Abstract
MicroRNAs (miRNAs) are conserved small non-coding RNAs that play an important role in the regulation of gene expression and participate in a variety of biological processes. The biogenesis of miRNAs is tightly controlled at multiple steps, such as transcription of miRNA genes, processing by Drosha and Dicer, and transportation of precursor miRNAs (pre-miRNAs) from the nucleus to the cytoplasm by exportin-5 (XPO5). Given the critical role of nuclear export of pre-miRNAs in miRNA biogenesis, any alterations of XPO5, resulting from either genetic mutation, epigenetic change, abnormal expression level or posttranslational modification, could affect miRNA expression and thus have profound effects on tumorigenesis. Importantly, XPO5 phosphorylation by ERK kinase and its cis/trans isomerization by the prolyl isomerase Pin1 impair XPO5′s nucleo-to-cytoplasmic transport ability of pre-miRNAs, leading to downregulation of mature miRNAs in hepatocellular carcinoma. In this review, we focus on how XPO5 transports pre-miRNAs in the cells and summarize the dysregulation of XPO5 in human tumors.
Keywords: Exportin-5, MicroRNA, Biogenesis, Dysregulation, Cancer
Introduction
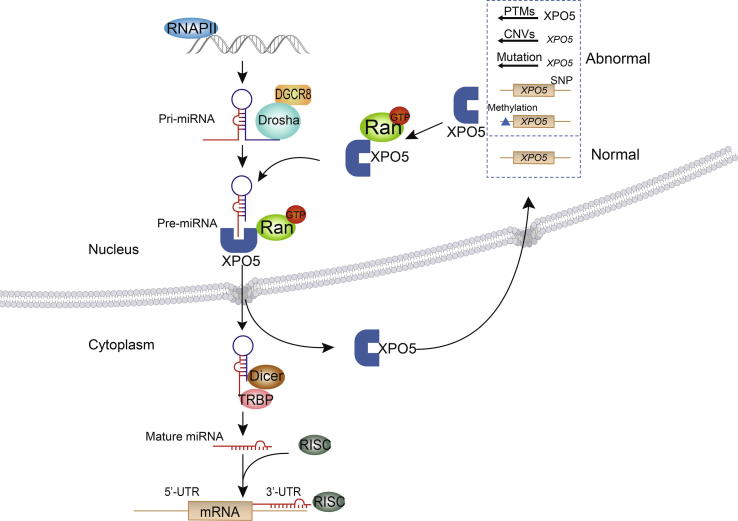
MicroRNAs (miRNAs) are endogenous small non-coding RNAs, which are about 20–22 nucleotides (nt) in length and evolutionarily conserved among different species. Until now, 38,589 miRNA molecules in 271 species have been identified, including 1982 human precursor-miRNAs (pre-miRNAs) and 2694 human mature miRNAs (Release 22, http://www.mirbase.org). The miRNA biogenesis in mammals involves multiple stages (Figure 1). First, miRNA genes are transcribed, usually by RNA polymerase II (RNAPII), into primary transcripts (pri-miRNAs), which are 5′-capped and 3′-polyadenylated and contain imperfect hairpin structures. Then, the pri-miRNAs are cleaved by Drosha/DiGeorge syndrome chromosomal region 8 (DGCR8) complex into the pre-miRNAs with a ∼70-nt stem-loop structure. Following nuclear transport by GTP-binding nuclear protein Ran (RanGTP)/exportin-5 (XPO5) complex to the cytoplasm, the pre-miRNAs are processed by Dicer to produce the mature miRNAs. The mature miRNAs are then ready to be incorporated into the RNA induced silencing complex (RISC) and regulate gene expression through translational repression or mRNA degradation [1].
Figure 1.
The schematic diagram of miRNA biogenesis
miRNA genes are transcribed into pri-miRNAs by RNAPII. Then, the pri-miRNAs are cleaved by Drosha/DGCR8 complex into the pre-miRNAs. Following nuclear transport by Ran/GTP/XPO5 complex to the cytoplasm, the pre-miRNAs are processed by Dicer to produce mature miRNAs, which are ready to be incorporated into RISCs and regulate gene expression. Abnormality of XPO5 or the encoded protein would affect the process of pre-miRNAs transport. RNAPII, RNA polymerase II; RISC, RNA induced silencing complex; XPO5, exportin-5; DGCR8, DiGeorge syndrome chromosomal region 8; PTM, post-translational modification; CNV, copy number variation; SNP, single nucleotide polymorphism.
