Abstract
A directed evolution approach was used to select for Adeno-associated virus (AAV) capsids that would exhibit more tropism toward an HIV-1 producer T cell line with the long-term goal of developing improved gene transfer vectors. A library of AAV variants was used to infect H9 T cells previously infected or uninfected by HIV-1 followed by AAV amplification with wild-type adenovirus. Six rounds of biological selection were performed, including negative selection and diversification after round three. The H9 T cells were successfully infected with all three wild-type viruses (AAV, adenovirus, and HIV-1). Four AAV cap mutants best representing the small number of variants emerging after six rounds of selection were chosen for further study. These mutant capsids were used to package an AAV vector and subsequently used to infect H9 cells that were previously infected or uninfected by HIV-1. A quantitative polymerase chain reaction assay was performed to measure cell-associated AAV genomes. Two of the four cap mutants showed a significant increase in the amount of cell-associated genomes as compared to wild-type AAV2. This study shows that directed evolution can be performed successfully to select for mutants with improved tropism for a T cell line in the presence of HIV-1.
Keywords: directed evolution, AAV, gene transfer, T cell, HIV-1
1. Introduction
Adeno-associated virus (AAV) is the third most popular gene transfer vector used today ranking just after adenovirus and retrovirus (Edelstein, 2017). Despite some recent controversy regarding its role in cancer (Nault et al., 2015; Park et al., 2016; Srivastava and Carter, 2017), AAV is generally considered to be a nonpathogenic virus. As of April 2017, AAV has been used in 183 clinical trials (Edelstein, 2017) with no reported cancers. AAV exhibits broad tropism, persistence, high transduction capability, lack of superinfection immunity, and high stability. It may be grown and purified to high titers, and it has the ability to infect dividing or nondividing cells (Daya and Berns, 2008). These characteristics make AAV an appealing choice for gene transfer applications. Recombinant AAVs are being investigated as vectors for clinical gene transfer to a wide variety of cells and tissues (Hasbrouck and High, 2008; Kota et al., 2009; Maguire et al., 2009; Maguire et al., 2010; Muzyczka, 1992; Srivastava, 2008; Wagner et al., 1999). Transduction with AAV vectors has been shown to result in long-term transgene expression in vivo in several cell types including skeletal muscle, photoreceptors, liver, and neuronal cells (Daya and Berns, 2008; Hasbrouck and High, 2008; Liu et al., 2007). As a result, AAV was used in the first regulatory approval of a gene therapy product in Western nations, in 2012 within the European Union.
AAV is a member of the Parvoviridae family. As a member of the Dependoparvovirus genus, this virus requires the help of another virus in order to replicate. Viruses known to help AAV include adenovirus, herpesvirus, and poxvirus. In the absence of a helper virus, AAV cannot fully replicate, but it can infect cells and deliver a foreign gene of interest (a transgene), which is the desired function of a viral vector. The wild-type virus is a 4.7 kb single-stranded DNA virus that contains two genes: rep, which encodes four proteins required for genome replication; and cap, which encodes four proteins required for capsid formation, namely VP1, VP2, VP3, and assembly activating protein (Sonntag, Schmidt, and Kleinschmidt, 2010; Trempe and Carter, 1988).
AAV infection efficiency on human lymphocytes and their precursors has been examined (Gardner et al., 1997; Horster et al., 1999; Mendelson et al., 1992; Ponnazhagan et al., 1997; Schuhmann et al., 2010; Song et al., 2013a; Song et al., 2013b; Srivastava, 2008; Veldwijk et al., 2010; Zhang and Fuleihan, 1999). AAV2 transduction of primary human T lymphocytes showed significant donor variability with efficiencies ranging from less than 1% to over 50% (Horster et al., 1999; Ponnazhagan et al., 1997). A recent survey on transduction efficiencies of nine naturally occurring AAV serotypes plus one engineered type showed the overall efficiency on immortalized Jurkat T cells (similar to the H9 T cells used in the current study) was 2% or less for 8 of the 10 serotypes. Only AAV1 and AAV6 scored above 2% with efficiencies of only 29% and 20%, respectively, and very high multiplicities of infection (100,000) were required to achieve these efficiencies (Ellis et al., 2013).
With regard to primary stem cells, serotypes 1-10 of AAV were shown to transduce CD34+ hematopoietic stem cells with efficiencies up to 26% (Maguire et al., 2010; Schuhmann et al., 2010; Song et al., 2013a; Song et al., 2013b; Veldwijk et al., 2010; Wang et al., 2015). An enrichment scheme involving fluorescence-activated cell sorting or magnetic beads was used to achieve greater than 85% of targeted gene integration for AAV6 in CD34+ hematopoietic stem cells, as compared with 4-10% efficiency without enrichment (Dever et al., 2016). While this new discovery is promising, ex vivo enrichment may not be feasible for many gene transfer applications, and significantly improved AAV vectors are still needed to meet the required therapeutic index for direct in vivo applications.
Since the AAV capsid is a primary determinant of transduction, altering and engineering the capsid could conceivably overcome many of the existing transduction and targeting issues, including binding, entry, endosomal escape, and trafficking (Buning et al., 2008; Michelfelder and Trepel, 2009). However, due to the molecular and cellular complexity of each step, rational design of improved AAV-based therapeutics is challenging. For instance, several investigators have attempted to re-target the AAV capsid through the insertion of candidate receptor binding peptides (Perabo et al., 2003; Perabo et al., 2006) and peptides derived from phage libraries (Muller et al., 2003). Viral capsids have also been modified with antibodies (Bartlett et al., 1999) or fusion proteins (Ponnazhagan et al., 2002). Although these approaches have enjoyed some success in re-targeting AAV, they can be accompanied by reductions in titer, and these approaches do not address pre- or post-binding barriers to infection.
Another way to re-target AAV would be to select for mutant capsids that are better able to recognize specific cell surface changes that occur under certain disease conditions. In the case of HIV-1 infection, specific cell surface changes are known to occur upon HIV-1 infection. These surface changes could be exploited to develop a targeted gene therapy vector carrying an anti-HIV-1 payload. For example, the viral envelope protein gp120 is expressed on infected cell membranes, while the viral receptors CD4, CXCR4, and CCR5 are down-regulated (Chenine, Sattentau, and Moulard, 2000; Choi et al., 2008; Hoxie et al., 1986). Major histocompatibility complex class I and II molecules and several cluster of differentiation (CD) antigens, including CD2, CD3, CD5, and CD8 (Meerloo et al., 1992; Stevenson, Zhang, and Volsky, 1987), are also down-regulated. Up-regulation of several CD antigens has also been observed, including CD11a, CD54, and CD63 (Meerloo et al., 1992; Meerloo et al., 1993). These cell surface changes were documented in HIV-1-infected H9 cells, the target cell of the current study (Meerloo et al., 1992; Meerloo et al., 1993).
