Abstract
Precision oncology aims to offer the most appropriate treatments to cancer patients mainly based on their individual genetic information. Genomics has provided numerous valuable data on driver mutations and risk loci; however, it remains a formidable challenge to transform these data into therapeutic agents. Transcriptomics describes the multifarious expression patterns of both mRNAs and non-coding RNAs (ncRNAs), which facilitates the deciphering of genomic codes. In this review, we take breast cancer as an example to demonstrate the applications of these rich RNA resources in precision medicine exploration. These include the use of mRNA profiles in triple-negative breast cancer (TNBC) subtyping to inform corresponding candidate targeted therapies; current advancements and achievements of high-throughput RNA interference (RNAi) screening technologies in breast cancer; and microRNAs as functional signatures for defining cell identities and regulating the biological activities of breast cancer cells. We summarize the benefits of transcriptomic analyses in breast cancer management and propose that unscrambling the core signaling networks of cancer may be an important task of multiple-omic data integration for precision oncology.
Keywords: Precision oncology, Transcriptomics, RNA interference, microRNA, Breast cancer
Introduction
The fundamental mission of precision medicine is to confer the most appropriate management to patients within an appropriate time based on the clinical and molecular characteristics of their diseases [1], [2], [3]. The Precision Medicine Initiative was proposed in 2015, which consists of two main objectives, i.e., a short-term goal aimed to improve cancer management, and a long-term vision promised to provide a better and healthier quality of life [4]. Oncology is considered to be “the clear choice for enhancing the near-term impact of precision medicine” [5]. Recent advancements in sequencing technologies and big data analytics have provided an unprecedented insight into the detailed molecular information of different tumor types, consequently promoting the development of potential targeted therapies and the innovation of clinical-trial strategies [6], [7], [8], [9], [10]. Comprehensive transcriptomic analyses present a global view of the RNA-based variants and contribute to the decoding of genomic data into actual gene expression patterns [11]. Transcriptomics is now regarded as an impactful approach to improving the application of genomic information to the identification, confirmation, evaluation and implementation in precision medicine exploration. In this review, we use breast cancer as a model to summarize the groundbreaking advancements and achievements of transcriptomics in cancer management in recent years.
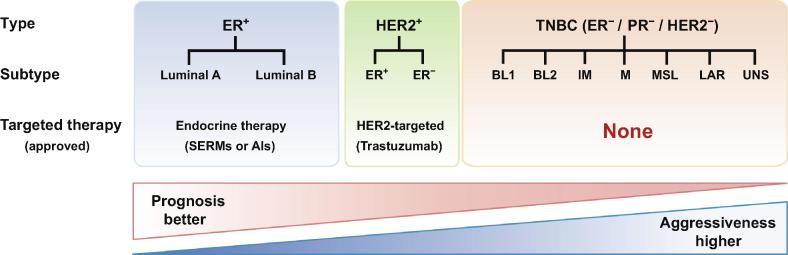
Breast cancer is the most common malignant tumor in women [12]. This extremely heterogeneous disease is clinically classified into three types (Figure 1), mainly depending on the expression status of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2). The ER+ group is the most common type of breast cancers (accounting for approximately 70% of all breast cancer cases), and endocrine therapies with selective ER modulators (SERMs) or aromatase inhibitors (AIs) have been adopted as the standard adjuvant treatments for ER+ tumors [13], [14]. The HER2-overexpressed group has achieved favorable therapeutic effects from the humanized monoclonal antibody trastuzumab that targets HER2 [15], [16]. Triple-negative breast cancer (TNBC) represents a distinctive subset of breast cancers with neither ER/PR expression nor HER2 amplification [17], [18]. TNBC accounts for approximately 15% of all types of breast cancers and is more malignant than the ER+ or HER2 highly-amplified breast cancers [18], [19], [20], [21], [22]. Current treatment of TNBC largely relies on chemotherapy and radiation therapy, with no targeted drugs approved for TNBC yet.
Figure 1.
Schema of the clinical classifications of breast cancers and the corresponding targeted therapies approved
Breast cancers are clinically classified into three types: ER+, HER2+, and TNBC, according to the expression status of ER, PR, and HER2, which can be further divided into several subtypes as illustrated. TNBC is associated with the worst prognosis and is more aggressive than the other two types, and currently there are no targeted agents approved for TNBC yet. ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer; SERM, selective estrogen receptor modulator; AI, aromatase inhibitor; BL, basal-like; IM, immunomodulatory; M, mesenchymal; MSL, mesenchymal stem-like; LAR, luminal androgen receptor; UNS, unstable.