Although initially discovered in Caenorhabditis elegans, miRNAs have been a hot topic in biomedical field since Croce’s group for the first time revealed the frequent deletions and downregulation of MIR-15 and MIR-16 genes in human B cell chronic lymphocytic leukemia [2]. Moreover, human MIR genes are found to be frequently located at fragile sites and genomic regions that are involved in cancers. For example, MIR genes for miR-29a, miR-29b, miR-96, miR-182s, miR-182as, miR-183, and miR-129-1 were found in or close to the fragile site FRA7H [3]. High-throughput technologies have demonstrated that compared to normal tissues, there is a general downregulation of miRNA expression in multiple human tumors, which may be caused by defects in the miRNA biogenesis machinery [4], [5]. For instance, the expression of both Drosha and Dicer proteins, two key components of miRNA biogenesis, is significantly decreased in ovarian cancer specimens, and their expression levels are correlated with outcomes in patients with ovarian cancer [6]. Given that pre-miRNA transportation has been recently demonstrated to be critical for the generation of mature miRNAs, the impact of XPO5 on miRNA biogenesis and its role in tumorigenesis have attracted considerable attention in the field.
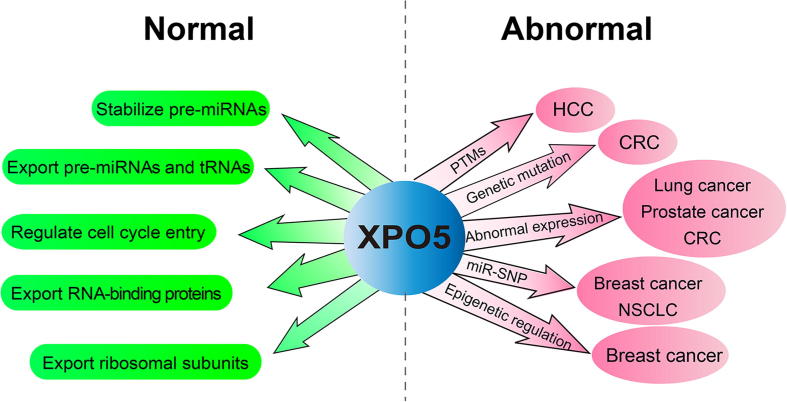
Increasing evidence indicates that XPO5 plays an important role in nuclear export of various substrates (Figure 2). During cell cycle entry, XPO5 is promptly induced to increase global miRNA expression through a PI3K-dependent post-transcriptional mechanism. Inhibition of XPO5 induction leads to a proliferation defect, which is associated with delayed G1/S transition [7]. Therefore, XPO5 acts as a critical molecular hub to control gene expression through regulating miRNA expression. Besides pre-miRNAs, XPO5 has been demonstrated to be involved in the export of some RNA-binding proteins (RBPs), such as Staufen homolog 2 (STAU2) and interleukin enhancer-binding factor 3 (ILF3), dependent on its double-stranded RNA-binding domain [8], [9], [10]. In addition, XPO5 is also shown to be required for nuclear export of 60S ribosomal subunit in a RanGTP-dependent process [11]. In this review, we briefly describe the role of XPO5 in miRNA biogenesis and discuss its dysregulation in cancer.
Figure 2.
Cellular function of XPO5
XPO5 participates in different cellular biological processes and is associated with various cancers. HCC, hepatocellular carcinoma; CRC, colorectal cancer; NSCLC, non-small cell lung cancer.
Role of XPO5 in miRNA biogenesis
Exportin belongs to the karyopherin β family of transport factors, which mediate protein shuttling from the nucleus to the cytoplasm by recognizing specific transport signals in the cargo proteins. XPO5 is one of nucleo-cytoplasmic exportins and uses Ran-GTPase to control cargo association. XPO5 binds to pre-miRNAs at high Ran-GTP levels in the nucleus and transports the pre-miRNAs through the nuclear pore complex into the cytoplasm, where the pre-miRNAs are released from the Ran/GTP/XPO5 complex upon GTP hydrolysis. Subsequently, the free XPO5 in the cytoplasm returns to the nuclear compartment and mediates another round of pre-miRNA transport [12]. Since XPO5 is responsible for transporting pre-miRNAs from the nucleus to the cytoplasm, overexpression of XPO5 is found to improve transport efficiency and further enhance mature miRNA expression [13]. Therefore, XPO5-mediated nuclear export of pre-miRNAs could be a rate-limiting step during miRNA biogenesis.
