. IL-1β secretion was measured by ELISA. The results were analyzed using the Mann-Whitney U non-parametric test, and all statistical analyses were performed using Graph Pad Prism 5.
Results
EVs were isolated and shown to be spherical structures (about 100 nm) surrounded by a membrane. Total EV-associated protein was found to be significantly increased (p = 0.02) in patients as compared to normotypic controls. EVs (5 μg/mL) isolated from the serum of patients with ASD stimulated cultured human microglia to secrete significantly more of the pro-inflammatory cytokine interleukin IL-1β (163.5 ± 13.34 pg/mL) as compared to the control (117.7 ± 3.96 pg/mL, p < 0.0001). The amount of mitochondrial DNA (mtDNA7S) contained in EVs from children with ASD was found to be increased (p = 0.046) compared to the normotypic controls.
Conclusions
These findings provide novel information that may help explain what triggers inflammation in the brain of children with ASD and could lead to novel effective treatments.
Keywords: Autism spectrum disorder, Brain, Exosomes, Extracellular vesicles, Inflammation, IL-1β, Microglia, Mitochondrial DNA
Introduction
Autism spectrum disorder (ASD) is a pervasive neurodevelopmental disorder characterized by deficits in communication, as well as the presence of restricted, repetitive behaviors [1–3]. The prevalence of ASD in the USA was recently estimated to be 1 in about 59 children [4]. Numerous gene mutations have been identified in patients with ASD, but no direct link has so far been uncovered [5–7]. The lack of reliable biomarkers [8] and specific pathogenesis for ASD [9], as well as the existence of subgroups or comorbidities [10], makes the diagnosis, staging, and treatment of ASD difficult. As a result, the annual economic burden for ASD was estimated at $268 billion for 2015 and projected at $416 billion in 2025 [11].
Abnormal microglial growth and activation indicating inflammation has been reported in the brain of patients with ASD since 2005 [12–16]. However, what stimulates microglia remains unknown. Some of the triggers for microglia activation could be mediators secreted from mast cells (MC), [17, 18] interaction of which with microglia is considered to have an important role in neuroinflammation [19]. One trigger of microglia could be mitochondrial DNA (mtDNA), which we had previously reported to be secreted from stimulated MC [20], and shown to be increased in the serum of children with ASD [20].
Triggers for microglia activation could be carried in the extracellular vesicles (EVs) that are secreted in the intercellular space by diverse cell types [21]. EVs are generated from the cell either when multivesicular bodies (MVBs) fuse with the plasma membrane or they are released directly from the plasma membrane. EVs can be isolated from the serum, plasma, urine, and other biological fluids and can be separated depending on their size (50–1000 nm) with exosomes being on the smaller size range (50–100) [22, 23]. EVs have been shown to contain RNA, DNA, lipids, or proteins [24] that are delivered to the surrounding cells or carried to distal sites [25, 26]. EVs can also be directed to specific cells via targeting proteins in their envelope [27, 28]. Consequently, EVs have the potential to transmit, worsen, or improve disease [29, 30].
Here, we report that serum from children with ASD contains significantly increased EVs as compared to healthy normotypic controls, that these EVs contain mtDNA, and that they can stimulate human-cultured microglia to secrete the pro-inflammatory cytokine interleukin-1β (IL-1β). We chose to study IL-1β because we previously reported that this cytokine is secreted from human-cultured microglia in response to the peptide neurotensin (NT), which we had shown to be increased in the serum of children with ASD [31, 32].
Methods
Human subjects
Caucasian children (n = 20, 16 males and 4 females, 4-12 years old) diagnosed with ASD were evaluated as part of a clinical trial that was conducted at the Attikon General Hospital, Athens Medical School, Athens, Greece [33]. Children were diagnosed with ASD based on clinical assessment and corroborated by meeting the cutoff scores on both the DSM-IV-TR symptom list and the Autism Diagnostic Observation Schedule (ADOS) algorithm. Subjects were medication-free prior to a blood draw for at least 2 weeks for all psychotropic medications (4 weeks for fluoxetine or depot neuroleptics).
