Abstract
Echinochloa phyllopogon proliferation seriously threatens rice production worldwide. We combined a restriction-site associated DNA (RAD) approach with Illumina DNA sequencing for rapid and mass discovery of simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers for E. phyllopogon. RAD tags were generated from the genomic DNA of two E. phyllopogon plants, and sequenced to produce 5197.7 Mb and 5242.9 Mb high quality sequences, respectively. The GC content of E. phyllopogon was 45.8%, which is high for monocots. In total, 4710 putative SSRs were identified in 4132 contigs, which permitted the design of PCR primers for E. phyllopogon. Most repeat motifs among the SSRs identified were dinucleotide (>82%), and most of these SSRs were four motif-repeats (>75%). The most frequent motif was AT, accounting for 36.3%–37.2%, followed by AG and AC. In total, 78 putative polymorphic SSR loci were found. A total of 49,179 SNPs were discovered between the two samples of E. phyllopogon, 67.1% of which were transversions and 32.9% were transitions. We used eight SSRs to study the genetic diversity of four E. phyllopogon populations collected from rice fields in China and all eight loci tested were polymorphic.
Keywords: Echinochloa phyllopogon, Polymorphic, RAD sequencing, SNP, SSR
Highlights
-
•
RAD-sequencing was applied for rapid and mass discovery of SSRs and SNPs in E. phyllopogon.
-
•
Totally 4710 putative SSRs were identified, and AT was the most frequent motif.
-
•
Totally 49179 SNPs were discovered between the two samples of E. phyllopogon.
1. Introduction
Echinochloa phyllopogon (= Echinochloa oryzicola) proliferation seriously threatens rice production worldwide. As a C4-photosynthetic weed, E. phyllopogon is highly adapted to rice (C3-photosynthesis type) planting environments, where it causes significant rice yield loss (Holm et al., 1979, Rao et al., 2007, Yamasue, 2001). Furthermore, E. phyllopogon has evolved resistance to various herbicides in different areas (Heap, 2015). Understanding the genetic diversity of agricultural pests, such as E. phyllopogon, is important for both evolutionary and population biology, and critical for agricultural management (Sun et al., 2015).
Microsatellite markers (simple sequence repeats, SSR) and single-nucleotide polymorphisms (SNP) are useful tools for studying genetic diversity and evolution (Zhang et al., 2011), and for developing high density genetic maps (Zhang et al., 2012). SSRs are short tandem repetitive sequences, which are co-dominant, abundant, multi-allelic, uniformly distributed, and can be detected by simple reproducible assays (Wang et al., 2015). SNPs are usually bi-allelic and characterized by low mutation rates; and thus, SNPS are stable from generation to generation across the genome (Kruglyak, 1997). This stability coupled with the abundance of SNPs makes them very useful both for linkage and genetic diversity studies (Talukder et al., 2014). To date, there are only eight SSR markers available for E. phyllopogon (Osuna et al., 2011, Lee et al., 2015), and an even more limited number of SNPs.
One promising approach to reduced-representation genomics is restriction site-associated DNA (RAD) sequencing, which sequences short DNA fragments flanking restriction enzyme cut sites, allowing orthologous sequences to be targeted across multiple samples to identify and score thousands of genetic markers (Miller et al., 2007). Therefore, a RAD sequencing approach can be successfully used to identify genome-wide SSRs (Gupta et al., 2015, Orjuela et al., 2010) and SNPs (Baird et al., 2008, Talukder et al., 2014, Vandepitte et al., 2013) in different species. In this study, we describe the generation of genomic RAD tags from E. phyllopogon plants. The RAD tags were sequenced using the Illumina platform and then annotated/categorized. These data allowed the discovery of a large number of SSR and SNP markers.
2. Material and methods
2.1. DNA isolation
Seeds from E. phyllopogon individuals were collected and cultivated to fruiting stage in a greenhouse at Nanjing Agricultural University. Two E. phyllopogon plants with typical characteristics were used for SSR identification. Total genomic DNA was extracted from young leaves using DNeasy Plant Mini Kits (Qiagen, USA) according to the manufacturer's protocol.
2.2. RAD library preparation, sequencing and assembly
The RAD library was constructed at Hengchuang Inc. (China), according to the protocol described by Baird et al. (2008). Briefly, genomic DNA (300 ng) was digested for 60 min at 37 °C in a 50 μL reaction containing 20 U each of SgrAI and PstI (New England Biolabs, Beverly MA, USA). Reactions were stopped by incubating at 65 °C for 20 min. The P1 adapter (a modified Illumina adapter, see Baird et al., 2008) was ligated to the products of the restriction reaction, and the “barcoding” of the various samples was achieved with a set of index nucleotides in the P1 adapter sequence. A 2.5 μL aliquot of 100 nM P1 adapter was added to each sample, along with 1 μL 10 mM ATP (Promega), 1 μL 10× NEBBuffer4, 1 μL (equivalent to 1000 U) T4 DNA ligase (Enzymatics, Inc) and 5 μL water, then incubated at room temperature for 20 min, before heat-inactivated (20 min at 65 °C). The reactions were then pooled and the products randomly sheared to a mean size of 500 bp using a Bioruptor (Diagenode). The material was electrophoresed through a 1.5% agarose gel, and the DNA in the range 300–800 bp isolated using a MinElute Gel Extraction Kit (Qiagen). dsDNA ends were treated with end blunting enzymes (Enzymatics, Inc) to remove overhangs, and the samples purified using a MinElute column (Qiagen). 3′-adenine overhangs were then added by the addition of 15 U Klenow exo-(Enzymatics), followed by incubation at 37 °C for 10 min. Following re-purification, 1 μL 10 μM P2 adapter (a modified Illumina adapter, see Baird et al., 2008) was ligated, as described above for P1. The samples were then purified as above, and eluted in a volume of 50 μL. Following quantification (Qubit fluorimeter), 20 ng were taken as the template for a 100 μL PCR containing 20 μL Phusion Master Mix (NEB), 5 μL 10 μM P1 adapter primer (Illumina), 5 μL 10 μM P2 adapter primer (Illumina) and water. The Phusion PCR settings followed product guidelines (NEB) over 18 cycles. The amplicons were gel purified, the size range 300–700 bp was excised from the gel, with the DNA content adjusted to 3 ng/μL. The constructed RAD libraries were sequenced on the NGS Illumina platform PE150 at Hengchuang Inc. (China), following the manufacturer's protocol.