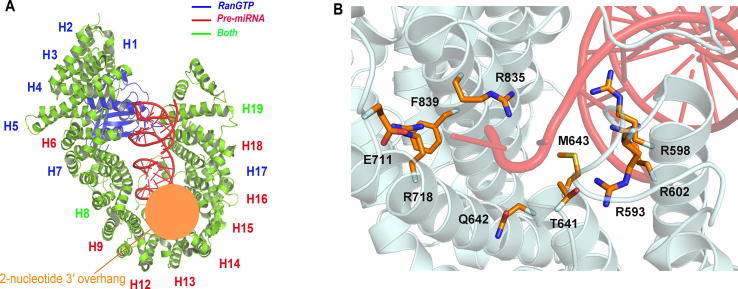
Using X-ray technology, Okada et al. has reported a 2.9 angstrom structure of the nuclear export machinery formed by pre-miRNA with complex of XPO5 and RanGTP [14]. XPO5 recognizes the double-stranded stem structure of the pre-miRNAs via the XPO5 tunnel-like structure comprising HEAT repeats (a tandem repeat protein structural motif composed of two alpha helices linked by a short loop) 8, 9, 12-16, 18, and 19. In detail, the tunnel-like structure of XPO5, shaped by R593, R598, R602, T641, Q642, M643, E711, R718, R835, and F839, interacts with 3′ overhang of a pre-miRNA via hydrogen bonds or salt bridges, protecting the pre-miRNA stem from hydrolysis by nucleases (Figure 3). Since the RNA binding by XPO5/RanGTP is not sequence-dependent, it is suggested that this nuclear export machinery can recognize a variety of pre-miRNAs [14].
Figure 3.
The structure of the XPO5:RanGTP:pre-miRNA complex
A. Structure showing human pre-miRNA (red) bound to XPO5 (green) with RanGTP (blue). The stem moiety of a pre-miRNA is mostly covered by XPO5, and the 2-nt 3′overhang structure (highlighted in the orange circle) has many interactions with HEAT repeats 12-15 of XPO5. HEAT repeats 1-19 are indicated with a letter “H” and the corresponding numbers, which are shown in different colors based on their interacting partners (blue: interacting with RanGTP; red: interacting with pre-miRNA; green: interacting with both RanGTP and pre-miRNA). B. Intermolecular interaction details of the 2-nt 3′overhang structure of pre-miRNA(red) with HEAT repeats 12-15 of XPO5 (green). XPO5 PDB ID: 3A6P.
Other than transporting pre-miRNAs from the nucleus to the cytoplasm, XPO5 also mediates nuclear export of eukaryotic elongation factor 1A (eEF1A) via aminoacylated tRNAs [15]. Intriguingly, XPO5 has been recently shown to regulate the expression of Dicer, an RNase III required for pre-miRNA maturation [16]. Mechanistically, XPO5 directly interacts with human Dicer mRNA, and knockdown of XPO5 leads to an increased nuclear accumulation of Dicer mRNA in HeLa cells, thus decreasing Dicer protein expression [16]. Therefore, XPO5 plays an important role in miRNA biogenesis through both exporting pre-miRNAs and modulating Dicer expression.
Dysregulation of XPO5 in cancers
Nuclear export of pre-miRNAs is accurately regulated in normal cells and its dysregulation could cause abnormal expression of mature miRNAs in cancer cells. Strikingly, more pre-miRNAs are found to be retained in the nucleus of both cancer cells and tumors, when compared with normal cells and normal tissues [17]. Using real-time PCR, Lee et al have profiled the expression of 225 pre-miRNAs and mature miRNAs in 22 different human tissues, 37 human cancer cell lines, as well as 16 pancreas and liver tissues/tumors. They find that a large number of MIR genes are transcribed and processed into pre-miRNAs, but not processed to mature miRNAs in cancer cells, indicating defects in the pre-miRNA nuclear export by XPO5 in human tumors [17]. The abnormal function of XPO5 could be caused by genetic or epigenetic change of XPO5 as well as abnormal expression level or post-translational modifications (PTMs) of XPO5 protein (Table 1).