The exclusion criteria were (a) any medical condition likely to be etiological for ASD (e.g., Rett syndrome, fragile X syndrome, or tuberous sclerosis); (b) any neurologic disorder involving pathology above the brain stem, other than uncomplicated non-focal epilepsy; (c) contemporaneous evidence, or unequivocal retrospective evidence, of probable neonatal brain damage; (d) any genetic syndrome involving the CNS, even if the link with autism was uncertain; (e) clinically significant visual or auditory impairment, even after correction; (f) any circumstances that might possibly account for the picture of autism (e.g., severe nutritional or psychological deprivation); (g) mastocytosis (including urticaria pigmentosa); (h) history of upper airway diseases; (i) history of inflammatory diseases; and (j) history of any allergies [33]. This protocol was approved by the Attikon Hospital Human Investigation Review Committee, and all parents or legal guardians provided written informed consent.
Serum collection
Fasting blood was collected in serum separator vacutainer tubes (BD Biosciences, Rockville, MD). Whole blood was allowed to clot at room temperature for about 15–30 min. The samples were then centrifuged at 1000–2000×g for 10 min at 4 °C. The upper clear fraction (serum) was carefully removed and aliquoted (0.5 mL/tube) into clean plastic capped tubes. Serum was also collected from normally developing, healthy Caucasian children (n = 20, 16 males and 4 females, 4–12 years old), unrelated to the ASD subjects, who were seen for routine health visits at the Pediatric Department of the Social Security Administration (IKA) polyclinic. All ASD and control blood samples were prepared immediately and stored at − 80 °C. They were later shipped on dry ice to Tufts University for further analysis.
All blood samples were labeled only with a code number, as well as the age and sex of the respective subject. They were sent to Tufts blind without any other identifiers, such as weight, or severity of ASD in the case of patients.
EV isolation and purification
Total EVs were isolated and purified from 1 mL of serum using the exoEasy Maxi Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Briefly, serum samples were filtered to exclude particles larger than 0.8 μm using syringe filters (EMD Millipore, Burlington, MA). Pre-filtered samples were then mixed with Buffer XBP (EMD) and were bound to an exoEasy membrane affinity spin column. The bound EVs were washed with Buffer XWP (EMD), were eluted with 400 μL Buffer XE (an aqueous buffer containing primarily inorganic salts, EMD), and were then ready for further analysis.
Transmission electron microscopy
One drop (5 μL) of a sample containing EVs was floated on the grid storage box for 1 min. The grid was then moved to a drop of double-distilled water, and the excess liquid was removed with a filter paper and stained by floating on a small drop of uranyl formate 0.75% for 30 s. After removing the excess uranyl formate with a filter paper, the grids were examined in a TecnaiG2 Spirit BioTWIN TEM (FEI Company, Hillsboro, OR, USA), and images were recorded with an AMT 2k CCD camera at a primary magnification of × 20,000–50,000 (Harvard Medical School’s Electron Microscopy Core Facility).
BCA assay
The concentration of EV total protein was quantified by the bicinchoninic acid (BCA) assay (Thermo Fisher Inc., Rockford, IL) using bovine serum albumin (BSA) as standard.
Western blot analysis
Extracellular vesicle-associated markers (CD9 and CD81) were determined by Western blot analysis. Protein samples of 10 μg were loaded, separated on 4–12% NuPAGE Bis-Tris gels under SDS-denaturing conditions (Invitrogen Life Technologies, Grand Island, NY) starting by 65 V for 45 min, and then increased to 90 V for another 30 min. Proteins were then electrotransferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA) followed by blocking for 1 h using 5% BSA in Tris-buffered saline containing 0.05% Tween-20. The membranes were then incubated overnight at 4 °C with the following primary antibodies at 1:1000 dilutions: CD9 and CD81 (System Biosciences, Mountain View, CA). For detection, the membranes were incubated with the appropriate secondary horseradish peroxidase (HRP)-conjugated antibody (System Biosciences) at 1:20,000 dilutions for 1 h at room temperature, and the blots were visualized by enhanced Super Signal West Pico Chemiluminescence (Fisher Scientific, Pittsburgh, PA).
Cell culture
The immortalized human microglia-SV40 cell line derived from primary human microglia was purchased from Applied Biological Materials Inc. (ABM Inc., Richmond, BC, Canada) and cultured in Prigrow III medium (ABM Inc., Richmond, BC, Canada) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in type-I collagen-coated T25-flasks (ABM Inc.). Microglia-SV40 maintained a specific phenotype and proliferation rate for over ten passages, during which all experiments were performed using multiple microglia thaws and sub-cultured cells. Experiments were carried out in type-I collagen-coated plates (BD PureCoat™ ECM Mimetic Cultureware Collagen I peptide plates (Becton Dickinson, Bedford, MA)). Microglia-SV40 were seeded in 6-well plates (1.0 × 105 cells/well) for 24 h before the stimulation with EVs. Lipopolysaccharide (LPS) or the peptide neurotensin (NT) was used as “positive” controls. Cells were stimulated for 24–48 h, and secreted IL-1β was measured in supernatant fluids using ELISA. Cell viability was determined by Trypan blue (0.4%) exclusion.