To obtain clean, high quality reads, we discarded low quality raw sequences with adapter contamination or N content >10%. We used Stacks software for RAD tag clustering for each sample (ustacks). The Reads group (Read1 and Read2) at a same enzyme loci RAD were assembled by using the ABYSS software (Catchen et al., 2011).
2.3. SSR identification
SSR motifs were identified by SSRIT software (http://www.gramene.org/db/markers/ssrtool) using default parameters (Temnykh et al., 2001). Both perfect and imperfect di-, tri-, tetra-, penta- and hexa-nucleotide motifs were targeted. Di-nucleotide motifs with at least 4 repeats and other motifs with at least 3 repeats were selected. We used Primer3 software (http://sourceforge.net/projects/primer3/) to design primers in the flank regions of SSR sequences (SSR sequences were not contained in the primers), the replicated primers were removed and unique primers and relative loci were retained.
To analyze the frequency of SSR motifs, SSRs were first standardized (Wang et al., 2015). For example, SSRs with motifs of AT and TA were analyzed as AT, and motifs of ATG, TGA, GAT, TAC, ACT and CAT are analyzed as ATG.
2.4. Sequence annotation
For the contigs with SSR loci, sequence annotation and Gene Ontology analyses were further conducted. BlastN searches were performed against the Gene Ontology database (http://www.geneontology.org/), using 90% identity and a minimum alignment of 100 bp as cut-off parameters. A threshold E-value of e−15 was adopted for each annotation. The annotated sequences were assigned a function based on the Gene Ontology database (http://www.geneontology.org/); GO terms were determined with respect to cellular component, biological process and molecular function (Barchi et al., 2011).
2.5. SNP discovery
SNPs were detected by Stacks pipeline, ustacks software was used to build loci, cstacks software was used to create a catalog of loci, and sstacks software was used to match samples back against the catalog (Catchen et al., 2011). Default settings were used in Stacks.
2.6. Microsatellites amplification
To test the validity of the SSRs identified by RAD sequencing here, we used eight SSRs (Table 1) to study the genetic diversity of four E. phyllopogon populations collected from rice fields in China. We extracted total genomic DNA from four-leaf stage plants using a DNeasy Plant Mini Kit (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions. Isolated DNA concentration and relative purity were checked using Nanodrop ND-1000 (Thermo Scientific), and adjusted to 30–40 ng/μL. Forward primers of SSRs were labeled with fluorescent tags (Table 1). PCR amplification was conducted in a total volume of 10 μL. The PCR mixture contained 0.2 μL of DNA, 0.4 μL of each primer (10 μM), 5 μL of 2× PCR Taq Mix (Dongsheng Biotech, China), and ddH2O to a final volume of 10 μL. The amplifications were performed using the following cycling program: initial denaturation at 94 °C for 4 min, followed by 35 cycles of 94 °C for 30 s, relative annealing temperatures for 30 s, and 72 °C for 1 min, with a final extension step at 72 °C for 10 min. The amplification products were combined with formamide and a size standard GeneScan-500 LIZ (Applied Biosystems, Foster City, California, USA), and separated on a 3730 ABI automated sequencer (Applied Biosystems). Sample profiles were scored manually using GeneMarker v. 2.4 (Applied Biosystems).
Table 1.
Characteristics of the eight primers tested for E. phyllopogon genotyping: locus name, forward (F) and reverse (R) primer sequences, motif, annealing temperature (Tm), fluorescent dye used (Fl. dye), allele size range (ASR), number of alleles amplified per sample, and number of alleles amplified among the plants of four populations sampled (Allele. total).
| Marker | Sequence | Motif | ASR (bp) | Fl. dye | Tm (°C) | No. of alleles per sample |
Allele. total | ||
|---|---|---|---|---|---|---|---|---|---|
| Min. | Mean | Max. | |||||||
| EG _1 | F: GCTCCTGAACTGTGTACATTCTTGC | TG | 123–153 | TAM | 49 | 0 | 0.7 | 2 | 5 |
| R: TCGATTCACCCTTCAGCTTCTC | |||||||||
| EG_2 | F: CATCGGATTCAGATTGAAAGGG | TA | 131–159 | FAM | 51.5 | 1 | 1.7 | 3 | 7 |
| R: GGTCGTAGGTCTATAGTCCGTAGAGTCA | |||||||||
| EG_301 | F: GCGTCGTCAAGTCGTTCTTCTA | AT | 147–173 | TAM | 57 | 0 | 2.4 | 3 | 8 |
| R: TGTATTCAGCTGTCGTGCATGT | |||||||||
| EG_302 | F: ATTCGAACACCCATCAACCAAC | ATTT | 133–293 | FAM | 57 | 1 | 2.8 | 5 | 12 |
| R: GAAACAGAAGGGAGGTGTGCTG | |||||||||
| EG_305 | F: AGCCGTTCCTCTAGTCGGATTTCT | AT | 100–162 | ROX | 57 | 3 | 4.1 | 6 | 14 |
| R: TATTCAGCTGCCGTGCATGTAGTA | |||||||||
| EG_306 | F: TAAAACAAAACGACCGGCGTAA | CT | 146–167 | HEX | 57 | 1 | 1.25 | 2 | 7 |
| R: TCAATCATTTCAGCCTTCGGAT | |||||||||
| EG_307 | F: AACATTGTCATCACAAATATCATCATCA | ATC | 108–134 | TAM | 57 | 2 | 3.5 | 5 | 8 |
| R: AATCAAGGAAGCCCCTTCACTC | |||||||||
| EG_320 | F: CAACTCATAAGACAATTCAAAGGGTTT | TA | 136–153 | FAM | 57 | 2 | 3.0 | 4 | 5 |
| R: GCATCATTTAAGCATCAAAATGACA | |||||||||
FAM: 6-carboxyfluorescein, HEX: hexachloro-fluoresceine, ROX: carboxy-X-rhodamine, and TAM: 5-TAMRA (5-Carboxytetramethylrhodamine).
