Abstract
Background
Tubercidin (TBN), an adenosine analog with potent antimycobacteria and antitumor bioactivities, highlights an intriguing structure, in which a 7-deazapurine core is linked to the ribose moiety by an N-glycosidic bond. However, the molecular logic underlying the biosynthesis of this antibiotic has remained poorly understood.
Results
Here, we report the discovery and characterization of the TBN biosynthetic pathway from Streptomyces tubercidicus NBRC 13090 via reconstitution of its production in a heterologous host. We demonstrated that TubE specifically utilizes phosphoribosylpyrophosphate and 7-carboxy-7-deazaguanine for the precise construction of the deazapurine nucleoside scaffold. Moreover, we provided biochemical evidence that TubD functions as an NADPH-dependent reductase, catalyzing irreversible reductive deamination. Finally, we verified that TubG acts as a Nudix hydrolase, preferring Co2+ for the maintenance of maximal activity, and is responsible for the tailoring hydrolysis step leading to TBN.
Conclusions
These findings lay a foundation for the rational generation of TBN analogs through synthetic biology strategy, and also open the way for the target-directed search of TBN-related antibiotics.
Electronic supplementary material
The online version of this article (10.1186/s12934-018-0978-8) contains supplementary material, which is available to authorized users.
Keywords: Biosynthesis, Tubercidin, 7-deazapurine, Phosphoribosylpyrophosphate, NADPH-dependent reductase, Nudix hydrolase, Synthetic biology
Background
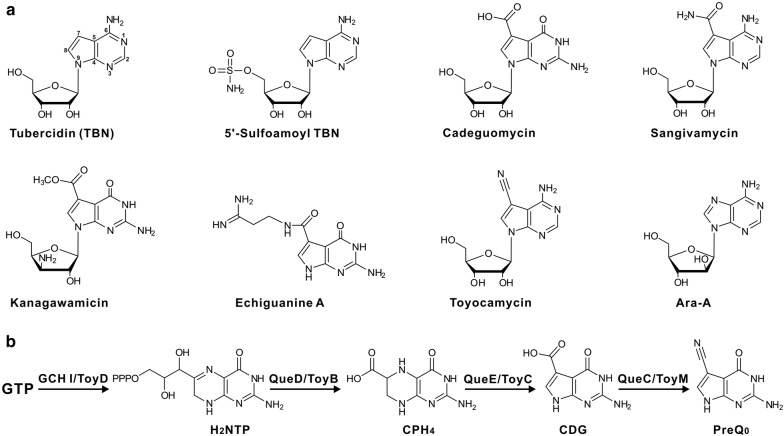
Pyrrolopyrimidine (also known as 7-deazapurine) containing compounds have been discovered to be widely distributed in nature as microbial secondary metabolites (SMs) and also as hypermodified base (queuosine) in RNA [1, 2]. Usually, this family of SMs shows diverse biological functions, ranging from cofactors (involved in the biosynthesis of tetracycline antibiotics) as well as DNA repair to antibiotics with herbicidal, antibacterial, antiviral, antifungal, antitumor, and antineoplastic activities [1, 2]. Previously, toyocamycin/sangivamycin (produced by Streptomyces rimosus ATCC 14673) and tubercidin (TBN, produced by Streptomyces tubercidicus NBRC 13090) served as prototypes for the deazapurines (Fig. 1a) [2–4]. Since their discovery, a series of structurally-related antibiotics has been continuously isolated from terrestrial and marine sources with guidance of bioactivity assays (Fig. 1a) [2].
Fig. 1.
Structures of TBN and related antibiotics and the confirmed biosynthetic pathway to PreQ0. a Chemical structures of TBN and related purine nucleoside antibiotics. TBN is produced by S. tubercidicus NBRC 13090; 5′-sulfamoyl TBN is produced by S. mirabilis; cadeguomycin is produced by S. hygroscopicus; sangivamycin/toyocamycin is produced by S. rimosus ATCC 14673; kanagawamicin is produced by Actinoplanes kanagawaensis; echiguanine A is produced by Streptomyces sp. MI698-50F1; Ara-A (arabinofuranosyladenine) is produced by S. antibioticus NRRL 3238. b The confirmed biosynthetic pathway leading to PreQ0. A four-enzyme cascade for the synthesis of PreQ0 is identified in both queuosine and sangivamycin/toyocamycin biosynthetic pathways. GCH I, GTP cyclohydrolase I (ToyD); QueD/ToyB, 6-carboxy-5,6,7,8-tetrahydropterin (CPH4) synthase; H2NTP, 7, 8-dihydroneopterin-3′-triphosphate; QueE/ToyC, 7-carboxy-7-deazaguanine (CDG) synthase; QueC/ToyM, 7-cyano-7-deazaguanine (PreQ0) synthetase
Tubercidin is susceptible to phosphorylation by adenosine kinase to mono-, di-, and tri-phosphorylated forms accounting for its structural resemblance to adenosine [5, 6]. Accordingly, it is capable of acting as a powerful inhibitor of RNA and DNA polymerase, and shows a relatively-broad spectrum of bioactivities [2]. TBN is bioactive against Candida albicans, Mycobacterium tuberculosis, and Streptococcus faecalis, however, it exhibits no inhibition of other Gram positive bacteria and fungi [2, 5]. In addition, TBN shows antiviral and antitumor activities [5], and more interestingly, it can also kill trypanosomes by targeting glycolysis, especially by inhibition of phosphoglycerate kinase [7].