Enzyme-linked immunosorbent assay
IL-1β secretion in supernatant fluids was determined in duplicate by using commercially available ELISA kits (R&D Systems, Minneapolis, MN) according to the manufacturer’s instructions. For all experiments, the control cells were treated with an equal volume of culture medium. The minimum detectable level for IL-1β by ELISA was 5 pg/mL.
Total DNA isolation and mitochondrial DNA analysis
Total DNA was extracted from EVs using Qiagen DNA Micro extraction kit (Qiagen, CA). Mitochondrial-specific DNA for 7S (mtDNA7S) was detected and quantified by real-time PCR (RT-PCR) using TaqMan gene expression assays (Mt-7S: Hs02596861_s1; GAPDH: Hu, VIC, TAMRA, Applied Biosystems, Carlsbad, CA). Samples were run at 45 cycles using Applied Biosystems 7300 Real-Time PCR System. GAPDH DNA was used to exclude any genomic “contamination.” The same amount of EV-associated DNA (50 ng/μL) from each individual ASD and control sample was used for the analysis of mtDNA.
Statistical analysis
The concentration of EV-associated proteins was compared between the control and ASD samples using the Mann-Whitney U non-parametric test following the examination of normality of distribution using Shapiro-Wilk’s test.
All in vitro conditions were performed in triplicate, and all experiments were repeated at least three times (n = 3). Results are presented as mean ± SD. Data from stimulated and control samples were compared using the unpaired two-tailed Student’s t test. The significance of comparisons is denoted by p < 0.05. All analyses were performed using Graph Pad Prism 5.
Results
EV characterization
The morphology of EVs was evaluated using TEM. EVs had a spherical shape with a size of approximately 100 nm surrounded by a membrane (Fig. 1a). The identity of EVs was further validated by quantitating the EV membrane-associated markers CD9 and CD81 using Western blot analysis (Fig. 1b). There was no apparent difference in the size or shape of EVs between the control and ASD samples.
Fig. 1.
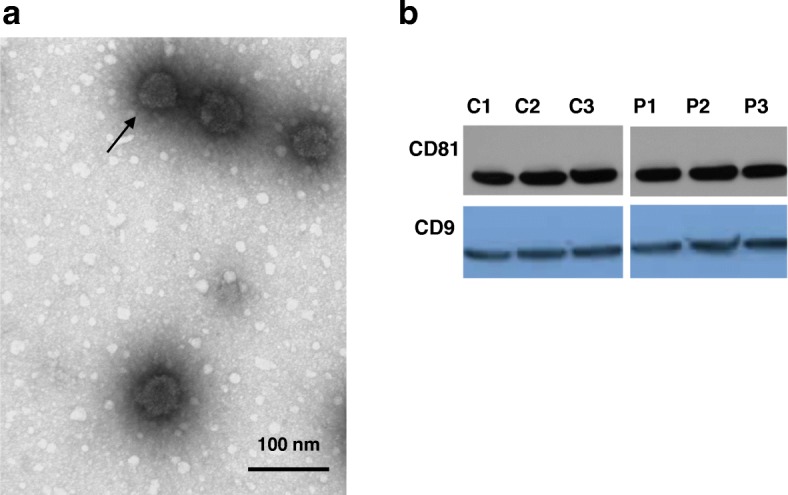
Characterization of EVs. a The morphology of EVs from the serum of patients with ASD was evaluated using TEM (scale bar, 100 nm); the arrow points to the EV membrane. b EVs collected from the serum samples of normotypic controls (n = 3) and ASD children (n = 3) were characterized by Western blot analysis for the presence of the EV-associated proteins, CD81 and CD9. C = controls, P = patients
Serum EV-associated protein is increased in children with ASD
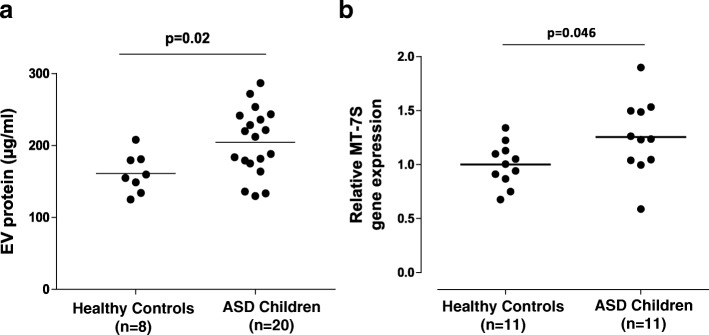
The total protein concentration in EV-enriched serum samples was significantly higher in children with ASD (204.6 ± 10.6 μg/mL) as compared to the normotypic controls (161.4 ± 9.6 μg/mL), p = 0.02 (Fig. 2a).