In general, proteomic analysis of plasma membrane proteins of T cell lines latently infected with HIV-1 revealed the presence of 17 different proteins not found on uninfected cells. Of these, 47% were integral membrane proteins, 18% were membrane-associated proteins, and 35% were cytoplasmic- or organelle-associated proteins (Berro et al., 2007). In addition to the novel proteins, surface glycosylation changes have also been documented in HIV-1-infected T cells, including decreases in sialylation, increases in core 2 O-glycans, and elongation of lactosamine sequence (Lanteri et al., 2003). These cell surface changes, including the expression of HIV-1 gp120 protein on the surface of the cells, could be exploited for targeting AAV to HIV-1-infected cells for research or clinical purposes.
HIV-1 proteins have been used to target cells infected by HIV-1 itself. Rabies virus, Vesicular stomatitis virus, and Murine leukemia virus were all pseudotyped with CD4 and CXCR4 proteins and shown to bind successfully to the gp120 protein on the surface of HIV-1-infected cells (Mebatsion et al., 1997; Schnell et al., 1997; Somia et al., 2000). Cell killing and reduction of HIV-1 production was demonstrated in the Vesicular stomatitis virus system by taking advantage of the natural lytic ability of the virus (Schnell et al., 1997).
The current paper describes the application of a directed evolution approach to select for AAVs that are better able to target HIV-1-infected or uninfected H9 T cells. Reported here is the successful use of this new method to isolate novel AAV capsids.
2. Methods
2.1 Cells
The H9 T cell line, hereafter referred to as H9 cells, was derived from a single cell clone of a specific HUT 78 cell line, HT (Mann et al., 1989; Popovic, Read-Connole, and Gallo, 1984a; Popovic et al., 1984b). HUT 78 cells were derived originally from the peripheral blood of a patient with Sezary syndrome (Gazdar et al., 1980). The H9 cell line was obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH: H9 from R. Gallo). The 293T cell line is a highly transfectable derivative of the human embryonic kidney cell line, 293 (DuBridge et al., 1987; Pear et al., 1993) and was obtained from the American Type Culture Collection (Manassas, VA). The 293T cell line expresses the SV40 T antigen and contains an insertion of the E1A and E1B adenovirus sequences into chromosome 19 at 19q13.2 (Kovesdi and Hedley, 2010).
The H9 cell line was maintained in RPMI 1640 medium supplemented with 8% heat-inactivated fetal calf serum, and the 293T cell line was maintained in Dulbecco’s Modified Eagle’s Medium with 10% heat-inactivated fetal calf serum. Both cell lines were also supplemented with 2mM L-glutamine, penicillin (100 U/ml), streptomycin (100 mg/ml). Cell cultures were split at ratios of 1:9 (H9) and 1:8 (293T) twice per week and grown at 37°C and 5% CO2. The medium, serum, and cell culture supplements were obtained from ThermoFisher.
Cell viabilities were determined using 0.4% trypan blue solution to measure the number of cells capable of excluding the dye.
A chronically infected H9 cell line was established that maintained long-term viability and expression of HIV-1. The H9 cells were chosen because they were pivotal to the discovery of HIV-1, have been used as producer line for HIV-1 ever since its discovery, and have been very well characterized over the past decades. Cells were infected with HIV-1 at a multiplicity of infection (MOI) of 500. After an initial crisis phase of cytopathic effects and cell death, a stable cell line emerged by the eighth passage with a viability of approximately 90%, similar to the parental H9 line. One hundred percent of the cells in this cell line, designated H9-HIV, were HIV-1-positive and produced over 100 ng/ml of HIV-1 p24 protein on a continual basis but otherwise appeared similar to the uninfected H9 cells as viewed under the phase contrast microscope. A seed stock of H9-HIV cells was frozen in liquid nitrogen, and thawed cells were used between passages 5 and 20 for all experiments. A total of 2.5 × 105 total cells plated at a density of 5 × 105 cells/ml in 48-well plates were used for the biological selection experiments. H9 and H9-HIV cells plated under these conditions showed a doubling time of 40 h.
2.2 Virus
The T-tropic HIV-1 strain HTLV-IIIB/H9 was derived from the peripheral blood of a patient with AIDS (Popovic et al., 1984a; Popovic et al., 1984b) and was obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH: HTLV-IIIB/H9 from R. Gallo). The stock of HIV-1 was prepared from infected H9 cells on day 10 post-infection. On the day prior to stock preparation, infected culture medium was completely changed, such that virus stocks contained freshly made virions. Infected cells were centrifuged at 1430 x g for 10 min in sealed GH 3.8 rotor buckets of a Beckman GS-6R tabletop centrifuge (Fullerton, CA). Culture supernatants were filtered through a 0.22-μm pore size Steriflip-GP filter unit (Millipore, Bedford, MA), and filtered virus stocks were stored in aliquots at liquid nitrogen vapor phase (approximately -150°C). HIV-1 p24 was quantified using the RETROtek HIV-1 p24 Antigen ELISA from Zeptometrix Corporation (Buffalo, NY).
The AAV library consisted of approximately 107 variants generated by a combination of error-prone PCR mutagenesis, peptide insertion, and capsid shuffling (Koerber et al., 2006; Li et al., 2008; Muller et al., 2003). An overrepresentation of AAV2 was present in the library since the mutagenesis and peptide insertions were performed with the AAV2 serotype. The number of variants was determined by quantifying the number of independent bacterial transformants after cloning the original library. Serotypes 1, 2, 4-6, 8, and 9 were chosen for the shuffling because they have been shown to have a diverse range of gene-delivery properties. The AAV library was packaged by using 70 ng of a plasmid encoding the full length AAV genome (psub2cap2) and 25 μg of a plasmid encoding adenovirus helper genes (pHelper).
Wild-type human adenovirus type 5 was obtained from the Viral Vector Core Facility, University of Iowa (Iowa City, IA).
Wild-type AAV2 was prepared by transfecting 293T cells with psub2cap2 and pHelper in a 1:1 ratio.
Recombinant AAVs were made using the HindIII and NotI restriction sites to replace the wild-type AAV2 cap gene with variant cap genes that emerged from the H9 selection. The recombinant AAVs were then produced in 293T cells transfected in a 1:1:1 ratio with plasmids encoding: 1) an AAV vector genome; 2) the AAV rep and cap genes; and 3) a plasmid containing adenovirus helper genes as described previously (Xiao, Li, and Samulski, 1998). The AAVs were purified by iodixanol gradient and were quantified by qPCR to obtain genomic titers.