2.7. Data analysis
The multilocus data were transformed to a binary matrix of presence/absence of each allele for each individual, which was used for further analysis with GenAlex 6.5 (Peakall and Smouse, 2012, Teixeira et al., 2014). Total number of alleles and the number of private alleles for each population were determined using GenAlex 6.5, and genetic diversity was determined using GenoDive2.0b23 (Teixeira et al., 2014), according to the tutorials (www.patrickmeirmans.com/software/GenoDive.html). GenoDive allows analyzing polyploids with unknown dosage of alleles (Meirmans and Van Tienderen, 2004).
3. Results
3.1. Sequencing and contig assembly
The sequencing procedure generated 71.45 million reads for the two E. phyllopogon samples (Table 2). After editing/trimming, 10,440.6 Mb of high quality sequences were available, which were assembled into 37,662 contigs. Average contig lengths for the two samples were 334 and 346 bp. The GC content of E. phyllopogon was 45.8%.
Table 2.
Summary statistics of the RAD tags sequencing via Illumina for E. phyllopogon.
| Feature | Total |
|---|---|
| Illumina reads (million) | 71.45 |
| Total base (million) | 10,440.6 |
| GC% | 45.8% |
| Q20 (%) | 94.0% |
| No. of contigs | 37,662 |
| Total length (bp) | 12,789,629 |
| Contig length range (bp) | 200–588 |
| Average contig length (bp) | 339.5 |
3.2. Identification of SSRs
A screen of the dataset resulted in the identification of 4710 putative SSRs that permitted PCR primer design for E. phyllopogon. Tables S1 and S2 show motifs, number of repeats, sequence of 5′- and 3′-flanking, sequences and annealing temperatures of primers, sequence of PCR products and the potential relative genes for each SSR loci.
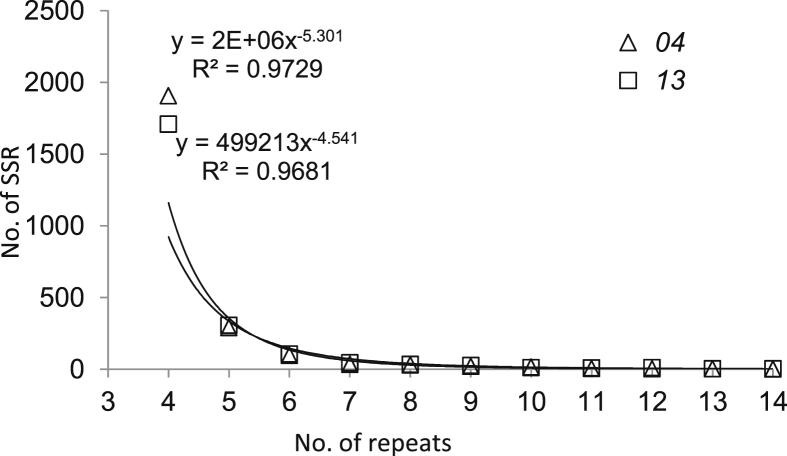
The majority of motifs among the RAD SSRs were dinucleotide (>82%) for both samples, and 14%–15% of the SSR motifs were trinucleotide (Table 3). The majority of SSRs were four motif-repeats. The abundance of SSRs decreased significantly (P < 0.01) with increasing motif-repeats for E. phyllopogon (Fig. 1).
Table 3.
Length distributions of SSR motifs identified for the two samples of E. phyllopogon tested.
| Motif length | 13 | 04 |
|---|---|---|
| Dinucleotide | 1908 (83.0%) | 1998 (82.4%) |
| Trinucleotide | 329 (14.3%) | 360 (14.9%) |
| Tetranucleotide | 40 (1.7%) | 50 (2.1%) |
| Pentanucleotide | 15 (0.7%) | 13 (0.5%) |
| Hexanucleotide | 6 (0.3%) | 3 (0.1%) |
| Total | 2298 | 2424 |
Fig. 1.
SSR motifs with different repeat numbers for the two samples of E. phyllopogon.
Nearly all (97.3%) E. phyllopogon SSR motifs consisted of dinucleotide plus trinucleotide repeats. Thus, we further analyzed dinucleotide and trinucleotide motifs. Before the analysis, SSRs were standardized. For example, SSRs with motifs of AT and TA were analyzed as AT, and motifs of ATG, TGA, GAT, TAC, ACT and CAT were analyzed as ATG. AT was the most frequent, accounting for 36.3%–37.5%, followed by AG and AC (Table 4). Among the four kinds of dinucleotide motifs, CG dinucleotide repeats represented the lowest percentage of all SSRs (<6%). CCG was the most frequent kind of trinucleotide motif for both samples (Table 4), accounting for about 4% of the total SSRs for E. phyllopogon. The predicted length of PCR products amplified by SSR primers designed in this study are shown in Table 4.
Table 4.