TBN features an unusual deazapurine core, in which N-7 is substituted for C-7 (Fig. 1a) [4]. Previous metabolic labeling experiments indicated that the deazapurine core is derived from a purine precursor, likely GTP, and C-1′, 2′, and 3′ of ribose are utilized to construct the pyrrole ring with elimination of C-8 in GTP [8, 9]. Subsequently, the enzymatic logic for the construction of deazapurine core has been deciphered by independent studies, which were focused on the identification of the biosynthetic pathway of queuosine and toyocamycin/sangivamycin (Fig. 1b) [2, 10]. A four-enzyme cascade has been demonstrated to be responsible for the biosynthetic steps leading to PreQ0 (7-cyano-7-deazaguanine) (Fig. 1b). Among them, GTP cyclohydrolase I (GCH I, ToyD) catalyzes the first step from GTP to H2NTP (7, 8-dihydroneopterin-3′-triphosphate) with opening the ribose ring, followed by a series of Amadori rearrangement and subsequent recyclization. The intermediate H2NTP is then sequentially recognized by CPH4 (6-carboxy-5, 6, 7, 8-tetrahydropterin) synthase (QueD/ToyB) and CDG synthase (QueE/ToyC) [2, 10]. The enzymatic mechanism of QueE/ToyC, a member of the radical S-adenosyl-l-methionine (SAM) protein, has been elucidated by elegant studies of the Bandarian group. This enzyme catalyzes a key SAM- and Mg2+-dependent radical-mediated ring contraction step [11, 12], which is common to the biosynthetic pathways of all deazapurine-containing compounds. More recently, QueC/ToyM has been verified as an amide synthetase, as well as a nitrile synthetase, which could accept both the acid and amide forms of CDG enabling sequential amidation and dehydration to the nitrile [13, 14].
Although the distinguished activities and unusual structure of TBN are well known, nature’s strategy for the building of this molecule has as yet remained poorly understood. In the present study, we have identified the TBN biosynthetic gene cluster from S. tubercidicus NBRC 13090 (S. tubercidicus hereafter) by engineered production of TBN in a heterologous host, and have further elucidated that TBN biosynthesis involves a PRPP-dependent assembly logic associated with tailoring reduction and phosphohydrolysis steps. Our deciphering of the TBN biosynthetic pathway provides a solid basis for the further combinatorial biosynthesis of this group of nucleoside antibiotics towards improved features, and opens the way for the target-directed genome mining of novel TBN-related antibiotics from the available microbial genome reservoirs.
Results
Identification of the TBN biosynthetic gene cluster
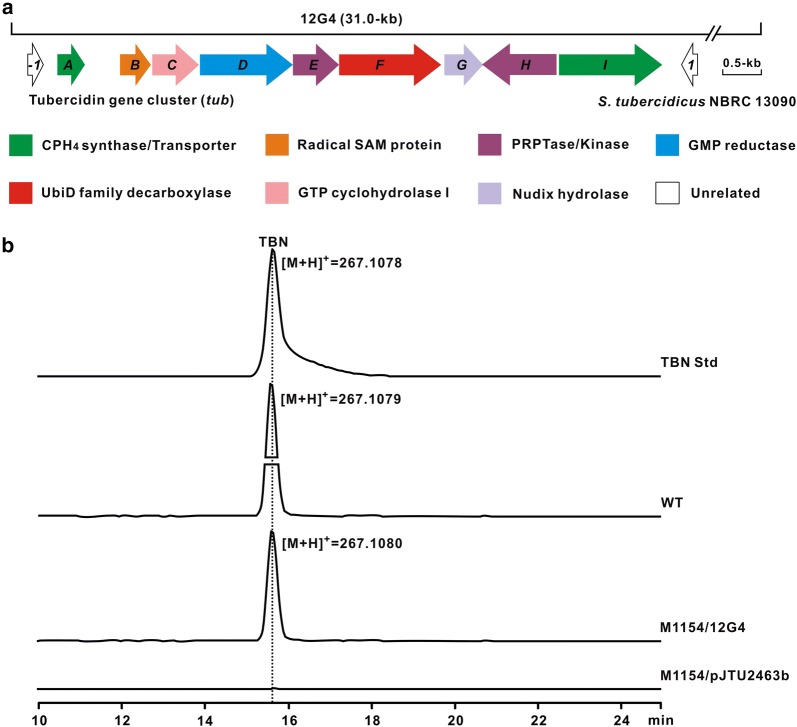
To identify the gene cluster for TBN biosynthesis, the genome of S. tubercidicus was sequenced by an Illumina Hiseq method, rendering appr. 7.88-Mb of non-redundant bases after assembly of clean reads (Additional file 1: Table S1). The genomic data was then annotated by Glimmer 3.02 software, affording 7263 open reading frames (ORFs) (Additional file 1: Table S1). TBN features the deazapurine core, which is also existed in toyocamycin, sangivamycin, and other related antibiotics; we therefore deduced that the initial enzymatic steps for the building of the core of them follow an identical logic. We accordingly utilized ToyD (GTP cyclohydrolase I) and ToyB (CPH4 synthase) as the enzyme probes to conduct BlastP analysis against the genome of S. tubercidicus. As expected, a candidate gene cluster (tub) encoding enzymes involving TubC (64% identity to ToyD) and TubA (65% identity to ToyB) was identified from the genome (Fig. 2a, Table 1). Further looking through the surrounding region resulted in the identification of the genes coding for a radical SAM enzyme (TubB) and a GMP reductase (TubD) (Fig. 2a, Table 1). These results suggest that the tub gene cluster is very likely responsible for the biosynthesis of TBN.
Fig. 2.
Genetic organization and validation of the gene cluster (tub) for TBN biosynthesis. a Genetic organization of the tub gene cluster. b LC–MS analysis of the target metabolite produced by the recombinants of S. coelicolor M1154. TBN Std, the authentic standard of TBN; WT, the metabolites of the wild type TBN producer S. tubercidicus; M1154/12G4, the metabolites of S. coelicolor M1154 derivative harboring the cosmid 12G4; M1154/pJTU2463b, the metabolites of S. coelicolor M1154 derivative harboring the plasmid pJTU2463b as negative control
Table 1.