Fig. 2.
Scattergrams presenting EV-associated protein and mtDNA in the serum of children with ASD compared to normotypic controls. a Total EV-associated protein isolated from the serum samples of children with ASD (n = 20) compared to healthy normotypic controls (n = 8). b Serum EV-associated mtDNA7S gene expression values. Levels of mtDNA7S were assayed by qPCR and normalized to the mean of all control samples. GAPDH was undetectable
Serum EV-associated mitochondrial DNA
We also measured the EV-associated mtDNA7S in ASD children and normotypic controls. The amount of mtDNA7S contained in EVs from children with ASD was found to be increased (p = 0.046) compared to the normotypic controls (Fig. 2b).
EVs stimulate secretion of IL-1β from human microglia
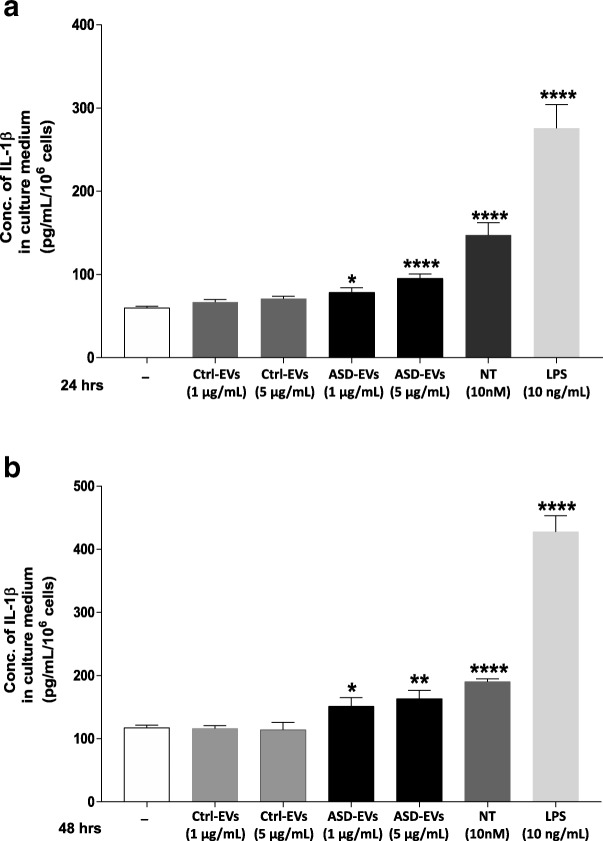
We then evaluated the effect of EVs derived from the serum of patients with ASD on human-cultured microglia secretion of IL-1β. We chose to measure this pro-inflammatory cytokine because we recently reported that microglia could secrete IL-1β in response to NT, which we had shown to be increased in the serum of children with ASD [31, 32]. Human-immortalized microglia SV40 were stimulated with EVs (1 or 5 μg/mL), quantitated as total EV-associated protein, for 24 or 48 h, and IL-1β secretion was measured by ELISA. EVs from each ASD and control sample were used separately and were not pooled. Both 1 and 5 μg/mL stimulated significant secretions of IL-1β (Fig. 3). Stimulation with 5 μg/mL for 24 h and 48 h significantly increased the secretion of IL-1β to 95.36 ± 5.31 pg/mL and 163.5 ± 13.34 pg/mL, respectively, as compared to control microglia (59.76 ± 2.03 pg/mL, p < 0.0001, and 117.7 ± 3.96 pg/mL, p = 0.008, respectively) (Fig. 3a–b). Lower amount of EVs (1 μg/mL) also stimulated IL-1β secretion at 24 h (78.47 ± 5.46 pg/mL, p = 0.02) and at 48 h (151.3 ± 13.63 pg/mL, p < 0.04), respectively (Fig. 3a–b).