SSR motifs with a frequency > 0.5% and the ranges of PCR product length (mean length) of the relative motifs for the two samples tested for E. phyllopogon.
| Motif | Count (% of total SSRs) |
PCR product length (average length, bp) |
||
|---|---|---|---|---|
| 13 | 04 | 13 | 04 | |
| AT | 854 (37.2) | 880 (36.3) | 80–234 (133.5) | 80–239 (131.0) |
| AG | 562 (24.5) | 617 (25.5) | 80–208 (126.5) | 80–208 (127.1) |
| AC | 372 (16.2) | 395 (16.3) | 80–225 (130.3) | 80–234 (126.6) |
| CG | 120 (5.2) | 106 (4.4) | 80–204 (131.6) | 80–237 (124.1) |
| CCG | 99 (4.3) | 103 (4.2) | 80–172 (132.0) | 80–160 (126.9) |
| AAG | 45 (2.0) | 43 (1.9) | 85–159 (128.8) | 81–153 (121.0) |
| AAT | 28 (1.2) | 30 (1.2) | 80–160 (130.3) | 80–160 (127.5) |
| ACC | 27 (1.2) | 14 (0.6) | 80–157 (122.9) | 80–220 (134.2) |
| AAC | 25 (1.1) | 47 (1.9) | 85–155 (120.3) | 122–155 (136.9) |
| AGG | 24 (1.0) | 25 (1.0) | 81–188 (128.3) | 80–159 (132.3) |
| AGC | 23 (1.0) | 29 (1.2) | 80–157 (122.6) | 89–159 (134.2) |
| ACG | 22 (1.0) | 15 (0.6) | 86–160 (136.1) | 83–159 (121.4) |
| AGT | 22 (1.0) | 32 (1.3) | 80–160 (133.9) | 81–160 (130.8) |
| ATG | 14 (0.6) | 22 (0.9) | 91–160 (134.5) | 87–159 (127.9) |
Note: motifs with dinucleotide plus trinucleotide contributed to 97.3% of the total SSRs for both samples. Thus motifs with length >3 were not shown in this table.
In total, 78 putative polymorphic SSR loci were found by RAD sequencing (Table 5). These 78 SSRs include 65 SSRs with dinucleotide motifs, 10 SSRs with trinucleotide motifs, two with tetranucleotide motifs and one with a pentanucleotide motif. The AT dinucleotide repeat, which accounts for 49.4% of all motifs, was the most frequent kind.
Table 5.
The 78 putative polymorphic SSR loci found by RAD sequencing.
| Marker | Motif | Primer_F | Primer_R | Marker | Motif | Primer_F | Primer_R |
|---|---|---|---|---|---|---|---|
| EG_1 | TG | gctcctgaactgtgtacattcttgc | tcgattcacccttcagcttctc | EG_40 | GAA | aacagacaaaatacaaaagaaagcaca | gtttttcagcatcatcctgtgg |
| EG_2 | TA | catcggattcagattgaaaggg | ggtcgtaggtctatagtccgtagagtca | EG_41 | AT | tcactacgaaattatcgtttatggacaa | gcccgctccgtgtttagattat |
| EG_3 | TA | ttgctttctgcaatgccaatta | gtccatgtggagtcagggagtt | EG_42 | TA | atgggcgacaagcaagtatgat | gacggacgaaggtttgaagattt |
| EG_4 | TA | ccgttgatgattaactcgttgattt | tgatggtagctacaagcgttgg | EG_43 | GA | catcctctggctgcttctctct | gaatgtgagaatctccgctgct |
| EG_5 | TA | ttcactatgctgaaccagcagc | ctgagtccggtatcgctcctta | EG_44 | GA | acacctttctccatcctctggc | ccgctgctgctactactcttgg |
| EG_6 | AT | ccatggtcaagtcactttgtctg | tctggatctcccaaattcatgtc | EG_45 | TG | ttgtacaagcttctgagataacctga | atttcagaaactgtttgaattaggattt |
| EG_7 | AAG | catttcttaccgtcccatctgc | cctttttcagggagaagccact | EG_46 | TA | aaatggatatggcaaacgcatc | ccaagtccatcatgccaagttt |
| EG_8 | AT | ttttgtaggcctaacctgttgtgg | tttttgctatgcatgtgtctactcg | EG_47 | AT | tttgggattgtttatgaggtttga | cacacggcaaaatgaccaata |
| EG_9 | TG | tataacatccctttcgttgccatc | tgcaatgaaattcagatattcggac | EG_48 | AAG | tgctatgcatgaggagatgcag | ccttataccttggaggctcgct |
| EG_10 | AAG | taaattgcccaaacaagaaagagg | atcggagtcccactcaacaaagta | EG_49 | TA | aattctagtttgcgacgggttatt | ttgagtgaatgggatcgaaaaa |
| EG_11 | TGCA | agccggtgcaggaagacag | aagaagggaaaaggtagtcgttgg | EG_50 | TC | aaggacaaagtcgcagcgttt | atgggatttggttttggcttct |
| EG_12 | CTCTC | tttgaagccttttcggtcttga | aacaagcagtggaagacgaagg | EG_51 | GC | gccgggtgattaacggattagt | agactagctagccagcgggttg |
| EG_13 | AT | ggcccaatataatatccatgcc | ctatcaagggcagctatttggg | EG_52 | AT | aattcaacacaaccaaaggtaaaaa | tcaatgccatattgattctccc |
| EG_14 | AT | ggtggtgtgtcctgatgtgtgt | tgtttccttttgtttttgttttgtttc | EG_53 | TG | tcaaatggcaaagtatggaactca | tcattttctcaagaagcagtggtc |
| EG_15 | TA | catgaactgttctgactccaacaac | aagcattgcagctctgtcttgt | EG_54 | AT | aatattaacgtacccttgacaaatgaa | tttttgttggtacgtaagataaacaatc |
| EG_16 | TA | tcagttgagctccatcatttgttt | tcactggctgttctttaccgtact | EG_55 | AG | ccaagaaaccaactaagagccaaa | atttgtgcatgatgtgctttgc |