Deduced functions of the open reading frames in the tub gene cluster
| Protein | aa | Proposed function | Homolog, Origin | Identity, similarity (%) | Accession no. |
|---|---|---|---|---|---|
| Orf-1 | 58 | (2Fe-2S)-binding protein | AWI43_15300, Streptomyces sp. WAC04657 | 78, 83 | KYG55615.1 |
| TubA | 125 | CPH4 synthase | ToyB, Streptomyces rimosus ATCC 14673 | 65, 79 | ACF06634.1 |
| TubB | 139 | Radical SAM family protein | SPAR_33961, Streptomyces sparsogenes DSM 40356 | 71, 79 | OMI34937.1 |
| TubC | 199 | GTP cyclohydrolase I (GCH I) | ToyD, Streptomyces rimosus ATCC 14673 | 64, 75 | ACF06635.1 |
| TubD | 385 | GMP reductase | ToyE, Streptomyces rimosus ATCC 14673 | 61, 75 | ACF06636.1 |
| TubE | 194 | Phosphoribosylpyrophosphate transferase | ToyH, Streptomyces rimosus ATCC 14673 | 39, 53 | ACF06639.1 |
| TubF | 422 | UbiD family decarboxylase | ASE41_09730, Streptomyces sp. Root264 | 90, 93 | KRD23282.1 |
| TubG | 168 | Nudix hydrolase | ASE41_09725, Streptomyces sp. Root264 | 96, 96 | KRD23281.1 |
| TubH | 314 | Carbohydrate kinase family protein | ASE41_09720, Streptomyces sp. Root264 | 82, 86 | KRD23280.1 |
| TubI | 374 | MFS transporter | ASE41_09715, Streptomyces sp. Root264 | 83, 88 | KRD23279.1 |
| Orf1 | 68 | Hypothetical protein | ASE41_09630, Streptomyces sp. Root264 | 41, 60 | KRD23264.1 |
Engineered production of TBN in the heterologous host S. coelicolor M1154
To correlate the tub gene cluster to TBN production, a cosmid 12G4 (with appr. 30.0-kb insertion DNA, Additional file 1: Table S2) harboring the TBN gene cluster was screened from the pJTU2463b-derived genomic library of S. tubercidicus (Additional file 1: Table S2), and then it was introduced into the heterologous host S. coelicolor M1154 [15] via conjugation. The validated conjugants were then fermented for further metabolite analysis, and LC–MS analysis indicated that the sample (M1154/12G4) is capable of producing the distinctive [M+H]+ ion of TBN at m/z 267.1080 (Fig. 2a). In addition, MS/MS analysis showed that the main fragments were generated at 134.8841, 248.9537, 231.1378, and so forth, fully consistent with those of the TBN authentic standard (Additional file 1: Figure S1). These combined data unambiguously demonstrate that the cosmid 12G4 confers the heterologous host S. coelicolor M1154 with the capability of TBN production.
In silico analysis of the TBN biosynthetic gene cluster
In silico analysis showed that the TBN gene cluster spans appr. 8.0-kb continuous chromosomal region, and consists of 9 genes (tubA-I) (Fig. 2a, Table 1). Genes tubCBA are proposed to be responsible for the initial biosynthetic steps to CDG. The gene tubC codes for GTP cyclohydrolase I which shows 64% identity to ToyD of toyocamycin biosynthesis, and the product of tubB (radical SAM enzyme) exhibits 71% identity to SPAR_33961 of S. sparsogenes DSM 40356. tubA encodes CPH4 synthase that indicates significant similarity (65% identity) to ToyB in toyocamycin biosynthetic pathway. tubD encodes a GMP reductase (61% identity to ToyE) that is proposed to be responsible for IMP (Inosine-5′-monophosphate) biosynthesis, whereas TubE has 39% identity to ToyH, an enzyme hypothetically responsible for the assembly step during toyocamycin biosynthesis. TubF exhibits high similarity (90% identity to ASE41_09730 from Streptomyces sp. Root264) to UbiD family decaboxylases, which bear a recently identified cofactor prFMN (prenylated-FMN) [16–19], but the roles of most UbiD family decaboxylases are functionally unassigned. TubG is annotated as a Nudix hydrolase, and this family of enzymes usually plays “house-cleaning” role to sanitize nucleotide pool in primary metabolism [20]. tubHI individually code for carbohydrate kinase family protein (82% identity to ASE41_09720) and MFS (Major facilitator superfamily) transporter.
TubE functions as a CDG-PRPP phosphoribosyltransferase
BlastP analysis indicates that tubE encodes a purine phosphoribosylpyrophosphate transferase (Additional file 1: Figure S2), which likely governs the assembly step during TBN biosynthesis. To determine if TubE fulfills such enzymatic role, we overexpressed and purified the protein to near homogeneity from E. coli (Additional file 1: Figure S3A), and tested its activity in vitro first, using deazaguanine and PRPP as substrates, and LC–MS results indicated that the TubE reaction could not generate the expected product (Additional file 1: Figure S3B, C), suggesting that this enzyme does not recognize deazaguanine as a substrate. As a result, we ponder whether the assembly step is prior to decarboxylation during TBN biosynthesis (Fig. 1b), namely, CDG is the prime substrate of TubE. We then re-examined TubE activity using this compound as substrate, and HPLC analysis showed that the TubE reaction is capable of generating a new peak at RT = 24.7 min, which is absent in the negative control (Fig. 3b). Further LC–MS analysis indicated that the target peak (RT = 24.7 min) could produce a characteristic [M+H]+ ion at m/z 407.0591 (Fig. 3b) with main fragment ions at 177.9070, 194.9660, 294.0352, and 389.0653, well matched to the theoretical fragmentation pattern of compound 1 (Additional file 1: Figure S3D). To finally clarify the chemical identity of the product (1) formed by TubE, it was purified and analyzed by nuclear magnetic resonance (NMR). The results showed that 1H, 13C, and 2D NMR signals of the compound are highly consistent with those of 1, confirming that the TubE-catalyzed product is 1 (Additional file 1: Figures S4, S5). Taken together, all of the data demonstrate that TubE functions as a CDG-specific phosphoribosyltransferase.
Fig. 3.