Fig. 3.
Serum-derived EVs stimulate IL-1β secretion from human microglia. Immortalized HM-SV40 microglia (1.0 × 105 cells) were stimulated with serum-derived EVs, quantitated as total EV-associated protein (1 or 5 μg/mL), from patients with ASD for a 24 h and b 48 h. Secretion of IL-1β was measured in the supernatant fluid by specific enzyme-linked immunosorbent assay (ELISA). All conditions were performed in triplicates for each data set and repeated three times (n = 3). Microglia were also stimulated with NT (10 nM) or LPS (10 ng/mL) used as “positive” controls. The significance of comparisons of each experimental condition to the control unstimulated cells is denoted by *p < 0.05, **p < 0.001, or ***p < 0.0001. Conc = concentration; ctrl = control
EVs from normotypic children did not have a significant effect (p = 0.09) at either 1 or 5 μg/mL, as compared to the unstimulated control cells. Neurotensin (10 nM) and LPS (10 ng/mL) used as “positive” controls also increased (p < 0.0001) the secretion of IL-1β (Fig. 3).
Discussion
This is the first report, to the best of our knowledge, that serum from children with ASD contains significantly increased total EV-associated protein as compared to the healthy normotypic controls. The size of the EVs qualifies them as exosomes. There was no apparent difference in shape, size, or the EV-associated proteins CD9 and CD81 between ASD and normotypic controls.
We also show that serum-derived EVs from children with ASD stimulate the secretion of the pro-inflammatory cytokine IL-1β from human SV40-immortalized microglia. We chose to measure IL-1β because we recently reported that the neuropeptide neurotensin (NT) stimulates human microglia to secrete IL-1β, [18] which has been shown to be increased in the brains of children with ASD, [34] and in a mouse model of autism [35].
EVs from children with ASD contain a significant amount of mtDNA compared to the normotypic controls. We had previously reported that mtDNA is increased in children with ASD [20]. We also reported that mtDNA can be secreted extracellularly from mast cell stimulated by the neuropeptide substance P (SP) [20], an important finding given that atopic diseases have been associated with increased risk of ASD [36]. Extracellular mtDNA serves as an alarmin and leads to pro-inflammatory mediator secretion from immune cells [37, 38]. In the case of ASD, mtDNA may serve as an “innate” pathogen [39] that would be protected from degradation inside EVs and could reach the microglia through the brain lymphatics [40] or through the blood-brain barrier [41].
IL-1β synthesis occurs via activation of the Nod-like receptor pyrin domain-containing protein 3 (NLRP3) inflammasome which requires two different signals [42]. In our findings, signal 1 could be mtDNA, but EVs must contain some other molecule that could serve as signal 2. A possible candidate could be the peptide NT. Our laboratory previously reported NT to be increased in the serum of children with ASD [31, 32] and which we recently reported can stimulate secretion of IL-1β from human microglia [18].
The cellular origin of the increased serum EVs is presently unknown. Astrocytes and glioblastoma cells have been reported to release EVs containing mtDNA [24], but there is no evidence at present that the serum EVs derive from the brain. Serum EVs could derive from MC [43] since MC-microglia interactions are considered important in the inflammation of the brain [19, 44].
Microglia-derived “microparticles,” a specific type of extracellular vesicles, are also released after a traumatic brain injury (TBI) and can activate microglia in vitro [45]. LPS can activate microglia to release “microparticles” with increased content of pro-inflammatory mediators IL-1β and miRNA-155 in vitro [45]. EVs could also potentially transport environmental triggers [46] or derive from other comorbid conditions, [10, 47] but such were not present in the patients with ASD analyzed.
In addition to mtDNA, mRNA and miRNA can be transported by EVs and have been associated with brain inflammation and multiple sclerosis [48]. In fact, a number of miRNAs such as miR-27a, miR-23a, and miR-628-5p were detected in the saliva of children with ASD [49]. One study reported differential expression of miR-497 in EVs isolated from postmortem pre-cortex from patients with schizophrenia and bipolar disorder [50]. EVs were also shown to contain several proteins or miRNA [51] that may be involved in Alzheimer’s and in Parkinson’s disease with dementia [52, 53]. EVs are increasingly discussed in the context of neurodegenerative diseases [22, 54].