| EG_17 | CGG | gatagcgactcgagcgtggt | tctcgagcatggggagagac | EG_56 | AG | agcaagaaaccaactaagagccaa | aattcgtgcatgatgtgctttg |
| EG_18 | ATG | agccatattgccttgtgaccaa | ttttccttgcgcaatttttcat | EG_57 | TC | tgaaaagccagtggacagtcag | gagttcctcctgatggcaagaa |
| EG_19 | AC | ccttcagctgatgtaatcttggtaag | tccatctctcagcacctgaaaa | EG_58 | TC | tctccctccaaactttactattcacc | gctcaaaagatttgtctcgtcg |
| EG_20 | AT | gaaggtcgtgcactatggtgag | agcaagttgaagcaatccaagg | EG_59 | AT | cgtcaagtcgttcctctagtcg | tgtattcagctgtcgtgcatgt |
| EG_21 | AT | cgccgtcaagtcattcctcta | tcagctgccgtgcatgtagta | EG_60 | AT | tgccagacagtccaacaagcta | ggccgactctatattcatattagctgac |
| EG_22 | CT | cacatgatacatccgttgcgtc | atcggaggagggggaagag | EG_61 | TA | aatgcagtcaggcccttgttta | gcacgggcacatttcctagt |
| EG_23 | AG | aaaacgccgcaaaaacaaaag | cccctctaggattctcgctgtt | EG_62 | TC | cttcttcctcgcctccaattc | aaacaagttattacccggcgct |
| EG_24 | TA | acgagcacccattatgttttgg | cgagatcccagagcaaagctac | EG_63 | TA | cgattgcttaagggaataaatgg | caacattttactggtaatcctttcttg |
| EG_25 | CT | atcaaaccccctcgaattcct | gagggagagaaagctgacaggc | EG_64 | GA | tcttggctgaaaaatctatttggG | acctctcccacttgaagaagca |
| EG_26 | TA | ttcaaaaattcgatctttgctgc | aaccttttccgtggcctacct | EG_65 | AT | cccctgagcaaatttcaatcat | agggacagggaaggatcttgac |
| EG_27 | GA | gctcagcatctccaacgaactt | caaaccaattctgaatcgaaaagc | EG_66 | AT | ttcatagaggtggtgtgtcctga | tggtttccttttgtttttattatgtttc |
| EG_28 | TA | gatgacgtggctagcttgcata | cgtaggacgaaggatgaaaacg | EG_67 | AT | cgcacactggctgtaattggta | ccgagctttcagatttactcctca |
| EG_29 | CT | cctccttcctttgctgagcC | ctgcagcatgccctttctattt | EG_68 | TA | aatgcaaaataggacaccacgg | ggaacccatgaataagctgcaa |
| EG_30 | GA | aggtcgtgcatgggctagag | cggagtagcttcacgcttcagt | EG_69 | AT | ggaaattgcatctgcatcaact | cccatgcagcatactaatgtgaa |
| EG_31 | TCT | ttgagatgatgatgcattcacttg | tgggaagccatgaagaatatgg | EG_70 | AT | ttcgttcatttcgctctcatca | ttggcaatagttttcaatcttgcat |
| EG_32 | TA | gtgggctcataccttaatgccc | ggggagccatctctcttctcat | EG_71 | GA | aggaagaaaagagaagtgaggcG | cgagcacctcctctaggaatca |
| EG_33 | AT | gccgtcaagtcgttcctctagt | cagctgccgtgcatgtaatact | EG_72 | TA | ctgcgggtgacatttgtacagt | gtctgaacacgttaccacaccg |
| EG_34 | TCT | gatgatgatgcattcacttgagttg | tggatgatgtgagaggtgatgg | EG_301 | AT | gcgtcgtcaagtcgttcttcta | tgtattcagctgtcgtgcatgt |
| EG_35 | AT | tcctctagtcggatttcttaatttgc | tgtattcagctgtcgtgcatgt | EG_302 | ATTT | attcgaacacccatcaaccaac | gaaacagaagggaggtgtgctg |
| EG_36 | AG | catgaccatcaggcatcatctc | atgaagaagctactccgccgat | EG_305 | AT | agccgttcctctagtcggatttct | tattcagctgccgtgcatgtagta |
| EG_37 | TCT | tcagaaacaatatgttcctcatcatca | caaatgggtcacaagacgagaa | EG_306 | CT | taaaacaaaacgaccggcgtaa | tcaatcatttcagccttcggat |
| EG_38 | TG | ggagctggagaaactgaaggaag | cacttcgttgagggctcgatag | EG_307 | ATC | aacattgtcatcacaaatatcatcatca | aatcaaggaagccccttcactc |
| EG_39 | CA | gtggcatgtgaattgtttccct | caatcttacctcccaccttccc | EG_320 | TA | caactcataagacaattcaaagggttt | gcatcatttaagcatcaaaatgaca |
To test the validity of the SSRs identified by RAD sequencing here, we used eight SSRs to study the genetic diversity of four E. phyllopogon populations collected from rice fields in China. We amplified 66 alleles from the eight microsatellite loci. The primer sequence EG_305 amplified 14 alleles, EG_302 amplified 12 alleles, and EG_320 and EG_1 amplified five alleles (Table 1). EG_305 amplified three to six alleles per sample, while EG_307 and EG_320 amplified two to five and two to four alleles per sample, respectively. Moreover, EG_305 amplified the most alleles on average (4.1). On average, 3.1–4.8 alleles were amplified from one locus per population (Table 6). All four populations showed private alleles, among which the populations EP13 and EP50 showed 13 and eight private alleles, respectively. The heterozygosity values of these populations ranged from 0.064 to 0.091, and their Shannon's information indices ranged from 0.087 to 0.381. Analysis of molecular variance (AMOVA) indicated that 39% of diversity occurs among populations, while 61% of diversity occurs within populations (Table 7).