2.3 Biological selection of AAV
For each of the six rounds of selection, cells were split 24 h prior to use with a complete change of media to ensure cells were in growth phase on the day of infection. Cells were washed three times with PBS pH 7.2. A total of 2.5 × 105 cells were used for the initial infection with the AAV library at a MOI of 104. Subsequent rounds of selection used an MOI of 102. The reduction in MOI points to the overall strategy for the biological selection in which one starts with a large MOI and then increases stringency in later rounds of selection in order to yield higher specificity for the successful AAVs.
For each such round, cells were infected for 1 h at 37°C, washed twice with culture medium (RPMI 1640 medium supplemented with 8% heat-inactivated fetal calf serum, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin), resuspended in 0.5 ml of medium, and placed in 48-well plates to yield a final cell density of 5.0 × 105 cells/ml. Cells were then incubated for 48 h at 37°C and 5% CO2. At 48 h post-infection, cells were infected with wild-type adenovirus 5 at an MOI of 103 to enrich for the population of AAV variants that were able to infect successfully. Cells were again resuspended in 0.5 ml of medium and placed in 48-well plates for an additional 24 h. Total cellular DNA was then isolated at 72 h post-infection using the DNeasy Blood and Tissue Kit.
For the final three rounds, a negative selection step was performed in the protocol. Cells were incubated for one h at 37°C with the opposing cell line, and the supernatant was then used to infect the cell line of choice for an additional one h.
In total, six rounds of biological selection were performed in which AAV variants were used to infect fresh H9 and H9-HIV cells. Total cell DNA was isolated from the infected cells after each of the six rounds, and the cap genes were cloned to create new libraries for subsequent rounds of selection. This process of cloning the cap genes for subsequent rounds of selection was performed to enrich for the population of AAV variants that were able to infect successfully. A small number of clones were chosen for sequencing after each round by the Sanger method of sequencing.
2.4. Quantitative PCR
For viral genome quantification, cells were lysed and total cell DNA was purified using the DNeasy Blood and Tissue kit according to the manufacturer’s instructions (QIAGEN, Valencia, CA). Quantitative PCR (qPCR) was performed using SYBR Green with low ROX (Quanta) in a Stratagene Real Time PCR System (Agilent Technologies) using primers against the AAV vector and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal standard control. The relative expression of target genes was quantified by comparative Ct analysis using Mx4000p V5 software for data analysis. Each individual qPCR reaction was performed in duplicate. Primers used were: AAV-F-5’-ACGTAAACGGCCACAAGTTC-3’ and AAV-R-5’-AAGTCGTGCTGCTTCATGTG-3’; GAPDH-F-5’CACCCTGTTGCTGTAGCCAAA-3’; GAPDH-R-5’-CAACAGCGACACCCACTCCT-3’. The qPCR efficiencies were comparable for both primer sets in the range of -3.0 to -3.4.
2.5 Computer and statistical analyses
All graphs were drawn and all statistical analyses were performed using Sigma plot version 11.0 (Systat Software, Inc., San Jose, CA). Images were processed for figure layout in Photoshop CC 2017 (Adobe Systems, Inc., San Jose, CA). Tests of statistical significance were performed by one-way analysis of variance (ANOVA). Post-hoc testing was performed using the Holm Sidak method. An alpha value of 0.05 was used for all statistical analyses.
3. Results
3.1 Biological selection strategy
A directed evolution approach was implemented to select for AAV variants selective for H9 or H9-HIV cells (Fig. 1). A library of 107 AAV variants with mutagenized cap genes was packaged and used to infect either H9 or H9-HIV cells. For the first three rounds of positive selection (Fig. 1A), H9 and H9-HIV cells were infected with AAV and allowed to grow for three days with help from wild-type adenovirus during the last day. The AAV cap genes were re-cloned and packaged for subsequent infections to enrich the selected virus population for subsequent rounds. Sequence analysis was performed on the cap genes after every selection to assess sequence convergence. After the first three rounds of selection, the sequences appeared to be converging, so the isolated cap genes were subjected to error-prone PCR mutagenesis to diversify the pool, followed by three more rounds of selection that each included a negative selection step (Fig. 1B). For the negative selections, virus was pre-incubated with the “opposite” cell line to remove nonspecific AAVs and then transferred back to the cell line of interest. That is, AAVs that resulted from the three rounds of selection on H9-HIV cells were negatively selected via pre-incubation with H9 cells followed by infection of H9-HIV cells with the resulting supernatant, or vice versa starting with H9-HIV cells.
Figure 1. Selection scheme for AAV variants.
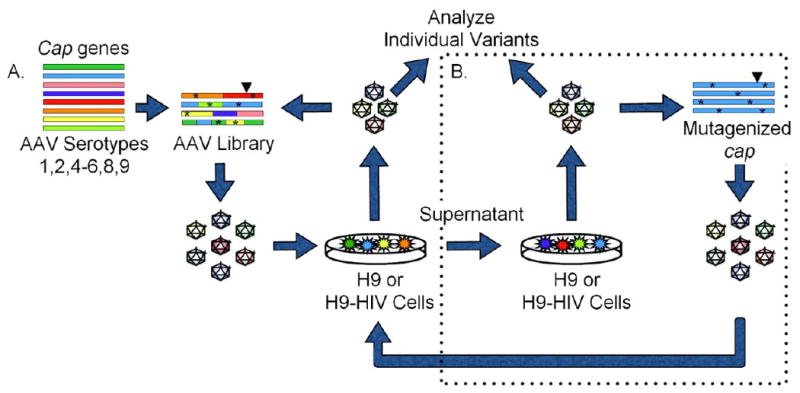
(A) A library of AAV mutants was packaged and used to infect either H9 or HIV-infected H9 cells. Wild-type adenovirus was added as a helper virus to induce replication of AAV that had infected the cells, and the resulting AAV clones were recovered by PCR amplification and cloning their cap genes, which were then re-packaged for two subsequent infections. (B) After three selections, isolated cap genes were subjected to error-prone mutagenesis and three additional rounds of selection were performed including a negative selection step, whereby virus was pre-incubated with the opposite cell line before infecting the cell line of interest. Asterisks (*) represent point mutations; Triangles (▼) represent peptide insertion.
3.2. Cell numbers and viabilities remained high during biological selection
We tracked absolute cell counts and viabilities during each round to ensure that we had adequate numbers of cells throughout our selection process. At both the 48 and 72 h time points, absolute cell counts averaged over 8 × 105 total cells with greater than 90% viability at 48 h and greater than 80% at 72 h. The 72 h time point showed slightly less viability as compared to the 48 h time point due to the wild-type adenovirus infection over the final 24 h of the experiment. The addition of wild-type adenovirus did not significantly affect cell viability at 24 h (p=0.319) but did reduce viability by 72 h (p<0.0001), as determined across multiple rounds by one-way ANOVA. All rounds of infection were performed similarly, and representative data are shown in Fig. 2A-D. As expected, the doubly and triply infected cultures showed lower cell counts and percent viabilities as compared to uninfected controls. At both time points, the H9-HIV cells produced HIV-1 (as measured by p24 production) in amounts equal to uninfected (mock) cells, despite being infected with two additional viruses (Fig. 2E and F). Thus, AAV and adenovirus did not inhibit HIV-1 production.