Conclusion
To the best of our knowledge, this is the first report describing that serum from children with ASD contains significantly increased total EV-associated protein as compared to healthy normotypic controls, that these EVs contain mtDNA, and that they can stimulate human-cultured microglia to secrete the pro-inflammatory cytokine IL-1β. Our results, although preliminary, provide novel information that may help explain the inflammation of the brain and ASD pathogenesis. Further studies on a larger sample size are needed to extend these findings and validate their usefulness for diagnosis and use as a target for novel effective treatments for ASD.
Acknowledgements
We appreciate the support and time given by the families who were involved in this study. We thank Drs. K. Francis and A. Taliou for the collection and processing of the serum from children with ASD and Dr. A. Theoharides for the collection and processing of the normotypic control samples. We also thank Dr. Alexandra Taracanova for the help with the Western blot analysis and Ms. Maria Ericsson, Manager of the Electron Microscopy Core Facility at Harvard University School of Medicine, for helping with the EV characterization. Finally, we thank Dr. P. W. Askenase (Yale University, New Haven, CT) for his encouragement and useful discussions.
Funding
This study was funded in part by an anonymous grant to TCT.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- ADOS
Autism Diagnostic Observation Schedule
- ASD
Autism spectrum disorder
- EVs
Extracellular vesicles
- IL-1β
Interleukin-1β
- MC
Mast cells
- mtDNA
Mitochondrial DNA
- MVBs
Multivesicular bodies
- NLRP3
Nod-like receptor pyrin domain-containing protein 3
- NT
Neurotensin
- SP
Substance P
- TEM
Transmission electron microscopy
Authors’ contributions
TCT and IT conceived the concept. IT performed the experiments, the data collection, and their analysis. TCT reviewed the results and interpretation. TCT and IT prepared the manuscript. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Attikon Hospital Human Investigation Review Committee, and all parents or legal guardians provided written informed consent.
Consent for publication
Not applicable.
Competing interests
Both authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Irene Tsilioni, Email: eirini.tsilioni@tufts.edu.
Theoharis C. Theoharides, Phone: (617) 636-6866, Email: theoharis.theoharides@tufts.edu
References
- 1.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- 2.McPartland J, Volkmar FR. Autism and related disorders. Handb Clin Neurol. 2012;106:407–418. doi: 10.1016/B978-0-444-52002-9.00023-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383:896–910. doi: 10.1016/S0140-6736(13)61539-1. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (2018) CDC estimates 1 in 59 children has been identified with autism spectrum disorder, https://www.cdc.gov/features/new-autism-data/index.html Access date July 11 2018.
- 5.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;11:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willsey AJ, State MW. Autism spectrum disorders: from genes to neurobiology. Curr Opin Neurobiol. 2015;30:92–99. doi: 10.1016/j.conb.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruggeri B, Sarkans U, Schumann G, Persico AM. Biomarkers in autism spectrum disorder: the old and the new. Psychopharmacology (Berl) 2014;231:1201–1216. doi: 10.1007/s00213-013-3290-7. [DOI] [PubMed] [Google Scholar]
- 9.Theoharides TC, Doyle R, Francis K, Conti P, Kalogeromitros D. Novel therapeutic targets for autism. Trends Pharmacol Sci. 2008;29:375–382. doi: 10.1016/j.tips.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Bauman ML. Medical comorbidities in autism: challenges to diagnosis and treatment. Neurotherapeutics. 2010;7:320–327. doi: 10.1016/j.nurt.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leigh JP, Du J. Brief report: forecasting the economic burden of autism in 2015 and 2025 in the United States. J Autism Dev Disord. 2015;12:4135–4139. doi: 10.1007/s10803-015-2521-7. [DOI] [PubMed] [Google Scholar]
- 12.Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez JI, Kern JK. Evidence of microglial activation in autism and its possible role in brain underconnectivity. Neuron Glia Biol. 2011;7:205–213. doi: 10.1017/S1740925X12000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, West AB, Arking DE. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:5748. doi: 10.1038/ncomms6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koyama R, Ikegaya Y. Microglia in the pathogenesis of autism spectrum disorders. Neurosci Res. 2015;100:1–5. doi: 10.1016/j.neures.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Takano T. Role of microglia in autism: recent advances. Dev Neurosci. 2015;37:195–202. doi: 10.1159/000398791. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Wang Y, Dong H, Xu Y, Zhang S. Induction of microglial activation by mediators released from mast cells. Cell Physiol Biochem. 2016;38:1520–1531. doi: 10.1159/000443093. [DOI] [PubMed] [Google Scholar]