Table 1.
The role of dysregulated XPO5 in different cancers
| Type | Description | Mechanism | Diseases | Refs. |
|---|---|---|---|---|
| PTMs | Phosphorylation at T345/S416/S497 and cis/trans isomerization | Impair pre-miRNA export | HCC | [18], [19] |
| Genetic mutation | Generation of truncated XPO5 protein | Impair pre-miRNA export | CRC | [20] |
| Abnormal expression | Increase or decrease in XPO5 expression | Cause abnormal expression of mature miRNAs | Prostate adenocarcinoma, lung adenocarcinoma, CRC | [21], [22], [23], [24] |
| miR-SNP | Impact on individual’s response to certain drugs and risk of particular diseases | Unknown | Breast cancer, NSCLC | [25], [26], [27], [28] |
| Epigenetic regulation | Decrease in methylation of XPO5 promoter region | Promote transcription | Breast cancer | [25] |
Note: PTM, post-translational modification; SNP, single nucleotide polymorphism; HCC, hepatocellular carcinoma; CRC, colorectal cancer; NSCLC, non-small cell lung cancer.
Abnormal expression of XPO5 protein
Abnormal expression of XPO5 protein has been found in human cancers. For example, compared to normal tissues, XPO5 expression is downregulated in bronchioloalveolar carcinoma and stage I adenocarcinoma [21], whereas it is about 1.6-fold upregulated in prostate adenocarcinoma [22], especially in the metastatic patients [23].
The alterations in XPO5 protein level lead to dysregulation of mature miRNA expression. Recently, XPO5 is found to act as an oncoprotein in colorectal cancer (CRC) due to its high expression in CRC and anti-tumor effect after XPO5 knockdown [24]. Shigeyasu et al. reported an upregulated expression of XPO5 at both mRNA and protein levels in CRC samples when compared to normal tissues. Moreover, high XPO5 expression is associated with worse clinicopathological features and poor survival in CRC patients [24]. In SW480 and Caco-2 CRC cells, XPO5 knockdown decreased the expression of key oncogenic miRNAs, such as miR-21, miR-10b, miR-27a, miR-92a, miR-182, and miR-155, resulting in decreased cell proliferation and invasion. In a xenograft animal model, silencing XPO5 also significantly attenuates tumor growth [24]. Similarly, XPO5 expression is increased in breast tumors and urothelial carcinoma of the bladder [25], [29]. Therefore, XPO5 could act as an oncoprotein and may be a potential therapeutic target in certain cancers.
Interestingly, it is found that XPO5 expression can be regulated by miRNAs. For example, Li et al. report that overexpression of miR-138, a tumor suppressor miRNA, reduces XPO5 levels through downregulating the expression of required for meiotic nuclear division 5 homolog A (RMND5A), which is responsible for XPO5 stability [30]. Expression of miR-138 is often downregulated in tumors, such as head and neck squamous cell carcinoma, and nasopharyngeal carcinoma [31], [32], which may explain why XPO5 expression is upregulated in certain tumors.
XPO5 protein PTMs
Protein PTMs, such as acetylation and phosphorylation, are considered as essential mechanisms to diversify the protein functions and dynamically coordinate their signaling networks in eukaryotic cells. Defects in PTMs of some proteins have been demonstrated in human diseases [33]. For the first time, Sun et al. discovered that XPO5 predominantly locates in the nucleus of hepatocellular carcinoma (HCC) cells, whereas in normal tissues it is distributed in both nucleus and cytoplasm [18]. When ERK kinase is activated in HCC cells, XPO5 is phosphorylated at T345/S416/S497 sites, which usually stay unphosphorylated in normal hepatocytes. Moreover, the prolyl isomerase Pin1 interacts with the phosphorylated XPO5 and mediates the change of its protein conformation through cis/trans isomerization, thus impairing the nuclear export of pre-miRNAs and leading to the retention of XPO5 in the nucleus [19]. As a consequence, such PTMs of XPO5 cause the decreased miRNA expression in HCC cells. These studies provide a novel mechanism to explain the downregulation of global miRNA expression in HCC. Interestingly, using the computer-aided drug design, Pu et al. identify a highly specific Pin1 inhibitor, API-1, and demonstrate that API-1 can effectively inhibit HCC growth both in vitro and in vivo [34]. Mechanistically, Pin1 inhibition by API-1 can upregulate miRNA biogenesis via retaining active XPO5 conformation, and suppress HCC development, indicating API-1 as a novel drug candidate for HCC therapy [34].