Figure 2. Cell counts, viabilities, and p24 production at 48 and 72 h post infection with AAV.
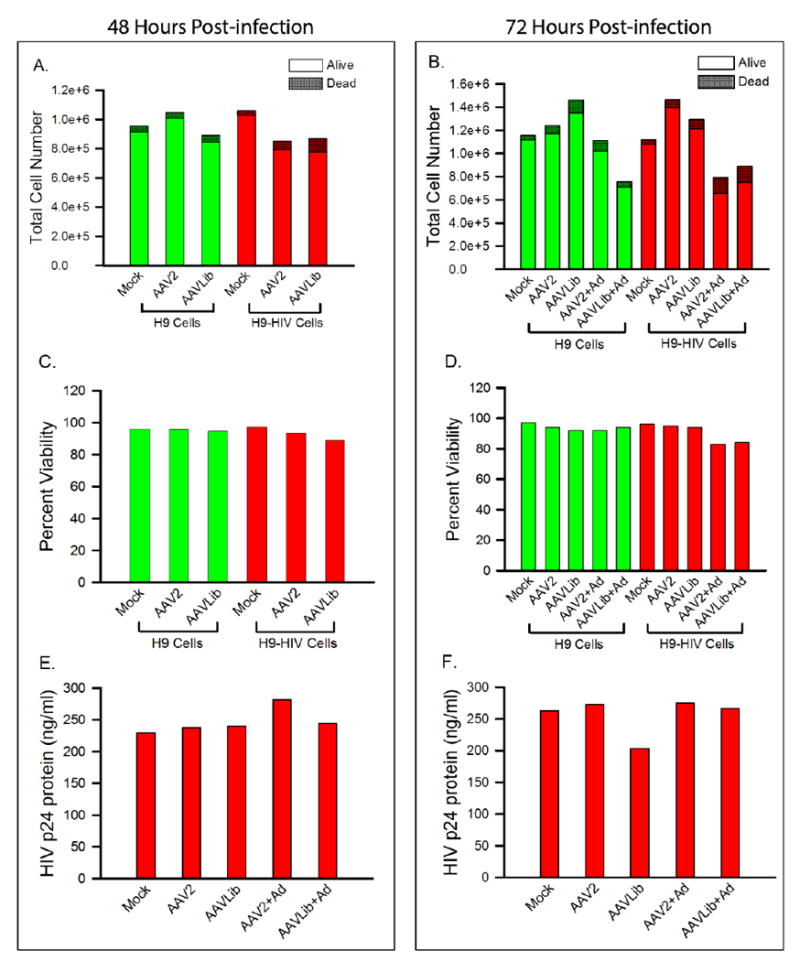
(A) Absolute cell counts at 48 h post-infection and (B) absolute cell counts at 72 h post-infection. (C) Viabilities at 48 h post-infection and (D) viabilities at 72 h post-infection. (E) HIV p24 production at 48 h post-infection and (F) HIV p24 production at 72 h post-infection. Mock: cells without virus; AAVLib: AAV library of variants; Ad: wild-type adenovirus.
3.3 Novel AAV variants emerged after six rounds of selection
After six rounds of selection as described above, including three rounds of negative selection at the end, a small number of AAV variants emerged that were all based on the AAV2 backbone. The variants were found to infect both H9 and H9-HIV cells. Four variants representative of the mutations and peptide insertions present after 6 rounds of selection were chosen for further study (Fig. 3). One of the variants, H9-5C (green horizontal bar), emerged from selection on H9 cells, while the other three variants, HIV-2B, 2C, and 4C (red horizontal bars), arose from selection on H9-HIV cells. The H9-5C, HIV-2B, and HIV-4C variants all contained distinct 10-mer peptide insertions in a key loop on the exterior capsid between amino acids 587 and 588, a position present in all three wild-type capsid proteins (VP1, VP2, and VP3; blue horizontal bars). In addition, H9-5C and HIV-4C both had a conservative V708I mutation. Variants HIV-2B and HIV-2C shared a non-conservative E67A mutation, while variants HIV-2C and HIV-4C shared a conservative S207G change. Finally, variants HIV-2B, HIV-2C, and HIV-4C each contained a different, conservative, single point mutation within the VP3 region (I698V, Q598L, and I648V, respectively) (Fig. 3; numbering based on the parent AAV2 VP1 sequence without the peptide insertion).
Figure 3. Selected AAV variants after six rounds of selection.
Wild-type AAV encodes three viral capsid proteins, (VP1, VP2, and VP3; blue), in the stoichiometric ratio of 1:1:10 through the use of alternative splice sites and nonconventional translational initiation codons. After six rounds of selection, the AAV library converged to a small number of sequences. One predominant sequence for H9 cells (green) and three predominant sequences for H9-HIV cells (red) are shown with percentages indicating prevalence within the population. The majority of sequence changes are located in the region encompassing all three of the viral capsid proteins. Triangles (▲) represent locations of point mutations or peptide insertions. Letters represent amino acid code. The letter before the number represents the wild-type amino acid, while the letter after the number represents the new amino acid. Peptides were inserted between amino acids 587 and 588 of the wild-type AAV 2 capsid protein. The first two and last amino acids were invariant amongst the peptides due to restriction site sequence. The figure is drawn approximately to scale.
3.4 Selected AAV variants showed increased cell-association by qPCR analysis
Variant capsids were packaged with an AAV vector and used to infect H9 and H9-HIV cells. Since the mutants that emerged from selection were based on the AAV2 serotype, this serotype was included in the qPCR experiment as a control. Compared to AAV2, variant HIV-2B showed a 9-fold increase in the relative number of cell-associated AAV genomes after infection of H9 (Fig. 4A) and a 4-fold increase after infection of H9-HIV cells (Fig. 4B). Upon infection of H9-HIV cells, variant HIV-4C showed a 3-fold increase in the number of genomes as compared to AAV2 (Fig. 4B). These results were statistically significant by one-way ANOVA, F(5, 18) = 28.573, p < 0.001 (for H9 cells) and F(5, 18) = 48.695, p < 0.001 (for H9-HIV cells). Post-hoc testing revealed statistically significant increases in cell-associated viral DNA for HIV-2B on both H9 and H9-HIV cells (p < 0.001) and for HIV-4C on H9-HIV cells (p < 0.001).