- 18.Patel AB, Tsilioni I, Leeman SE, Theoharides TC. Neurotensin stimulates sortilin and mTOR in human microglia inhibitable by methoxyluteolin, a potential therapeutic target for autism. Proc Natl Acad Sci U S A. 2016;113:E7049–E7058. doi: 10.1073/pnas.1604992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skaper SD, Giusti P, Facci L. Microglia and mast cells: two tracks on the road to neuroinflammation. FASEB J. 2012;26:3103–3117. doi: 10.1096/fj.11-197194. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Angelidou A, Alysandratos KD, Vasiadi M, Francis K, Asadi S, Theoharides A, Sideri k, Lykouras L, Kalogeromitros D, Theoharides TC. Mitochondrial DNA and anti-mitochondrial antibodies in serum of autistic children. J Neuroinflammation. 2010;7:80. doi: 10.1186/1742-2094-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porro C, Trotta T, Panaro MA. Microvesicles in the brain: biomarker, messenger or mediator? J Neuroimmunol. 2015;288:70–78. doi: 10.1016/j.jneuroim.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Levy E. Exosomes in the diseased brain: first insights from in vivo studies. Front Neurosci. 2017;11:142. doi: 10.3389/fnins.2017.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 25.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran S, Palanisamy V. Horizontal transfer of RNAs: exosomes as mediators of intercellular communication. Wiley Interdiscip Rev RNA. 2012;3:286–293. doi: 10.1002/wrna.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryniarski K, Ptak W, Jayakumar A, Pullmann K, Caplan MJ, Chairoungdua A, Lu J, Adams BD, Sikora E, Nazimek K, Marquez S, Kleinstein SH, Sangwung P, Iwakiri Y, Delgato E, Redegeld F, Blokhuis BR, Wojcikowski J, Daniel AW, Groot KT, Askenase PW. Antigen-specific, antibody-coated, exosome-like nanovesicles deliver suppressor T-cell microRNA-150 to effector T cells to inhibit contact sensitivity. J Allergy Clin Immunol. 2013;132:170–181. doi: 10.1016/j.jaci.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryniarski K, Ptak W, Martin E, Nazimek K, Szczepanik M, Sanak M, Askenase PW. Free extracellular miRNA functionally targets cells by transfecting exosomes from their companion cells. PLoS One. 2015;10:e0122991. doi: 10.1371/journal.pone.0122991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalani A, Tyagi A, Tyagi N. Exosomes: mediators of neurodegeneration, neuroprotection and therapeutics. Mol Neurobiol. 2014;49:590–600. doi: 10.1007/s12035-013-8544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turpin D, Truchetet ME, Faustin B, Augusto JF, Contin-Bordes C, Brisson A, Blanco P, Duffau P. Role of extracellular vesicles in autoimmune diseases. Autoimmun Rev. 2016;15:174–183. doi: 10.1016/j.autrev.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Angelidou A, Francis K, Vasiadi M, Alysandratos K-D, Zhang B, Theoharides A, Lykouras L, Kalogeromitros D, Theoharides T. Neurotensin is increased in serum of young children with autistic disorder. J Neuroinflam. 2010;7:48. doi: 10.1186/1742-2094-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsilioni I, Dodman N, Petra AI, Taliou A, Francis K, Moon-Fanelli A, Shuster L, Theoharides TC. Elevated serum neurotensin and CRH levels in children with autistic spectrum disorders and tail-chasing bull terriers with a phenotype similar to autism. Transl Psychiatry. 2014;4:e466. doi: 10.1038/tp.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taliou A, Zintzaras E, Lykouras L, Francis K. An open-label pilot study of a formulation containing the anti-inflammatory flavonoid luteolin and its effects on behavior in children with autism spectrum disorders. Clin Ther. 2013;35:592–602. doi: 10.1016/j.clinthera.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah I, Van de Water J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav Immun. 2011;25:40–45. doi: 10.1016/j.bbi.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Estes ML, McAllister AK. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci. 2015;16:469–486. doi: 10.1038/nrn3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theoharides TC, Tsilioni I, Patel AB, Doyle R. Atopic diseases and inflammation of the brain in the pathogenesis of autism spectrum disorders. Transl Psychiatry. 