Besides phosphorylation, acetylation is identified at K396 site of XPO5 protein [35]. But the function of XPO5 acetylation still remains unknown.
Genetic mutations of XPO5 gene
Inactivating mutations in XPO5 gene have been detected in a subset of human tumors with microsatellite instability, such as hereditary nonpolyposis colon cancer (HNPCC) [20]. In both HCT-15 and DLD-1 CRC cells, insertion of a single nucleotide “A” within XPO5 gene generates a premature termination codon in exon 32 of XPO5 mRNA. This frameshift mutation leads to the production of truncated proteins lacking the C-terminal region [20]. Because the C-terminal region of XPO5 is essential for the formation of pre-miRNA/XPO5/Ran-GTP ternary complex, the truncated XPO5 proteins can’t bind to and transport pre-miRNAs, thus leading to a nuclear accumulation of pre-miRNAs and a reduction in mature miRNA expression [20]. Interestingly, expression of wild-type XPO5 in these cells reverses the impaired export of pre-miRNAs and inhibits cancer cell growth, suggesting that XPO5 may function as a tumor suppressor in CRCs [20]. As mentioned above, XPO5 could also function as an oncoprotein in CRCs [24]. Therefore, whether XPO5 function as a tumor suppressor or oncoprotein in CRCs is still debatable and needs further investigation.
XPO5 SNPs and CNVs
Single nucleotide polymorphism (SNP) is the most common type of genetic variations, which could be used to predict an individual’s response to certain drugs and risk of particular diseases [26]. Increasing evidence reveals the presence of SNPs in XPO5 gene. For example, rs11544382 (M1115T) and rs34324334 (S241N), two missense SNPs, are reported in XPO5 gene. The variant genotypes of rs11544382 are significantly associated with high breast cancer risk when compared to the common homozygous genotype [25]. Moreover, rs11077 (GRCh38.p7 chr6:43523209), a potential miRNA-associated SNP (miR-SNP) located in the 3′UTR of XPO5 gene, has been extensively studied in various tumors. For example, rs11077 is associated with a better chemotherapeutic response in patients with metastatic colon cancer (AC genotype) [27]. The AC + CC genotypes of rs11077 are significantly associated with better survival and lower recurrence in NSCLC [28], whereas AA genotype of rs11077 is associated with better overall survival of SCLC patients [36]. Furthermore, multivariate analysis indicates that miR-SNP rs11077 of XPO5 gene is an independent prognostic marker to predict survival for advanced SCLC and NSCLC patients [36], [37]. Other studies have also reported that patients with AA genotype of rs11077 exhibit an increased risk of esophageal cancer and gastric cancer [38], [39], and are associated with short overall survival in multiple myeloma (after autologous stem cell transplantation), Hodgkin’s lymphoma, and renal cell carcinoma [40], [41], [42]. The rs11077 associated miRNAs need further investigation.
Copy number variations (CNVs) are usually categorized into two main groups: short repeats and long repeats. Duan et al. comprehensively analyze SNPs and CNVs of the genes involved in miRNA processing, and find that there exist CNVs within the XPO5 gene (Entrez ID:57510, Gene location(hg18):chr6:43598050-43651642(−), CNV ID:Variation_9530, CNV position(hg18):chr6:43583452-43604640, Observed CNVs:1 gain) [43]. In mammals, CNVs are the primary contributor to genome variability, thus modifying the diploid status of DNA [44]. To date, there is no direct evidence supporting that CNVs of XPO5 are associated with any human diseases.
Epigenetic regulation of XPO5
Epigenetic modifications including DNA methylation and histone modification are generally altered during tumorigenesis, which may trigger resistance to immune surveillance, chemotherapy, and targeted drugs [45], [46]. DNA hypermethylation at the CpG islands within gene promoter usually leads to gene silencing. Leaderer et al. have reported the decreased methylation in the CpG island spanning the promoter region, the first exon, and part of the first intron of the XPO5 gene (−600 to 808), which may explain why XPO5 expression is upregulated in breast cancer [25]. Whether RNA modification or histone acetylation could affect XPO5 gene expression still remains elusive.