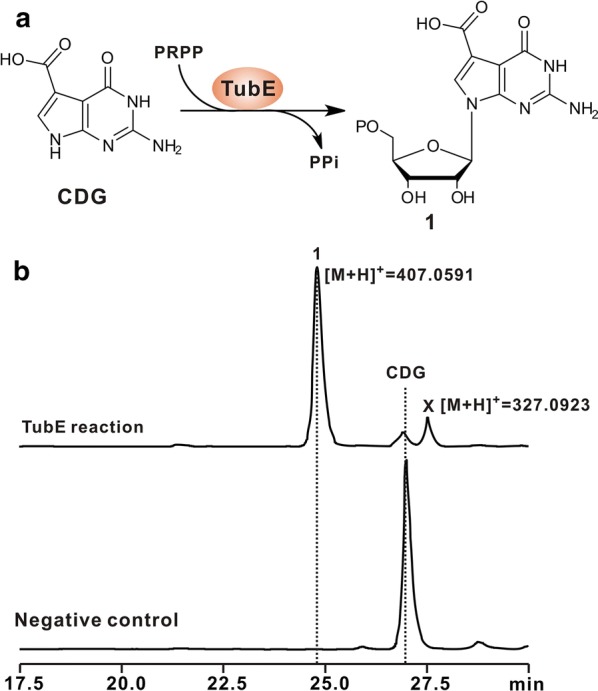
Biochemical characterization of TubE as a CDG-PRPP transferase. a Schematic of the TubE-catalyzed reaction in the presence of PRPP and CDG. b HPLC analysis of the TubE reaction. TubE reaction, HPLC profile of the TubE reaction using PRPP and CDG as substrates; Negative control, HPLC profile of the reaction using PRPP and CDG as substrates but without TubE added. Compound X shows [M+H]+ ion at m/z 327.0923, with main fragment ions at 177.9827, 194.9009, 292.0454, and 309.9970, which are fully consistent with the theoretical fragmentation pattern of dephosphorylated 1
Biochemical characterization of TubD as an NADPH-dependent reductase
In silico analysis exhibited that TubD bears an IMP dehydrogenase/GMP reductase domain (Additional file 1: Figure S6), implicating that TubD should play a similar role in TBN biosynthesis (presumably converting compound 2 to 3) (Fig. 4a). To see if TubD is capable of performing this function, it was expressed and purified from E. coli (Additional file 1: Figure S7A). We then tested its activity in vitro using GMP, an analog of 2, as a substrate (Fig. 4a). LC–MS analysis showed the target IMP [M+H]+ ion at m/z 349.0540 in the TubD reaction, while it was absent in the negative control (Fig. 4b). In addition, MS/MS analysis indicated that the predominant fragment ions were generated at 118.8544, 136.8826, 233.0000, and 331.0666, fully consistent with those of the IMP authentic standard (Additional file 1: Figure S7B, C). Subsequently, we tested cofactor specificity for TubD activity, and LC–MS analysis showed that TubD reaction with NADH as cofactor could not give rise to the target IMP [M+H]+ ion, suggesting that this cofactor could not replace NADPH to supply reducing equivalents for the TubD reaction (Additional file 1: Figure S7D). Moreover, we also tested the factors that potentially affect TubD activity, and found that the metal ion K+ plays a pivotal role for TubD activity (Additional file 1: Figure S7D). These combined data suggest that TubD is an NADPH-dependent reductase catalyzing reductive deamination of compound 2 to 3 in TBN biosynthesis.
Fig. 4.
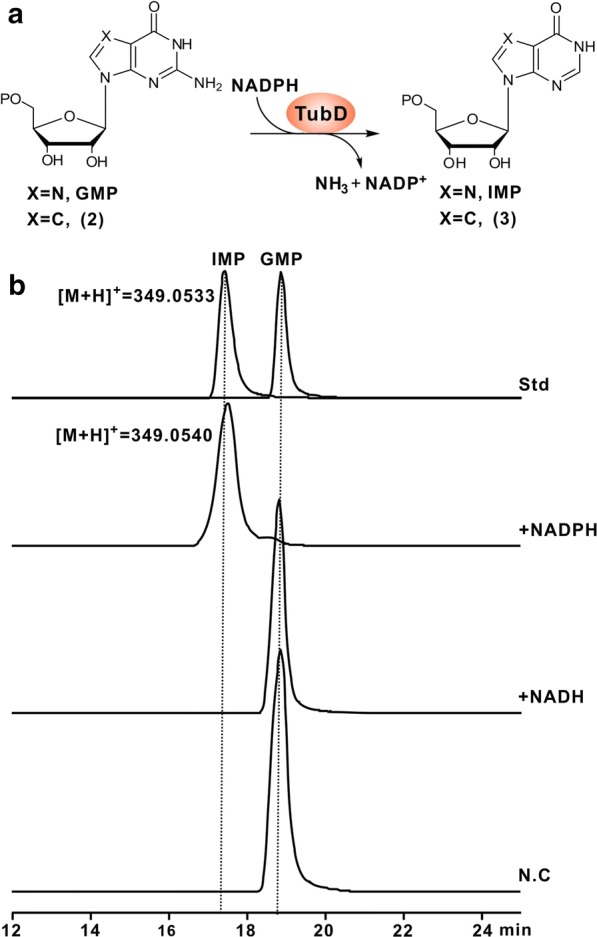
In vitro characterization of TubD as an NADPH-dependent reductase. a Schematic of the TubD-catalyzed reaction in the presence of the redox cofactor NADPH. b LC–MC analysis of the TubD reaction. Std, EIC analysis of the authentic standards of IMP and GMP; +NADPH, EIC analysis of the TubD reaction with NADPH as redox cofactor; +NADH, EIC analysis of the TubD reaction with NADH as redox cofactor; N.C, EIC analysis of the reaction with NADPH and substrate added but lacking the enzyme TubD
Biochemical characterization of TubG as a tailoring Nudix hydrolase
Bioinformatic analysis showed that TubG possesses a highly conserved 23-residue (GX5EX7REUXEEXGU) Nudix motif (Additional file 1: Figure S8) [21], which normally functions as a metal binding and catalytic site, accordingly, this protein is predicted to be a Nudix hydrolase for the final tailoring step during TBN biosynthesis. To determine if TubG executes the delineated functional role, it was overexpressed and purified from E. coli to near homogeneity (Additional file 1: Figure S9A). We then tested TubG activity using AMP (compound 5 analog) as an alternative substrate (Fig. 5a), and HPLC analysis indicated that the TubG catalyzed reaction produced the expected adenosine peak but not in the TubG-negative control (Fig. 5b). Further LC–MS results indicated that the peak could give rise to the target [M+H]+ ion at m/z 268.1034, whose fragmentation pattern is fully in accordance with that of the adenosine standard (Fig. 5b, Additional file 1: Figure S9B, C). In addition, we evaluated the divalent cations affecting enzymatic activity of TubG. Of all divalent cations (involving Mg2+, Ni2+, Mn2+, Co2+, Ca2+, Fe2+, Cu2+, and Zn2+) selected, we found that Co2+ is capable of maintaining the maximal enzymatic activity for TubG (Fig. 5c). All of this suggests that TubG is an atypical Nudix hydrolase using Co2+ as the most preferred divalent cation.