Table 6.
Diversity of four populations of E. phyllopogon using eight nuclear microsatellite loci.
| Population | EP13 | EP14 | EP53 | EP50 | Total |
|---|---|---|---|---|---|
| No. of alleles | 39 | 34 | 25 | 37 | 66 |
| No. of alleles per locus | 4.875 | 4.25 | 3.125 | 4.625 | 8.25 |
| No. of private alleles | 13 | 1 | 2 | 8 | / |
| Heterozygosity | 0.086 | 0.082 | 0.064 | 0.091 | 0.081 |
| Shannon's information index | 0.381 | 0.21 | 0.087 | 0.222 | 0.225 |
Table 7.
Analysis of molecular variance (AMOVA) showing the partitioning of genetic variation within and between regions of E. phyllopogon.
| Source | df | SS | MS | Est. var. | % | P |
|---|---|---|---|---|---|---|
| Among Pops | 3 | 135.563 | 45.188 | 3.328 | 39 | <0.01 |
| Within Pops | 44 | 231.250 | 5.256 | 5.256 | 61 | <0.01 |
df = degree of freedom, SS = sum of squares, MS mean squares, Est. var. = estimate of variance, % = percentage of total variation, P-value is based on 9999 permutations.
3.3. Annotation of contigs with SSR loci
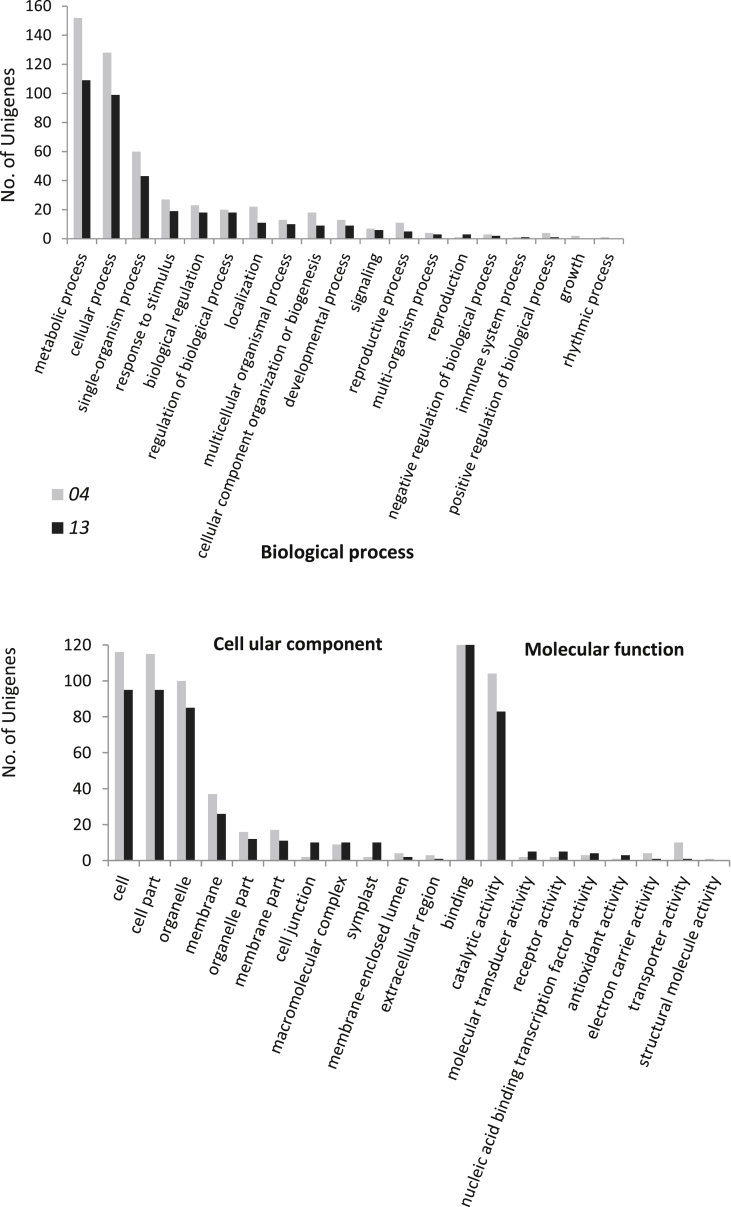
Using two E. phyllopogon individuals, we identified 4710 SSR loci in 4132 contigs, and annotated 643 contigs (Table S2). Among these 643 contigs, 8631 annotations, potentially referring to 2155 unigenes, were searched (a given gene product can be associated with more than one annotation). Annotated E. phyllopogon sequences with SSR loci were functionally assigned and arranged into Gene Ontology (GO) slim categories (Fig. 2). GO analyses suggested that contigs with SSR loci were mostly related to metabolic processes (12.1% of the total 2155 unigenes) and cellular processes (10.5%) among biological processes; cell (9.8%), cell part (9.7%) and organelle (8.6%) among cellular components; and binding (12.6%) and catalytic activity (8.7%) among molecular functions.
Fig. 2.
Functional annotation of assembled sequences with SSR loci for the two samples of E. phyllopogon based on gene ontology (GO) terms.