Concluding remarks
Nuclear export of pre-miRNA by XPO5 is a critical step for miRNA biogenesis, so XPO5 expression requires accurate regulation to maintain its normal function under physiological conditions. Recently, genetic or epigenetic change of XPO5 gene as well as abnormal expression level or PTMs of XPO5 protein, potentially leading to abnormal expression of mature miRNAs, have been found to be associated with certain tumors. However, the underlying mechanisms still remain elusive and need further investigation. For example, XPO5 expression is upregulated in CRCs and prostate cancer, but downregulated in lung adenocarcinoma. It seems that XPO5 may exert its function depending on tissue-specific expression of miRNAs. Such issues could be addressed by deep sequencing and proteomics technologies. Moreover, whether XPO5 functions as a tumor suppressor or oncoprotein in certain cancers should be investigated in animal model. Based on the findings that XPO5 can transport certain tRNAs and proteins besides pre-miRNAs, discovery of other XPO5 substrates could be helpful to understand the role of XPO5 in different tumors. Although PTMs of XPO5 such as acetylation [35] have been identified, their functional implications still remain unclear. Therefore, further investigations are needed to dissect the role of XPO5 during tumorigenesis and its underlying molecular mechanisms.
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81572739 and 81772960) awarded to YP.
Handled by Yangming Wang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Peng Y., Croce C.M. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Merritt W.M., Lin Y.G., Han L.Y., Kamat A.A., Spannuth W.A., Schmandt R. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasaki Y.W., Kiga K., Kayo H., Fukuda-Yuzawa Y., Weise J., Inada T. Global microRNA elevation by inducible Exportin 5 regulates cell cycle entry. RNA. 2013;19:490–497. doi: 10.1261/rna.036608.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leisegang M.S., Martin R., Ramírez A.S., Bohnsack M.T. Exportin T and exportin 5: tRNA and miRNA biogenesis – and beyond. Biol Chem. 2012;393:599–604. doi: 10.1515/hsz-2012-0146. [DOI] [PubMed] [Google Scholar]
- 9.Macchi P., Brownawell A.M., Grunewald B., DesGroseillers L., Macara I.G., Kiebler M.A. The brain-specific double-stranded RNA-binding protein Staufen2: nucleolar accumulation and isoform-specific exportin-5-dependent export. J Biol Chem. 2004;279:31440–31444. doi: 10.1074/jbc.C400226200. [DOI] [PubMed] [Google Scholar]
- 10.Gwizdek C., Ossareh-Nazari B., Brownawell A.M., Evers S., Macara I.G., Dargemont C. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem. 2004;279:884–891. doi: 10.1074/jbc.M306808200. [DOI] [PubMed] [Google Scholar]
- 11.Wild T., Horvath P., Wyler E., Widmann B., Badertscher L., Zemp I. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi R., Doehle B.P., Qin Y., Macara I.G., Cullen B.R. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada C., Yamashita E., Lee S.J., Shibata S., Katahira J., Nakagawa A. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 15.Bohnsack M.T., Regener K., Schwappach B., Saffrich R., Paraskeva E., Hartmann E. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennasser Y., Chable-Bessia C., Triboulet R., Gibbings D., Gwizdek C., Dargemont C. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat Struct Mol Biol. 2011;18:323–327. doi: 10.1038/nsmb.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee E.J., Baek M., Gusev Y., Brackett D.J., Nuovo G.J., Schmittgen T.D. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun H.L., Cui R., Zhou J., Teng K.Y., Hsiao Y.H., Nakanishi K. ERK activation globally downregulates miRNAs through phosphorylating exportin-5. Cancer Cell. 2016;30:723–736. doi: 10.1016/j.ccell.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Pu W., Sun H.L., Zhou J.K., Fan X., Zheng Y. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ. 2018 doi: 10.1038/s41418-018-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melo S.A., Moutinho C., Ropero S., Calin G.A., Rossi S., Spizzo R. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Chiosea S., Jelezcova E., Chandran U., Luo J., Mantha G., Sobol R.W. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 22.Chiosea S., Jelezcova E., Chandran U., Acquafondata M., McHale T., Sobol R.W. Up-regulation of dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varambally S., Yu J., Laxman B., Rhodes D.R., Mehra R., Tomlins S.A. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Shigeyasu K., Okugawa Y., Toden S., Boland C.R., Goel A. Exportin-5 functions as an oncogene and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:1312–1322. doi: 10.1158/1078-0432.CCR-16-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leaderer D.H.A., Zheng T., Fu A., Weidhaas J., Paranjape T., Zhu Y. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int J Mol Epidemiol Genet. 2011;2:9–18. [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein D.B., Ahmadi K.R., Weale M.E., Wood N.W. Genome scans and candidate gene approaches in the study of common diseases and variable drug responses. Trends Genet. 2003;19:615–622. doi: 10.1016/j.tig.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Boni V., Zarate R., Villa J.C., Bandres E., Gomez M.A., Maiello E. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics J. 2011;11:429–436. doi: 10.1038/tpj.2010.58. [DOI] [PubMed] [Google Scholar]
- 28.Ding C., Li C., Wang H., Li B., Guo Z. A miR-SNP of the XPO5 gene is associated with advanced non-small-cell lung cancer. Onco Targets Ther. 2013;6:877–881. doi: 10.2147/OTT.S48284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y., Liu Y., Gui Y., Cai Z. Inducing cell proliferation inhibition and apoptosis via silencing Dicer, Drosha, and Exportin 5 in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:201–205. doi: 10.1002/jso.23214. [DOI] [PubMed] [Google Scholar]
- 30.Li J., Chen Y., Qin X., Wen J., Ding H., Xia W. miR-138 downregulates miRNA processing in HeLa cells by targeting RMND5A and decreasing Exportin-5 stability. Nucleic Acids Res. 2014;42:458–474. doi: 10.1093/nar/gkt839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Wang C., Chen Z., Jin Y., Wang Y., Kolokythas A. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Lv X.B., Wang X.P., Sang Y., Xu S., Hu K. miR-138 suppressed nasopharyngeal carcinoma growth and tumorigenesis by targeting the CCND1 oncogene. Cell Cycle. 2012;11:2495–2506. doi: 10.4161/cc.20898. [DOI] [PubMed] [Google Scholar]
- 33.Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol. 2018;19:59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 34.Pu W., Li J., Zheng Y., Shen X., Fan X., Zhou J.K. Targeting Pin1 by inhibitor API-1 regulates microRNA biogenesis and suppresses hepatocellular carcinoma development. Hepatology. 2018 doi: 10.1002/hep.29819. [DOI] [PubMed] [Google Scholar]
- 35.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 36.Guo Z., Wang H., Li Y., Li B., Li C., Ding C. A microRNA-related single nucleotide polymorphism of the XPO5 gene is associated with survival of small cell lung cancer patients. Biomed Rep. 2013;1:545–548. doi: 10.3892/br.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geng J.Q., Wang X.C., Li L.F., Zhao J., Wu S., Yu G.P. MicroRNA-related single-nucleotide polymorphism of XPO5 is strongly correlated with the prognosis and chemotherapy response in advanced non-small-cell lung cancer patients. Tumour Biol. 2016;37:2257–2265. doi: 10.1007/s13277-015-3980-3. [DOI] [PubMed] [Google Scholar]
- 38.Ye Y., Wang K.K., Gu J., Yang H., Lin J., Ajani J.A. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Y., Wang Y., Zhao Y., Guo Z. Single-nucleotide polymorphisms of microRNA processing machinery genes are associated with risk for gastric cancer. Onco Targets Ther. 2015;8:567–571. doi: 10.2147/OTT.S79150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Larrea C.F., Navarro A., Tejero R., Tovar N., Diaz T., Cibeira M.T. Impact of miRSNPs on survival and progression in patients with multiple myeloma undergoing autologous stem cell transplantation. Clin Cancer Res. 2012;18:3697–3704. doi: 10.1158/1078-0432.CCR-12-0191. [DOI] [PubMed] [Google Scholar]
- 41.Dzikiewicz-Krawczyk A. MicroRNA polymorphisms as markers of risk, prognosis and treatment response in hematological malignancies. Crit Rev Oncol Hematol. 2015;93:1–17. doi: 10.1016/j.critrevonc.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Horikawa Y., Wood C.G., Yang H., Zhao H., Ye Y., Gu J. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan S., Mi S., Zhang W., Dolan M.E. Comprehensive analysis of the impact of SNPs and CNVs on human microRNAs and their regulatory genes. RNA Biol. 2014;6:412–425. doi: 10.4161/rna.6.4.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 45.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 46.Jones P.A., Issa J.P., Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]