Fig. 5.
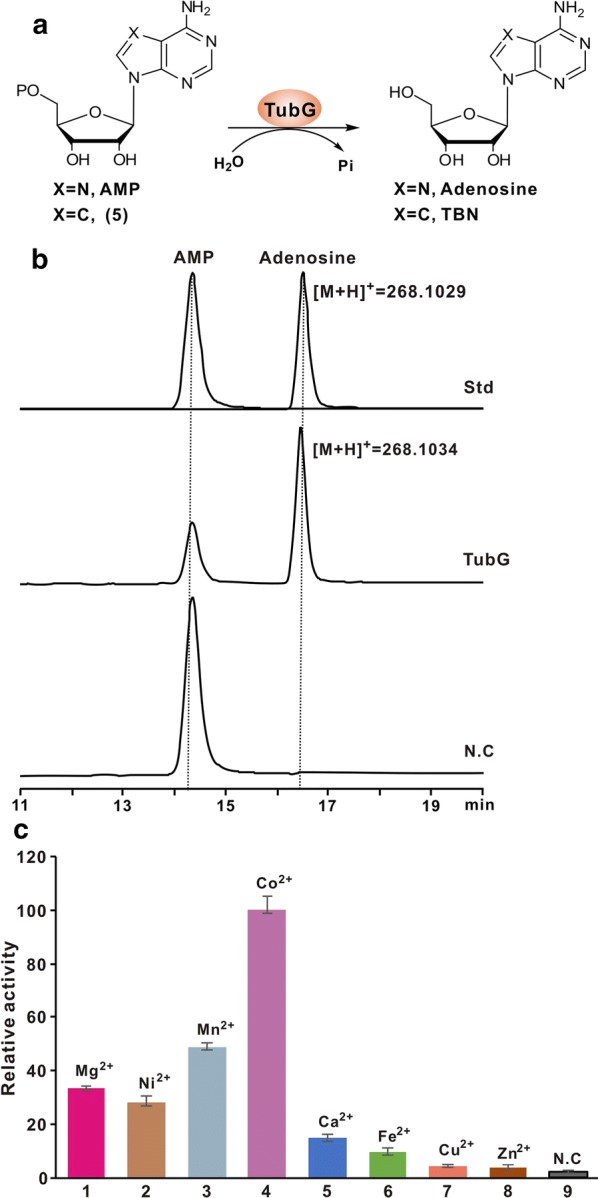
In vitro characterization of TubG as a Nudix hydrolase responsible for the final hydrolysis step. a Schematic of the TubG-catalyzed reaction using the analog of 5, AMP, as substrate. b HPLC traces of the TubG reaction. Std, the authentic standards of AMP and adenosine; TubG, HPLC traces of the TubG reaction with AMP as substrate; N.C, EIC analysis of the reaction with AMP as substrate added but without TubG added. c Relative activity of TubG with AMP as substrate and different divalent cations as metallic cofactor
Discussion
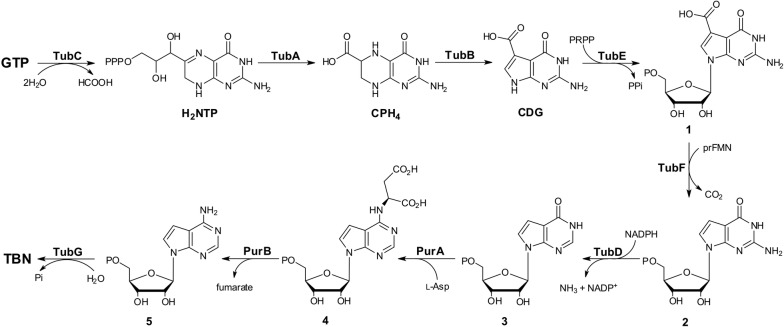
Previous labeling and biosynthetic studies revealed that GTP is the direct substrate for the biosynthesis of the 7-deazapurine core in toyocamycin/sangivamycin, and a three-enzyme cascade, including GCH I (GTP cyclohydrolase I), CPH4 synthase (6-carboxyl-5,6,7,8-tetrahydropterin synthase), and CDG synthase (7-carboxy-7-deazaguanine synthase), accomplishes the biosynthesis of the 7-deazapurine core [10]. In the present study, this mechanism is also shown to be suitable for the building of TBN scaffold. Three enzymes, TubC (GCH I), TubA (CPH4 synthase), and TubB (CDG synthase), are found in the biosynthetic pathway to TBN. TubC is proposed to govern the initial step, catalyzing GTP transformation to H2NTP, which is subsequently converted to CPH4 by TubA (Fig. 6). TubB (CDG synthase) is proposed to employ a radical and Mg2+ dependent strategy to realize the radical-mediated ring contraction step in the course of CDG biosynthesis (Fig. 6).
Fig. 6.