3.4. SNP discovery
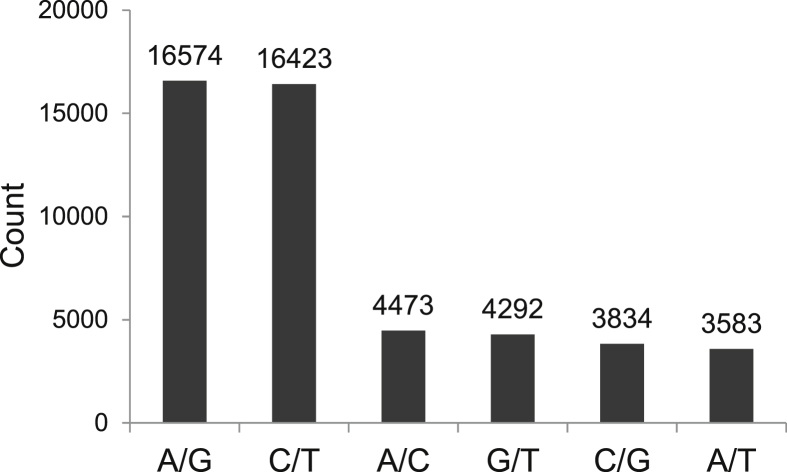
In total, 49,179 SNPs were discovered between the two samples of E. phyllopogon. Table S3 shows the kind, sequence and location of 49,179 SNPs discovered between two samples of E. phyllopogon. Among these SNPs, transversions (67.1% of total SNPs) were much more frequent than transitions (Fig. 3).
Fig. 3.
Transitions and transversions occurring within a set of 49,179 E. phyllopogon SNPs.
4. Discussion
4.1. High GC content of E. phyllopogon genome
Higher GC content in plant genomes possibly contributes to an increased ability to adapt to various arable lands that are mainly maintained and regulated by human disturbance. Šmarda et al. (2014) studied GC content in 239 different plant genomes, finding that the GC content of monocots varied between 33.6% and 48.9%, and increased GC content was documented in species able to grow in seasonally cold and/or dry climates, which possibly indicates GC-rich DNA may confer more stability during cell freezing and desiccation. The GC content of E. phyllopogon was higher than those of many monocots such as Juncus inflexus (33.7%), Luzula badia (33.6%), Carex acutiformis (35.6%), Schoenoplectus lacustris (35.8%), Canna indica (39.7%), Oryza sativa (43.6%) and Triticum aestivum (44.7%); and only lower than those of a few Poaceae species such as Stipa calamagrostis (47.5%) and Zea mays (47.4%) (Raats et al., 2013).
4.2. Characteristics on SSR motifs of E. phyllopogon
The majority of RAD SSR motifs were dinucleotide and with four motif-repeats. Gupta et al. (2015) identified SSR motifs in peanut (Arachis hypogaea) through RAD sequencing, and found that 67.6% of the motifs were dinucleotide, 14.6% were trinucleotide, 12.5% were tetranucleotide, 3.2% were pentanucleotide and 2.2% were hexanucleotide. Nevertheless, in eggplant (Solanum melongena), the percentages among total motifs with two to six nucleotides of dinucleotide, trinucleotide, tetranucleotide, pentanucleotide and hexanucleotide were 20.4%, 37.9%, 12.8%, 18.1% and 10.9% (Barchi et al., 2011). Using RAD sequencing in eggplant, Barchi et al. (2011) found that AAC was the most frequent kind of motif, accounting for 19.0% of the total SSRs, followed by AT (9.6%). Wang et al. (2015) analyzed the genomes of nine plant species from the Poaceae family, and found that among the genome SSRs of O. sativa ssp. indica, O. sativa ssp. japonica, Phyllostachys heterocycla, Sorghum bicolor and Z. mays, AT was the most frequent motif, and also very frequent in other Poaceae plants.
To test the validity of the SSRs identified by RAD sequencing here, we used eight SSRs to study the genetic diversity of four E. phyllopogon populations collected from rice fields in China. All eight loci were polymorphic, particularly when compared with the five SSRs that have been used for Echinochloa since 2002 (Danquah et al., 2002; Nozawa et al., 2006; Lee et al., 2015).
4.3. Potential usage of the SSRs and SNPs identified
A great number of Echinochloa species are aggressive invaders and managing crop lands requires unique strategies for each (Holm et al., 1979, Tabacchi et al., 2006). Thus, correctly identifying Echinochloa spp. is of agronomical and economic importance. The genus Echinochloa contains about 35 species that are widespread in both tropical and temperate regions and in dry or water-flooded soils (Flora of China, 2015). The taxonomy of this genus is complex, and Echinochloa species show wide variability in morphological, biological and physiological features (Danquah et al., 2002, Tabacchi et al., 2006, Vidotto et al., 2007). Conventionally, the identification of Echinochloa species has been attempted taxonomically using morphological assessment of plants, which has frequently been found to be difficult and uncertain (Tabacchi et al., 2006). Moreover, there are different taxonomic key systems for Echinochloa species, which may lead to misidentification (Flora of China, 2015, Tabacchi et al., 2006). Molecular identification of the Echinochloa species is not yet reliable and requires further study (Danquah et al., 2002, Kaya et al., 2014, Tabacchi et al., 2006). In addition, molecular markers may be very useful in studying the origin and distribution of herbicide-resistant populations (Okada et al., 2013, Osuna et al., 2011). SNPs and SSRs are ideal molecular tools for gene location and molecular breeding (Danquah et al., 2002, Gupta et al., 2015, Vandepitte et al., 2013, Zhang et al., 2011).
Acknowledgements
This research was supported by China Postdoctoral Science Foundation (2015M571763) and the Special Fund for Agroscientific Research in the Public Interest of China (201303022). We thank Kui Wu and Zhi-hui Yan (Nanjing Agricultural University, China) for providing helps on plant cultivation. Thanks are also due to the reviewers and editors for their helpful comments and English polish on earlier drafts of the manuscript.