2016;6:e844. doi: 10.1038/tp.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins LV, Hajizadeh S, Holme E, Jonsson IM, Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol. 2004;75:995–1000. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- 38.Sun S, Sursal T, Adibnia Y, Zhao C, Zheng Y, Li H, Otterbein LE, Hauser CJ, Itagaki K. Mitochondrial DAMPs increase endothelial permeability through neutrophil dependent and independent pathways. PLoS One. 2013;8:e59989. doi: 10.1371/journal.pone.0059989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theoharides TC, Asadi S, Panagiotidou S, Weng Z. The “missing link” in autoimmunity and autism: extracellular mitochondrial components secreted from activated live mast cells. Autoimmun Rev. 2013;12:1136–1142. doi: 10.1016/j.autrev.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 40.Louveau A, Harris TH, Kipnis J. Revisiting the mechanisms of CNS immune privilege. Trends Immunol. 2015;36:569–577. doi: 10.1016/j.it.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto J, Stewart T, Banks WA, Zhang J. The transport mechanism of extracellular vesicles at the blood-brain barrier. Curr Pharm Des. 2017;23:6206–6214. doi: 10.2174/1381612823666170913164738. [DOI] [PubMed] [Google Scholar]
- 42.Vanaja SK, Rathinam VA, Fitzgerald KA. Mechanisms of inflammasome activation: recent advances and novel insights. Trends Cell Biol. 2015;25:308–315. doi: 10.1016/j.tcb.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Inca F, Pucillo CE. Exosomes: tiny clues for mast cell communication. Front Immunol. 2015;6:73. doi: 10.3389/fimmu.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girolamo F, Coppola C, Ribatti D. Immunoregulatory effect of mast cells influenced by microbes in neurodegenerative diseases. Brain Behav Immun. 2017;65:68–89. doi: 10.1016/j.bbi.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 45.Kumar A, Stoica BA, Loane DJ, Yang M, Abulwerdi G, Khan N, Kumar A, Thom SR, Faden AI. Microglial-derived microparticles mediate neuroinflammation after traumatic brain injury. J Neuroinflammation. 2017;14:47. doi: 10.1186/s12974-017-0819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossignol DA, Genuis SJ, Frye RE. Environmental toxicants and autism spectrum disorders: a systematic review. Transl Psychiatry. 2014;4:e360. doi: 10.1038/tp.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujita Y, Yoshioka Y, Ito S, Araya J, Kuwano K, Ochiya T. Intercellular communication by extracellular vesicles and their microRNAs in asthma. Clin Ther. 2014;36:873–881. doi: 10.1016/j.clinthera.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Narahari A, Hussain M, Sreeram V. MicroRNAs as biomarkers for psychiatric conditions: a review of current research. Innov Clin Neurosci. 2017;14:53–55. [PMC free article] [PubMed] [Google Scholar]
- 49.Hicks SD, Ignacio C, Gentile K, Middleton FA. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016;16:52. doi: 10.1186/s12887-016-0586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banigan MG, Kao PF, Kozubek JA, Winslow AR, Medina J, Costa J, Schmitt A, Schneider A, Cabral H, Cagsal-Getkin O, Vanderburg CR, Delalle I. Differential expression of exosomal microRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PLoS One. 2013;8:e48814. doi: 10.1371/journal.pone.0048814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van GV, An SS. Emergence of exosomal miRNAs as a diagnostic biomarker for Alzheimer’s disease. J Neurol Sci. 2016;360:141–152. doi: 10.1016/j.jns.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Yuyama K, Sun H, Usuki S, Sakai S, Hanamatsu H, Mioka T, Kimura N, Okada M, Tahara H, Furukawa J, Fujitani N, Shinohara Y, Igarashi Y. A potential function for neuronal exosomes: sequestering intracerebral amyloid-beta peptide. FEBS Lett. 2015;589:84–88. doi: 10.1016/j.febslet.2014.11.027. [DOI] [PubMed] [Google Scholar]
- 53.Stuendl A, Kunadt M, Kruse N, Bartels C, Moebius W, Danzer KM, Mollenhauer B, Schneider A. Induction of alpha-synuclein aggregate formation by CSF exosomes from patients with Parkinson’s disease and dementia with Lewy bodies. Brain. 2016;139:481–494. doi: 10.1093/brain/awv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selmaj I, Mycko MP, Raine CS, Selmaj KW. The role of exosomes in CNS inflammation and their involvement in multiple sclerosis. J Neuroimmunol. 2017;306:1–10. doi: 10.1016/j.jneuroim.2017.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.