Proposed biosynthetic pathway to TBN. PurA, adenylosuccinate synthetase; PurB, adenylosuccinate lyase
Earlier bioinformatic analysis speculated that the 7-deazapurine core (7-cyano-7-deazaguanine/PreQ0) is biosynthesized preceding its assembly [2, 10]. In the present study, it seems to be logic and feasible in the TBN pathway. Two enzymes, TubE (PRPP transferase), and TubF (UbiD family decarboxylase), are demonstrated to be involved in the described enzymatic steps (Fig. 6). We initially deduced that CDG is likely to undergo decarboxylation prior to its condensation with PRPP for the accomplishment of assembly step, while TubE is not able to accept 7-deazaguanosine as the substrate. We therefore propose that CDG is first converted by TubE to form compound 1, which is then decarboxylated to compound 2 by TubF, an unusual decarboxylase likely using prenylated-FMN (prFMN) as cofactor (Fig. 6).
For the tailoring steps during TBN biosynthesis, TubD (NADPH-dependent reductase) and TubG (Nudix hydrolase), which were characterized in the present study using natural substrate analogs, are established to participate in the enzymatic reactions, compound 2 (GMP analog) is converted to 3 (IMP analog). We could imagine that these two enzymes definitely show the preference to the natural substrates, however, it is also commonly acceptable to explore the catalytic mechanisms of certain enzymes with substrate analogs [22]. Indeed, enzymes are often rationally evolved to accept artificial substrate analogs for industrial biocatalysis purposes [22]. It is very interesting to ask why the enzymes for the sequential transformation of 3 to 4, and to 5 are missing in the TBN pathway. Compound 3 is structurally similar to IMP; as a result, it is reasonable to propose that the missing two enzymes, adenylosuccinate synthetase (PurA), and adenylosuccinate lyase (PurB), could be certainly “borrowed” from the primary purine metabolic pathway (Fig. 6). As for compound 5, it is confirmed to be hydrolyzed by TubG with removal of a phosphate for the accomplishment of TBN biosynthesis (Fig. 6).
There are two other enzymes, TubH (kinase), and TubI (MFS transporter), also present in the TBN pathway. It is more reasonable to propose that TubI is responsible for transporting TBN out of the cell once synthesized. With respect to TubH, the assignment of its functional role in TBN biosynthesis is highly challenging. Previous studies showed that TBN is toxic to M. tuberculosis, and accordingly, it is acceptable to hypothesize that this compound is also potentially toxic to the host cell. Microbes have developed several intricate strategies for the self-resistance of its secondary metabolites during the long-term evolution, undoubtedly, targeted modification (e.g. phosphorylation) of the target antibiotic should be a more effective and convenient tactic, and a similar case for antibiotic self-resistance by host cell has been previously reported in capuramycin biosynthesis [23]. From this perspective, we tentatively propose that TubH is likely to play a self-resistance role by phosphorylating TBN to relieve its potential toxicity to host cells, and related research is now underway.
Notably, the advent of rapid and affordable next-generation DNA sequencing has revolutionized and accelerated the traditional programs for the discovery of chemical diversities [24]. Therefore, TBN could be used as a promising template for the target-directed genome mining of related antibiotics, and we have already been rewarded with uncovering several potential TBN-related antibiotics pathways from the currently-available reservoir of the microbial genomes (Additional file 1: Figure S10). As a consequence, it would be of great interest in the future to hunt for novel 7-deazapurine-containing antibiotics by targeted genome mining approach.
Conclusion
In summary, we report the discovery and functional analysis of the TBN biosynthetic pathway. We have determined that TBN biosynthesis employs a PRPP dependent strategy for the deazanucleoside scaffold assembly, and we have also provided the biochemical evidence that TubD and TubG are involved in the tailoring reduction and phosphohydrolysis steps. We anticipate that the deciphering of the biosynthetic puzzle of TBN will open the way for future combinatorial biosynthesis of this family of antibiotics for the rational generation of novel analogs with enhanced features.
Methods
Materials and general methods
Strains, plasmids used in this study are described in Additional file 1: Table S2, and PCR primers are listed in Additional file 1: Table S3. All of the enzymes (except for the DNA polymerase) were the products of New England Biolabs. The TBN standard was purchased from Medchem Express (MCE). Chemicals were purchased from Sigma-Aldrich, Thermo Scientific, J&K Scientific, or Sinopharm unless otherwise indicated. Standard protocols were employed to manipulate E. coli or Streptomyces according to those of Green et al. [25] or Kieser et al. [26].
Sequencing analysis of the genome of S. tubercidicus
Genomic DNA of S. tubercidicus was isolated on the basis of the standard protocol [26], and the genome sequencing was performed using the Illumina Hiseq4000 sequencing system, and the assembled sequence data was then annotated using the Glimmer 3.02 software. The online programs FramePlot 4.0beta (http://nocardia.nih.go.jp/fp4/) and 2ndFind (http://biosyn.nih.go.jp/2ndfind/) were utilized for the accurate analysis of the TBN gene cluster.
Accession numbers
The DNA sequence of the tub gene cluster is deposited in the GenBank database under accession number MG706975.
Genomic library construction and screening for S. tubercidicus
For the construction of pJTU2463b-derived genomic library for S. tubercidicus, standard method was adopted, using the EPI300-T1R as suitable host cells, and a narrow-down PCR screening strategy [27] with primers TubidF/R were employed to screen the positive cosmid 12G4 from the genomic library.
Fermentation of related Streptomyces strains for TBN production
For production of TBN, Streptomyces strains were inoculated in TSB medium and cultivated for 2 days, after that, the cultures (2%, V/V) were transferred to fermentation medium (including 20 g glucose, 30 g soluble starch, 10 g corn steep liquor, 10 g soybean meal, 5 g peptone, 2 g NaCl, 5 g CaCO3, pH 7.0) and fermented (180 r/min, 28 °C) for 5 days. Subsequently, the fermentation beer was processed (adding oxalic acid till pH 3.0) for LC–MS analysis.
Overexpression and purification of TubE, TubD, and TubG in E. coli Rosetta (DE3)/pLysS
For overexpression of these proteins, their structural genes were amplified by KOD-plus DNA polymerase (TOYOBO) using related primers listed in Additional file 1: Table S3. Then the NdeI–EcoRI engineered DNA fragments were individually cloned into the counterpart sites of pET28a. After confirmation by sequencing, the related constructs were subsequently transformed into E. coli Rosetta (DE3)/pLysS cells according to the standard protocols [25]. Expression and purification for the His6-tagged proteins were conducted according to the method by Wu et al. [28].