(Editor: Lianming Gao)
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.pld.2017.08.004.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Baird N.A., Etter P.D., Atwood T.S. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchi L., Lanteri S., Portis E. Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J.M., Amores A., Hohenlohe P. Stacks: building and genotyping loci De Novo from short-read sequences. G3 Genes Genomes Genet. 2011;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danquah E.Y., Hanley S.J., Brookes R.C. Isolation and characterization of microsatellites in Echinochloa (L.) Beauv. spp. Mol. Ecol. Notes. 2002;2:54–56. [Google Scholar]
- Flora of China. 2015. www.efloras.org Available from: [Google Scholar]
- Gupta S.K., Baek J., Carrasquilla-Garcia N. Genome-wide polymorphism detection in peanut using next-generation restriction-site-associated DNA (RAD) sequencing. Mol. Breed. 2015;35 [Google Scholar]
- Heap I. 2015. The International Survey of Herbicide Resistant Weeds, 2015.www.weedscience.org Available from: [Google Scholar]
- Holm L.G., Pancho J.V., Herberger J.P. John Wiley and Sons; New York: 1979. A Geographical Atlas of World Weeds. [Google Scholar]
- Kaya H.B., Demirci M., Tanyolac B. Genetic structure and diversity analysis revealed by AFLP on different Echinochloa spp. from northwest Turkey. Plant Syst. Evol. 2014;300:1337–1347. [Google Scholar]
- Kruglyak L. The use of a genetic map of biallelic markers in linkage studies. Nat. Genet. 1997;17:21–24. doi: 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- Lee J., Park K.W., Lee I.Y. Simple sequence repeat analysis of genetic diversity among acetyl-CoA carboxylase inhibitor-resistant and -susceptible Echinochloa crus-galli and E. oryzicola populations in Korea. Weed Res. 2015;55:90–100. [Google Scholar]
- Miller M.R., Dunham J.P., Amores A. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007;17:240–248. doi: 10.1101/gr.5681207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirmans P.G., Van Tienderen P.H. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes. 2004;4:792–794. [Google Scholar]
- Nozawa S., Takahashi M., Nakai H. Difference in SSR variations between japanese barnyard millet (Echinochloa esculenta) and its wild relative. E. crus-galli. Breed. Sci. 2006;56:335–340. [Google Scholar]
- Okada M., Hanson B.D., Hembree K.J. Evolution and spread of glyphosate resistance in Conyza canadensis in California. Evol. Appl. 2013;6:761–777. doi: 10.1111/eva.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjuela J., Garavito A., Bouniol M. A universal core genetic map for rice. Theor. Appl. Genet. 2010;120:563–572. doi: 10.1007/s00122-009-1176-1. [DOI] [PubMed] [Google Scholar]
- Osuna M.D., Okada M., Ahmad R. Genetic diversity and spread of thiobencarb resistant early watergrass (Echinochloa oryzoides) in California. Weed Sci. 2011;59:195–201. [Google Scholar]
- Peakall R., Smouse P.E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raats D., Frenkel Z., Krugman T. The physical map of wheat chromosome 1BS provides insights into its gene space organization and evolution. Genome Biol. 2013;14:19. doi: 10.1186/gb-2013-14-12-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A.N., Johnson D.E., Sivaprasad B. Weed management in direct-seeded rice. In: Donald L.S., editor. Advances in Agronomy. Academic Press; 2007. pp. 153–255. [Google Scholar]
- Šmarda P., Bures P., Horova L. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4096–E4102. doi: 10.1073/pnas.1321152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.T., Wang M.M., Zhang Y.K. Evidence for high dispersal ability and mito-nuclear discordance in the small brown planthopper, Laodelphax striatellus. Sci. Rep. 2015;5:8045. doi: 10.1038/srep08045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabacchi M., Mantegazza R., Spada A. Morphological traits and molecular markers for classification of Echinochloa species from Italian rice fields. Weed Sci. 2006;54:1086–1093. [Google Scholar]
- Talukder Z.I., Gong L., Hulke B.S. A high-density SNP map of sunflower derived from RAD-sequencing facilitating fine-mapping of the rust resistance gene R12. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira H., Rodríguez-Echeverría S., Nabais C. Genetic diversity and differentiation of Juniperus thurifera in Spain and Morocco as determined by SSR. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh S., DeClerck G., Lukashova A. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepitte K., Honnay O., Mergeay J. SNP discovery using Paired-End RAD-tag sequencing on pooled genomic DNA of Sisymbrium austriacum (Brassicaceae) Mol. Ecol. Resour. 2013;13:269–275. doi: 10.1111/1755-0998.12039. [DOI] [PubMed] [Google Scholar]
- Vidotto F., Tesio F., Tabacchi M. Herbicide sensitivity of Echinochloa spp. accessions in Italian rice fields. Crop Prot. 2007;26:285–293. [Google Scholar]
- Wang Y., Yang C., Jin Q.J. Genome-wide distribution comparative and composition analysis of the SSRs in Poaceae. BMC Genet. 2015;16:8. doi: 10.1186/s12863-015-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasue Y. Strategy of Echinochloa oryzicola Vasing. for survival in flooded rice. Weed Biol. Manag. 2001;1:28–36. [Google Scholar]
- Zhang Q., Ma B., Li H. Identification, characterization, and utilization of genome-wide simple sequence repeats to identify a QTL for acidity in apple. BMC Genomics. 2012;13 doi: 10.1186/1471-2164-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zalapa J., Jakubowski A. Post-glacial evolution of Panicum virgatum: centers of diversity and gene pools revealed by SSR markers and cpDNA sequences. Genetica. 2011;139:933–948. doi: 10.1007/s10709-011-9597-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.