Figure 4. Number of cell-associated genomes for selected AAV variants.
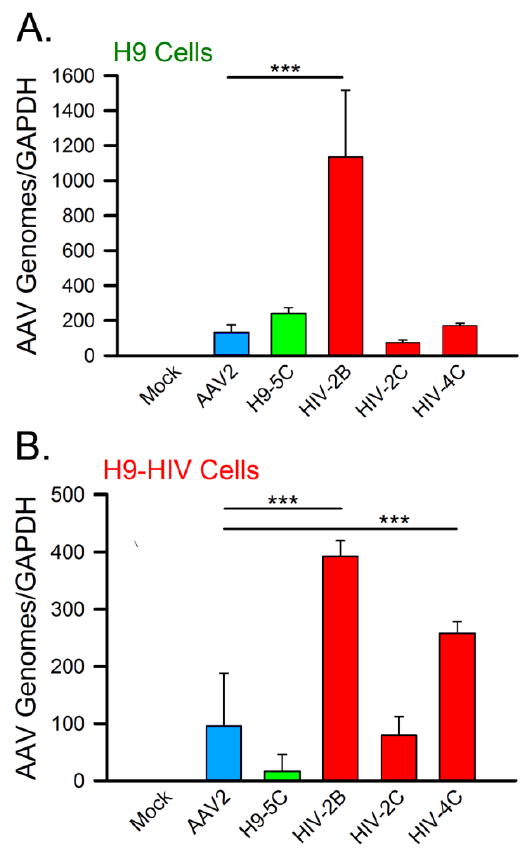
Specific AAV variants were packaged and used to infect either (A) H9 or (B) H9-HIV cells. qPCR was performed on total cell DNA isolated from the infected cells or control cells with normalization against the GAPDH gene. The bar graphs are color-coded as follows: blue = AAV2; green = variant isolated from H9 cells (H9-5C); red = variants isolated from H9-HIV cells (HIV-2B, HIV-2C, and HIV-4C). Mock: cells without virus. GAPDH: glyceraldehyde 3-phosphate dehydrogenase housekeeping gene for qPCR normalization. The horizontal bars and triple asterisks represent statistical significance between the designated conditions at a level of p < 0.001.
4. Discussion
In this study, a triple infection of H9 cells was performed to establish a system for selecting AAV variants with increased tropism for these cells. The results showed that all three viruses (AAV, adenovirus, and HIV-1) replicated in the presence of each other while maintaining sufficient cell viability to conduct selections (Fig. 2). The triply infected cells showed the greatest reduction in cell number, presumably due the cytotoxic effects of the wild-type adenovirus infection, which was used to enrich for successful AAVs in each round of selection. For a final gene delivery application, the wild-type adenovirus would no longer be needed because a highly transducing AAV would have already been chosen as the end product of a selection process.
Harnessing this system, a directed evolution approach was used to select for novel AAV capsids over six rounds of infection on H9 and H9-HIV cells, involving three rounds of positive selection followed by three rounds of coupled negative/positive selection (Fig. 1). Four mutants were chosen after the final selection step. Two of these mutants (HIV-2B and HIV-4C) were shown to have improved tropism for H9-HIV cells, and one of these two (HIV-2B) also had improved tropism for the parental H9 cells as evidenced by significant increases in cell-associated viral genomes measured by qPCR (Fig. 4). The fact that the selections converged on a small number of variants is not unusual. For example, one previous report on AAV selection described the isolation of a single, unique point mutation that exhibited enhanced binding to the apical surface of human airway epithelia as well as improved gene transfer in those cells (Excoffon et al., 2009). The observation that HIV-2B showed significant increases in cell-associated DNA on both H9 and H9-HIV cells (as compared with AAV2; Fig. 4) may indicate that this AAV variant utilizes a shared mechanism of cellular targeting between H9 and HIV-H9 cells. The two variants were used to transduce primary CD4+ T cells and showed an efficiency equal to that of parental AAV2 (data not shown).
In establishing and characterizing the system, we found that HIV-1 p24 production was not inhibited in H9 cells during co-infection (Fig. 2E and F), in contrast with previous reports indicating that AAV2 Rep protein may inhibit HIV-1 (Antoni et al., 1991; Mendelson et al., 1992; Oelze, Rittner, and Sczakiel, 1994; Rittner et al., 1992). These previous studies introduced the rep gene by microinjection or transfection, making it difficult to compare results with the current study. One of the studies used lower MOIs for both AAV (100 to 200 infectious units) and adenovirus helper (20 infectious units) as compared to the current study, and the duration of the experiment was longer (one week as compared to 24 h in the current study) (Mendelson et al., 1992). These experimental differences may likely explain the discrepancies between previous and current results with regard to inhibition of HIV-1 by the Rep protein.
One of the previous studies investigating Rep inhibition of HIV-1 used infectious viruses and described a triple infection of HUT78 cells with HIV-1, AAV2, and human adenovirus 2 (HUT78 is the parental line of H9 cells) (Mendelson et al., 1992). In this study, there was no reduction in HIV-1 production during infection by adenovirus alone, a 16% reduction by AAV alone, and a 51% reduction by both AAV and adenovirus, as measured by reverse transcriptase assay (Mendelson et al., 1992). AAV and adenovirus could exert opposing effects on HIV-1 during a triple infection, since adenovirus has been shown to activate HIV-1 (Antoni et al., 1991) whereas AAV has been shown to inhibit it via binding of viral proteins to the HIV-1 long terminal repeat (Oelze et al., 1994; Rice and Mathews, 1988; Rittner et al., 1992). Different serotypes of AAV may also exert different effects on HIV-1 replication, and this possibility could be explored further in future studies.
Although the original AAV library contained shuffled capsids from seven different serotypes of AAV (types 1, 2, 4–6, 8, and 9), the mutants that emerged after six rounds of selection were all based on the AAV2 serotype with a 98.6-99.9% sequence identity at the amino acid level. The fact that the selected variants had an AAV2 backbone does not necessarily mean that wild-type AAV2 is good, but rather only that small changes to a parental serotype like AAV2 are sufficient to positively shift tropism.
It is also important to consider the fact that DNA shuffling generally leads to smaller numbers of functional variants because of the modular nature by which the proteins are assembled; shuffled variants will only be functional if the structural modules are maintained. This is particularly important for a viral capsid since it is a multifunctional protein. As such, the shuffled clones must maintain functionality in multiple areas of the protein surface. All of the different classes of libraries (point mutants, peptide insertions, and shuffled capsids) were mixed together and allowed to compete with each other in this study. In previous studies, point mutants or peptide insertion variants have been observed to outcompete shuffled variants (Steines et al., 2016). However, one previous study showed positive selection for a shuffled variant with increased tropism for human airway epithelial cells, thus providing evidence that shuffled libraries are functional (Excoffon et al., 2009).