Enzymatic assays of TubE
For TubE activity, the reaction mixture (100 mM PBS buffer, pH 7.5; 1 mM CDG; 2 mM PRPP; 5 mM MgCl2; 1 mM DTT and 20 μg protein) was incubated at 30 °C for 8 h, and then terminated by the immediate addition of an equivalent volume methanol. After centrifugation to remove protein, the supernatant was filtrated by 0.22 μm filter. HPLC (Shimadzu LC-20A) analysis was performed on a reverse phase C18 column (Inertsil ODS-3, 4.6 × 250 mm, 5 µm) with the elution gradient of 5%–30% methanol:0.15% TFA over 35 min at a flow rate of 0.5 ml/min, and the elution was monitored at UV295 nm by a DAD detector.
Enzymatic assays of TubD
For TubD activity, the reaction containing 100 mM Tris–HCl buffer (pH 7.5), 1 mM GMP, 1 mM NADPH/NADH, 1 mM DTT, 50 mM KCl, 2 mM EDTA, and 20 μg protein, incubated at 30 °C for 4 h, then the supernatant was analyzed by LC–HRMS with the elution gradient of 5%–25% methanol:0.15% TFA over 20 min at a flow rate of 0.4 ml/min.
Enzymatic assays of TubG
The complete TubG reaction including 100 mM Tris–HCl buffer (pH 7.0), 1 mM AMP, 100 mM KCl, 5 mM divalent ion (Mg2+, Ni2+, Mn2+, Co2+, Ca2+, Fe2+, Cu2+, and Zn2+), and 20 μg protein, was incubated at 30 °C for 4 h. The protein in reaction was removed by adding an equal volume of methanol, then the supernatant was analyzed by HPLC (Shimadzu LC-20A) equipped with C-18 reversed-phase column (Inertsil ODS-3, 4.6 × 250 mm, 5 µm) with 5%–30% methanol:0.15% TFA over 15 min at a flow rate of 0.4 ml/min, the condition (30% methanol:0.15% TFA) was maintained for another 10 min.
The conditions for LC–MS analysis
LC–MS analysis was carried out on a Thermo Fisher Scientific ESI-LTQ Orbitrap (Scientific Inc.) equipped with a C-18 reversed-phase column (Inertsil ODS-3, 4.6 × 250 mm, 5 µm) in positive mode with an elution gradient of 5%–30% Methanol:0.15% TFA over 30 min at 0.5 ml/min, and the parameters for the LC–MS analysis are as follows: Dry gas at 275 °C, 10 l/ml, and nebulizer pressure of 30 psi.
Additional file
Additional file 1: Figure S1. LC-MS analysis of the target metabolite produced by S. coelicolor M1154/12G4. Figure S2. Bioinformatic analysis of the phosphoribosyltransferase TubE with its homologs. Figure S3. Biochemical characterization of TubE reaction. Figure S4. 1H and 13C NMR analysis of the compound 1. Figure S5. 1H-1H cosy, HMBC, and HSQC spectrums of the compound 1. Figure S6. Bioinformatic analysis of the GMP reductase TubD with its homologs. Figure S7. Biochemical characterization of TubD-catalyzed reaction. Figure S8. Bioinformatics analysis of the Nudix superfamily hydrolase TubG. Figure S9. Biochemical analysis of TubG and its reaction. Figure S10. Target-directed genome mining of the gene clusters for the potential TBN-related antibiotics. Table S1. Parameters for the genome sequencing and assembly of S. tubercidicus. Table S2. Strains, plasmids and cosmids used in this study. Table S3. PCR primers used in this study.
Authors’ contributions
YL, RG, XL, and PZ carried out experiments, QZ and YSC analyzed the primary data. YL, JW, and WC wrote the manuscript. WC and JW conceived the project and supervised the research. MW and ZD help with the critical reading of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We are very grateful to Prof. Guo-Qiang Chen (Tsinghua University) and Prof. Lili Zhang (Tarim University) for kindly providing us with the strains containing the potential TBN biosynthetic gene cluster.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset(s) supporting the conclusions of this article is (are) included within the article [and its additional file(s)].
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by grants of the National Natural Science Foundation of China (31770041).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yan Liu, Email: liuyan_hb@163.com.
Rong Gong, Email: gongrong@whu.edu.cn.
Xiaoqin Liu, Email: liuxiaoqin@whu.edu.cn.
Peichao Zhang, Email: zhangpeichao@stu.ouc.edu.cn.
Qi Zhang, Email: 151847114@qq.com.
You-Sheng Cai, Email: cysh2002@163.com.
Zixin Deng, Email: zxdeng@sjtu.edu.cn.
Margit Winkler, Email: margit.winkler@tugraz.at.
Jianguo Wu, Email: jwu@whu.edu.cn.
Wenqing Chen, Email: wqchen@whu.edu.cn.