Wild-type AAV2, which binds heparan sulfate proteoglycans as its primary receptor, has a natural tropism for liver, kidney, neurons in the central nervous system, and retinal pigment epithelium in the eye (Burger et al., 2004; Grimm and Kay, 2003; Kwon and Schaffer, 2008; Wu, Asokan, and Samulski, 2006). Each of the point mutations identified in the current study has been observed by others in previous selections (Koerber et al., 2009; Steines et al., 2016) but in different combinations and without the peptide insertions, raising interesting mechanistic questions for future investigation. These mutants could be used to explore mechanisms of AAV transduction to enable better vector development, as well as to create new AAV libraries for further selection on other cells, including primary and latent T cells.
5. Conclusion
The experiments described herein were aimed at establishing a system for selecting AAV variants on HIV-1-infected and uninfected H9 T cells. This initial, proof-of-concept study used a cell line to develop the method for this type of selection with three replicating viruses (AAV, adenovirus, and HIV-1). Future studies will be aimed at performing the described selection process on other cell types, such as primary T cells, which have additional technical challenges, such as cell viability and donor variability.
It is important to note that the entire AAV selection process (all six rounds or more) must be performed on the target cell of final interest. It would be highly unlikely for an AAV variant to be successful on primary T cells if it had been previously selected on an immortalized cell line, such as the H9 line described here. The AAV variants selected in the current study were not expected to translate directly into an in vivo application. Rather, they were intended to provide evidence that unique AAVs could be isolated in this way.
Using AAV to target HIV-1-infected cells offers a potential research tool or gene therapy approach aimed at delivering an anti-HIV-1 therapeutic molecule, such as a small interfering RNA molecule or a genome editing system (Bobbin, Burnett, and Rossi, 2015; Limsirichai, Gaj, and Schaffer, 2016; Yin et al., 2017). Alternatively, isolation of an AAV variant that is better able to transduce non-HIV-1 infected T cells could be used potentially to protect them from HIV-1 infection as part of a prophylactic strategy. The new method may also be applicable to other T cell disorders, such as cancer, autoimmunity, and other non-HIV-1 immune deficiencies.
Highlights.
A novel AAV directed evolution approach to target parental and HIV-1-infected cells
Triple infection of H9 T cells with AAV, adenovirus, and HIV-1 was successful
Novel AAV capsids were isolated that show increased tropism toward the target cells
Acknowledgments
Funding
This work was supported by Public Health Service Grant R21AI093268 from the National Institutes of Health and by the State of Ohio.
Footnotes
Conflicts of Interest
KJDAE is a member of the scientific advisory board, and DVS is a co-founder, the acting Chief Scientific Officer, and co-chairman of 4D Molecular Therapeutics, Emeryville, CA. DVS and JRW have patents related to AAV engineering.
Disclaimer
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, Wright State University, or the University of California, Berkeley
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antoni BA, Rabson AB, Miller IL, Trempe JP, Chejanovsky N, Carter BJ. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J Virol. 1991;65:396–404. doi: 10.1128/jvi.65.1.396-404.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JS, Kleinschmidt J, Boucher RC, Samulski RJ. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab’gamma)2 antibody. Nat Biotechnol. 1999;17:181–186. doi: 10.1038/6185. [DOI] [PubMed] [Google Scholar]
- Berro R, de la Fuente C, Klase Z, Kehn K, Parvin L, Pumfery A, Agbottah E, Vertes A, Nekhai S, Kashanchi F. Identifying the membrane proteome of HIV-1 latently infected cells. J Biol Chem. 2007;282:8207–8218. doi: 10.1074/jbc.M606324200. [DOI] [PubMed] [Google Scholar]
- Bobbin ML, Burnett JC, Rossi JJ. RNA interference approaches for treatment of HIV-1 infection. Genome Med. 2015;7:50. doi: 10.1186/s13073-015-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buning H, Perabo L, Coutelle O, Quadt-Humme S, Hallek M. Recent developments in adeno-associated virus vector technology. J Gene Med. 2008;10:717–733. doi: 10.1002/jgm.1205. [DOI] [PubMed] [Google Scholar]
- Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ, Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Chenine AL, Sattentau Q, Moulard M. Selective HIV-1-induced downmodulation of CD4 and coreceptors. Arch Virol. 2000;145:455–471. doi: 10.1007/s007050050039. [DOI] [PubMed] [Google Scholar]
- Choi B, Gatti PJ, Fermin CD, Vigh S, Haislip AM, Garry RF. Down-regulation of cell surface CXCR4 by HIV-1. Virol J. 2008;5:6. doi: 10.1186/1743-422X-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, Uchida N, Hendel A, Narla A, Majeti R, Weinberg KI, Porteus MH. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein M. J Gene Med. John Wiley & Sons, Inc; Hoboken, NJ: 2017. Vectors used in gene therapy clinical trials. [Google Scholar]
- Ellis BL, Hirsch ML, Barker JC, Connelly JP, Steininger RJ, 3rd, Porteus MH. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol J. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon KJ, Koerber JT, Dickey DD, Murtha M, Keshavjee S, Kaspar BK, Zabner J, Schaffer DV. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc Natl Acad Sci USA. 2009;106:3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JP, Zhu H, Colosi PC, Kurtzman GJ, Scadden DT. Robust, but transient expression of adeno-associated virus-transduced genes during human T lymphopoiesis. Blood. 1997;90:4854–4864. [PubMed] [Google Scholar]
- Gazdar AF, Carney DN, Bunn PA, Russell EK, Jaffe ES, Schechter GP, Guccion JG. Mitogen requirements for the in vitro propagation of cutaneous T-cell lymphomas. Blood. 1980;55:409–417. [PubMed] [Google Scholar]
- Grimm D, Kay MA. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr Gene Ther. 2003;3:281–304. doi: 10.2174/1566523034578285. [DOI] [PubMed] [Google Scholar]
- Hasbrouck NC, High KA. AAV-mediated gene transfer for the treatment of hemophilia B: problems and prospects. Gene Ther. 2008;15:870–875. doi: 10.1038/gt.2008.71. [DOI] [PubMed] [Google Scholar]
- Horster A, Teichmann B, Hormes R, Grimm D, Kleinschmidt J, Sczakiel G. Recombinant AAV-2 harboring gfp-antisense/ribozyme fusion sequences monitor transduction, gene expression, and show anti-HIV-1 efficacy. Gene Ther. 1999;6:1231–1238. doi: 10.1038/sj.gt.3300955. [DOI] [PubMed] [Google Scholar]
- Hoxie JA, Alpers JD, Rackowski JL, Huebner K, Haggarty BS, Cedarbaum AJ, Reed JC. Alterations in T4 (CD4) protein and mRNA synthesis in cells infected with HIV. Science. 1986;234:1123–1127. doi: 10.1126/science.3095925. [DOI] [PubMed] [Google Scholar]
- Koerber JT, Klimczak R, Jang JH, Dalkara D, Flannery JG, Schaffer DV. Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Mol Ther. 2009;17:2088–2095. doi: 10.1038/mt.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber JT, Maheshri N, Kaspar BK, Schaffer DV. Construction of diverse adeno-associated viral libraries for directed evolution of enhanced gene delivery vehicles. Nat Protoc. 2006;1:701–706. doi: 10.1038/nprot.2006.93. [DOI] [PubMed] [Google Scholar]
- Kota J, Handy CR, Haidet AM, Montgomery CL, Eagle A, Rodino-Klapac LR, Tucker D, Shilling CJ, Therlfall WR, Walker CM, Weisbrode SE, Janssen PM, Clark KR, Sahenk Z, Mendell JR, Kaspar BK. Follistatin gene delivery enhances muscle growth and strength in nonhuman primates. Sci Transl Med. 2009;1:6ra15. doi: 10.1126/scitranslmed.3000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovesdi I, Hedley SJ. Adenoviral producer cells. Viruses. 2010;2:1681–1703. doi: 10.3390/v2081681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Schaffer DV. Designer gene delivery vectors: molecular engineering and evolution of adeno-associated viral vectors for enhanced gene transfer. Pharm Res. 2008;25:489–499. doi: 10.1007/s11095-007-9431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanteri M, Giordanengo V, Hiraoka N, Fuzibet JG, Auberger P, Fukuda M, Baum LG, Lefebvre JC. Altered T cell surface glycosylation in HIV-1 infection results in increased susceptibility to galectin-1-induced cell death. Glycobiology. 2003;13:909–918. doi: 10.1093/glycob/cwg110. [DOI] [PubMed] [Google Scholar]
- Li W, Asokan A, Wu Z, Van Dyke T, DiPrimio N, Johnson JS, Govindaswamy L, Agbandje-McKenna M, Leichtle S, Redmond DE, Jr, McCown TJ, Petermann KB, Sharpless NE, Samulski RJ. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- Limsirichai P, Gaj T, Schaffer DV. CRISPR-mediated Activation of Latent HIV-1 Expression. Mol Ther. 2016;24:499–507. doi: 10.1038/mt.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen YH, He X, Martins I, Heth JA, Chiorini JA, Davidson BL. Adeno-associated virus type 5 reduces learning deficits and restores glutamate receptor subunit levels in MPS VII mice CNS. Mol Ther. 2007;15:242–247. doi: 10.1038/sj.mt.6300016. [DOI] [PubMed] [Google Scholar]
- Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, Mingozzi F, Bennicelli JL, Ying GS, Rossi S, Fulton A, Marshall KA, Banfi S, Chung DC, Morgan JI, Hauck B, Zelenaia O, Zhu X, Raffini L, Coppieters F, De Baere E, Shindler KS, Volpe NJ, Surace EM, Acerra C, Lyubarsky A, Redmond TM, Stone E, Sun J, McDonnell JW, Leroy BP, Simonelli F, Bennett J. Age-dependent effects of RPE65 gene therapy for Leber’s congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009;374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire CA, Gianni D, Meijer DH, Shaket LA, Wakimoto H, Rabkin SD, Gao G, Sena-Esteves M. Directed evolution of adeno-associated virus for glioma cell transduction. J Neurooncol. 2010;96:337–347. doi: 10.1007/s11060-009-9972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DL, O’Brien SJ, Gilbert DA, Reid Y, Popovic M, Read-Connole E, Gallo RC, Gazdar AF. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res Hum Retroviruses. 1989;5:253–255. doi: 10.1089/aid.1989.5.253. [DOI] [PubMed] [Google Scholar]
- Mebatsion T, Finke S, Weiland F, Conzelmann KK. A CXCR4/CD4 pseudotype rhabdovirus that selectively infects HIV-1 envelope protein-expressing cells. Cell. 1997;90:841–847. doi: 10.1016/s0092-8674(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Meerloo T, Parmentier HK, Osterhaus AD, Goudsmit J, Schuurman HJ. Modulation of cell surface molecules during HIV-1 infection of H9 cells. An immunoelectron microscopic study. AIDS. 1992;6:1105–1116. doi: 10.1097/00002030-199210000-00007. [DOI] [PubMed] [Google Scholar]
- Meerloo T, Sheikh MA, Bloem AC, de Ronde A, Schutten M, van Els CA, Roholl PJ, Joling P, Goudsmit J, Schuurman HJ. Host cell membrane proteins on human immunodeficiency virus type 1 after in vitro infection of H9 cells and blood mononuclear cells. An immuno-electron microscopic study. J Gen Virol. 1993;74(Pt 1):129–135. doi: 10.1099/0022-1317-74-1-129. [DOI] [PubMed] [Google Scholar]
- Mendelson E, Grossman Z, Mileguir F, Rechavi G, Carter BJ. Replication of adeno-associated virus type 2 in human lymphocytic cells and interaction with HIV-1. Virology. 1992;187:453–463. doi: 10.1016/0042-6822(92)90447-w. [DOI] [PubMed] [Google Scholar]
- Michelfelder S, Trepel M. Adeno-associated viral vectors and their redirection to cell-type specific receptors. Adv Genet. 2009;67:29–60. doi: 10.1016/S0065-2660(09)67002-4. [DOI] [PubMed] [Google Scholar]
- Muller OJ, Kaul F, Weitzman MD, Pasqualini R, Arap W, Kleinschmidt JA, Trepel M. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- Nault JC, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, Letouze E, Pilati C, Verret B, Blanc JF, Balabaud C, Calderaro J, Laurent A, Letexier M, Bioulac-Sage P, Calvo F, Zucman-Rossi J. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- Oelze I, Rittner K, Sczakiel G. Adeno-associated virus type 2 rep gene-mediated inhibition of basal gene expression of human immunodeficiency virus type 1 involves its negative regulatory functions. J Virol. 1994;68:1229–1233. doi: 10.1128/jvi.68.2.1229-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KJ, Lee J, Park JH, Joh JW, Kwon CH, Kim JW. Adeno-Associated Virus 2-Mediated Hepatocellular Carcinoma is Very Rare in Korean Patients. Ann Lab Med. 2016;36:469–474. doi: 10.3343/alm.2016.36.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perabo L, Buning H, Kofler DM, Ried MU, Girod A, Wendtner CM, Enssle J, Hallek M. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol Ther. 2003;8:151–157. doi: 10.1016/s1525-0016(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Perabo L, Goldnau D, White K, Endell J, Boucas J, Humme S, Work LM, Janicki H, Hallek M, Baker AH, Buning H. Heparan sulfate proteoglycan binding properties of adeno-associated virus retargeting mutants and consequences for their in vivo tropism. J Virol. 2006;80:7265–7269. doi: 10.1128/JVI.00076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnazhagan S, Mahendra G, Kumar S, Thompson JA, Castillas M., Jr Conjugate-based targeting of recombinant adeno-associated virus type 2 vectors by using avidin-linked ligands. J Virol. 2002;76:12900–12907. doi: 10.1128/JVI.76.24.12900-12907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnazhagan S, Mukherjee P, Wang XS, Qing K, Kube DM, Mah C, Kurpad C, Yoder MC, Srour EF, Srivastava A. Adeno-associated virus type 2-mediated transduction in primary human bone marrow-derived CD34+ hematopoietic progenitor cells: donor variation and correlation of transgene expression with cellular differentiation. J Virol. 1997;71:8262–8267. doi: 10.1128/jvi.71.11.8262-8267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M, Read-Connole E, Gallo RC. T4 positive human neoplastic cell lines susceptible to and permissive for HTLV-III. Lancet. 1984a;2:1472–1473. doi: 10.1016/s0140-6736(84)91666-0. [DOI] [PubMed] [Google Scholar]
- Popovic M, Sarngadharan MG, Read E, Gallo RC. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984b;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- Rice AP, Mathews MB. Trans-activation of the human immunodeficiency virus long terminal repeat sequences, expressed in an adenovirus vector, by the adenovirus E1A 13S protein. Proc Natl Acad Sci USA. 1988;85:4200–4204. doi: 10.1073/pnas.85.12.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittner K, Heilbronn R, Kleinschmidt JA, Sczakiel G. Adeno-associated virus type 2-mediated inhibition of human immunodeficiency virus type 1 (HIV-1) replication: involvement of p78rep/p68rep and the HIV-1 long terminal repeat. J Gen Virol. 1992;73(Pt 11):2977–2981. doi: 10.1099/0022-1317-73-11-2977. [DOI] [PubMed] [Google Scholar]
- Schnell MJ, Johnson JE, Buonocore L, Rose JK. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- Schuhmann NK, Pozzoli O, Sallach J, Huber A, Avitabile D, Perabo L, Rappl G, Capogrossi MC, Hallek M, Pesce M, Buning H. Gene transfer into human cord blood-derived CD34(+) cells by adeno-associated viral vectors. Exp Hematol. 2010;38:707–717. doi: 10.1016/j.exphem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Somia NV, Miyoshi H, Schmitt MJ, Verma IM. Retroviral vector targeting to human immunodeficiency virus type 1-infected cells by receptor pseudotyping. J Virol. 2000;74:4420–4424. doi: 10.1128/jvi.74.9.4420-4424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Kauss MA, Kopin E, Chandra M, Ul-Hasan T, Miller E, Jayandharan GR, Rivers AE, Aslanidi GV, Ling C, Li B, Ma W, Li X, Andino LM, Zhong L, Tarantal AF, Yoder MC, Wong KK, Jr, Tan M, Chatterjee S, Srivastava A. Optimizing the transduction efficiency of capsid-modified AAV6 serotype vectors in primary human hematopoietic stem cells in vitro and in a xenograft mouse model in vivo. Cytotherapy. 2013a;15:986–998. doi: 10.1016/j.jcyt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Li X, Jayandharan GR, Wang Y, Aslanidi GV, Ling C, Zhong L, Gao G, Yoder MC, Ling C, Tan M, Srivastava A. High-efficiency transduction of primary human hematopoietic stem cells and erythroid lineage-restricted expression by optimized AAV6 serotype vectors in vitro and in a murine xenograft model in vivo. PLoS One. 2013b;8:e58757. doi: 10.1371/journal.pone.0058757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci USA. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. Adeno-associated virus-mediated gene transfer. J Cell Biochem. 2008;105:17–24. doi: 10.1002/jcb.21819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Carter BJ. AAV Infection: Protection from Cancer. Hum Gene Ther. 2017;28:323–327. doi: 10.1089/hum.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steines B, Dickey DD, Bergen J, Excoffon KJ, Weinstein JR, Li X, Yan Z, Abou Alaiwa MH, Shah VS, Bouzek DC, Powers LS, Gansemer ND, Ostedgaard LS, Engelhardt JF, Stoltz DA, Welsh MJ, Sinn PL, Schaffer DV, Zabner J. CFTR gene transfer with AAV improves early cystic fibrosis pig phenotypes. JCI Insight. 2016;1:e88728. doi: 10.1172/jci.insight.88728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M, Zhang XH, Volsky DJ. Downregulation of cell surface molecules during noncytopathic infection of T cells with human immunodeficiency virus. J Virol. 1987;61:3741–3748. doi: 10.1128/jvi.61.12.3741-3748.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JP, Carter BJ. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J Virol. 1988;62:3356–3363. doi: 10.1128/jvi.62.9.3356-3363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldwijk MR, Sellner L, Stiefelhagen M, Kleinschmidt JA, Laufs S, Topaly J, Fruehauf S, Zeller WJ, Wenz F. Pseudotyped recombinant adeno-associated viral vectors mediate efficient gene transfer into primary human CD34(+) peripheral blood progenitor cells. Cytotherapy. 2010;12:107–112. doi: 10.3109/14653240903348293. [DOI] [PubMed] [Google Scholar]
- Wagner JA, Messner AH, Moran ML, Daifuku R, Kouyama K, Desch JK, Manley S, Norbash AM, Conrad CK, Friborg S, Reynolds T, Guggino WB, Moss RB, Carter BJ, Wine JJ, Flotte TR, Gardner P. Safety and biological efficacy of an adeno-associated virus vector-cystic fibrosis transmembrane regulator (AAV-CFTR) in the cystic fibrosis maxillary sinus. Laryngoscope. 1999;109:266–274. doi: 10.1097/00005537-199902000-00017. [DOI] [PubMed] [Google Scholar]
- Wang J, Exline CM, DeClercq JJ, Llewellyn GN, Hayward SB, Li PW, Shivak DA, Surosky RT, Gregory PD, Holmes MC, Cannon PM. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Zhang T, Qu X, Zhang Y, Putatunda R, Xiao X, Li F, Xiao W, Zhao H, Dai S, Qin X, Mo X, Young WB, Khalili K, Hu W. In Vivo Excision of HIV-1 Provirus by saCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol Ther. 2017;25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PX, Fuleihan RL. Transfer of activation-dependent gene expression into T cell lines by recombinant adeno-associated virus. Gene Ther. 1999;6:182–189. doi: 10.1038/sj.gt.3300803. [DOI] [PubMed] [Google Scholar]