References
- 1.Isono K. Nucleoside antibiotics: structure, biological activity, and biosynthesis. J Antibiot (Tokyo) 1988;41:1711–1739. doi: 10.7164/antibiotics.41.1711. [DOI] [PubMed] [Google Scholar]
- 2.McCarty RM, Bandarian V. Biosynthesis of pyrrolopyrimidines. Bioorg Chem. 2012;43:15–25. doi: 10.1016/j.bioorg.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anzai K, Nakamura G, Suzuki S. A new antibiotic, tubercidin. J Antibiot (Tokyo) 1957;10:201–204. [PubMed] [Google Scholar]
- 4.Mizuno Y, Ikehara M, Watanabe K, Suzaki S. Structural elucidation of tubercidin. Chem Pharm Bull (Tokyo) 1963;11:1091–1094. doi: 10.1248/cpb.11.1091. [DOI] [PubMed] [Google Scholar]
- 5.Bloch A, Mihich E, Leonard RJ, Nichol CA. Studies on the biologic activity and mode of action of 7-deazainosine. Cancer Res. 1969;29:110–115. [PubMed] [Google Scholar]
- 6.Lindberg B, Klenow H, Hansen K. Some properties of partially purified mammalian adenosine kinase. J Biol Chem. 1967;242:350–356. [PubMed] [Google Scholar]
- 7.Drew ME, Morris JC, Wang Z, Wells L, Sanchez M, Landfear SM, Englund PT. The adenosine analog tubercidin inhibits glycolysis in Trypanosoma brucei as revealed by an RNA interference library. J Biol Chem. 2003;278:46596–46600. doi: 10.1074/jbc.M309320200. [DOI] [PubMed] [Google Scholar]
- 8.Smulson ME, Suhadolnik RJ. The biosynthesis of the 7-deazaadenine ribonucleoside, tubercidin, by Streptomyces tubercidicus. J Biol Chem. 1967;242:2872–2876. [PubMed] [Google Scholar]
- 9.Yoo JC, Han JM, Ko OH, Bang HJ. Purification and characterization of GTP cyclohydrolase I from Streptomyces tubercidicus, a producer of tubercidin. Arch Pharm Res. 1998;21:692–697. doi: 10.1007/BF02976759. [DOI] [PubMed] [Google Scholar]
- 10.McCarty RM, Bandarian V. Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem Biol. 2008;15:790–798. doi: 10.1016/j.chembiol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowling DP, Bruender NA, Young AP, McCarty RM, Bandarian V, Drennan CL. Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nat Chem Biol. 2014;10:106–112. doi: 10.1038/nchembio.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandarian V, Drennan CL. Radical-mediated ring contraction in the biosynthesis of 7-deazapurines. Curr Opin Struct Biol. 2015;35:116–124. doi: 10.1016/j.sbi.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelp MT, Bandarian V. A single enzyme transforms a carboxylic acid into a nitrile through an amide intermediate. Angew Chem Int Ed Engl. 2015;54:10627–10629. doi: 10.1002/anie.201504505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkler M, Dokulil K, Weber H, Pavkov-Keller T, Wilding B. The nitrile-forming enzyme 7-cyano-7-deazaguanine synthase from Geobacillus kaustophilus: a reverse nitrilase? ChemBioChem. 2015;16:2373–2378. doi: 10.1002/cbic.201500335. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall SA, Payne KAP, Leys D. The UbiX-UbiD system: the biosynthesis and use of prenylated flavin (prFMN) Arch Biochem Biophys. 2017;632:209–221. doi: 10.1016/j.abb.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 17.White MD, Payne KA, Fisher K, Marshall SA, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Rigby SE, Scrutton NS, et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature. 2015;522:502–506. doi: 10.1038/nature14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payne KA, White MD, Fisher K, Khara B, Bailey SS, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Beveridge R, et al. New cofactor supports alpha, beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature. 2015;522:497–501. doi: 10.1038/nature14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin F, Ferguson KL, Boyer DR, Lin XN, Marsh EN. Isofunctional enzymes PAD1 and UbiX catalyze formation of a novel cofactor required by ferulic acid decarboxylase and 4-hydroxy-3-polyprenylbenzoic acid decarboxylase. ACS Chem Biol. 2015;10:1137–1144. doi: 10.1021/cb5008103. [DOI] [PubMed] [Google Scholar]
- 20.Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 21.Gao Y, Xu G, Wu P, Liu J, Cai YS, Deng Z, Chen W. Biosynthesis of 2′-chloropentostatin and 2′-amino-2′-deoxyadenosine highlights a single gene cluster responsible for two independent pathways in Actinomadura sp. Strain ATCC 39365. Appl Environ Microbiol. 2017 doi: 10.1128/AEM.00078-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler M. Carboxylic acid reductase enzymes (CARs) Curr Opin Chem Biol. 2018;43:23–29. doi: 10.1016/j.cbpa.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z, Funabashi M, Nonaka K, Hosobuchi M, Shibata T, Pahari P, Van Lanen SG. Functional and kinetic analysis of the phosphotransferase CapP conferring selective self-resistance to capuramycin antibiotics. J Biol Chem. 2010;285:12899–12905. doi: 10.1074/jbc.M110.104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachmann BO, Van Lanen SG, Baltz RH. Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol. 2014;41:175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green MR, Sambrook J. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 26.Kieser T, Bibb MJ, Chater KF, Butter MJ, Hopwood DA. Practical streptomyces genetics. 2. Norwich: John Innes Foundation; 2000. [Google Scholar]
- 27.Cheng L, Chen W, Zhai L, Xu D, Huang T, Lin S, Zhou X, Deng Z. Identification of the gene cluster involved in muraymycin biosynthesis from Streptomyces sp. NRRL 30471. Mol BioSyst. 2011;7:920–927. doi: 10.1039/C0MB00237B. [DOI] [PubMed] [Google Scholar]
- 28.Wu P, Wan D, Xu G, Wang G, Ma H, Wang T, Gao Y, Qi J, Chen X, Zhu J, et al. An unusual protector-protégé strategy for the biosynthesis of purine nucleoside antibiotics. Cell Chem Biol. 2017;24:171–181. doi: 10.1016/j.chembiol.2016.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. LC-MS analysis of the target metabolite produced by S. coelicolor M1154/12G4. Figure S2. Bioinformatic analysis of the phosphoribosyltransferase TubE with its homologs. Figure S3. Biochemical characterization of TubE reaction. Figure S4. 1H and 13C NMR analysis of the compound 1. Figure S5. 1H-1H cosy, HMBC, and HSQC spectrums of the compound 1. Figure S6. Bioinformatic analysis of the GMP reductase TubD with its homologs. Figure S7. Biochemical characterization of TubD-catalyzed reaction. Figure S8. Bioinformatics analysis of the Nudix superfamily hydrolase TubG. Figure S9. Biochemical analysis of TubG and its reaction. Figure S10. Target-directed genome mining of the gene clusters for the potential TBN-related antibiotics. Table S1. Parameters for the genome sequencing and assembly of S. tubercidicus. Table S2. Strains, plasmids and cosmids used in this study. Table S3. PCR primers used in this study.
Data Availability Statement
The dataset(s) supporting the conclusions of this article is (are) included within the article [and its additional file(s)].