Genomic architecture of breast cancer
The delightful advancements in whole-genome sequencing (WGS) technologies have provided an exhaustive description of the genomic landscape of breast cancer that includes rich information on DNA copy number aberrations (CNAs), driver mutations, and single nucleotide polymorphisms (SNPs) [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44]. A large number of CNAs, particularly deletions in PPP2R2A, MTAP, and MAP2K4 genes, have been identified in primary breast tumors [28]. Using the highly multiplexed single-nucleus sequencing approach, a study involving 1000 single cells from 12 TNBC patients reveals that most CNAs are detected as early as the onset of breast cancer [35]. Inactivating mutations of BRCA1 and BRCA2 frequently occur in breast cancer as well [30], [36], [42], while unique mutations in GATA3, PIK3CA, and MAP3K1 are enriched in the luminal A subtype of breast cancer [25], [45]. By analyzing the WGS data from 560 breast cancer samples, Nik-Zainal et al. further find numerous mutations in protein-coding genes [36]. TNBC exhibits a higher mutation rate than those observed in ER+ and HER2+ breast cancers, particularly in TP53, and an enrichment of the MAGI3–AKT3 fusion is also detected in TNBC [24]. Notably, Ding et al. discover that the metastatic breast cancer shares 20 mutations with the primary tumor [23]. Yates et al. further confirm that the majority of mutations detected in the metastatic samples are similar to those present in the primary breast tumors, indicating that the metastatic clones probably arise from the primary tumors [38]. Additionally, two ESR1 mutations (ESR1Y537C and ESR1Y537S) occur after the acquisition of endocrine resistance in response to long-term estrogen deprivation (LTED) [41]. In the last decade, genome-wide association studies (GWAS) have also discovered a series of novel breast cancer risk loci [29], [34], [39], [46], [47], [48], [49], [50]. These GWAS-identified loci contain abundant non-coding SNPs that could alter transcription factor (TF) binding sites and confer breast cancer-specific phenotypic variations [39], [51]. All of these studies have shed light on genetic susceptibilities to breast cancer and facilitated improvements in the prediction and assessment of breast cancer.
Genomic profiling has provided tremendously valuable information on genetic vulnerabilities in breast cancer; however, certain limitations remain. First, DNA variations may not reveal the actual activities of the corresponding biological pathways. In some cases, the master signaling pathways may be deregulated without any observed genomic alterations, and such cases are probably ignored in genomic analyses. Furthermore, even though the genetic variants have been discovered in genomics, it is often difficult to discriminate between “passenger” and “driver” mutations. Functional genomics needs to be replenished by large-scale data derived from other omic platforms. Transcriptomics is the most frequently used method for unscrambling genomic information [10], since it can comprehensively reflect the expression patterns of different kinds of RNAs [52], and is widely applied to investigate the genes that are differentially expressed under specific physiological or pathological conditions [53].
Novel insights into breast cancer arising from transcriptomic analyses
Technological advancements in transcriptome profiling
Transcriptomic analyses have been frequently utilized for exploring prospective biomarkers and potential therapeutic targets for human cancers [53]. Microarray analysis is useful in measuring the gene expression levels via complementary probe hybridization [54], [55], [56] and through which a large number of breast cancer-related genes have been discovered [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69]. Furthermore, the wide application of RNA sequencing (RNA-seq) technologies has greatly expanded our knowledge of breast cancer [27]. Using RNA-seq, we can quantify genes that are expressed at extremely low levels and hence might be neglected when using microarrays [70], [71], [72], [73], [74]. RNA-seq also facilitates the analysis of specific fusion transcripts that are usually enriched in TNBC [24], [75], [76], [77], [78], [79]. More importantly, through manipulating RNA isolation prior to RNA-seq experiments, the transcriptome of specific ncRNA species, e.g., microRNAs (miRNAs), can be enriched for better coverage [80], [81]. The applications of the transcriptomics to breast cancer studies are discussed below in more detail.
Utility of mRNA expression patterns in TNBC subtyping
TNBC is exceedingly heterogeneous, poorly differentiated and highly metastatic [17]. It frequently afflicts young women and is associated with poor prognosis. Due to the lack of ER, PR, and HER2 expression, TNBC is insensitive to hormonal therapies. Therefore, conventional cytotoxic chemotherapy and radiation therapy remain the mainstay of treatment for TNBC, whereas the outcomes are far from satisfactory [12], [13], [14], [18], [19], [20], [22], [82], [83]. Efficacious targeted strategies are thus in urgent need for TNBC patients. Revising the sub-classification of TNBC into distinct molecular subtypes with unique transcriptional features is helpful for therapeutic decision-making and prognostic prediction [82], [83], [84], [33], [85], [86], [87], [88], [89].
Using massively parallel mRNA sequencing, numerous transcripts that are differentially expressed between TNBC and non-TNBC have been identified [83]. Based on the comprehensive transcriptomic analysis of 21 breast cancer datasets, Lehmann et al. classify TNBC into seven subtypes [82]. These include two basal-like subtypes (BL1 and BL2), an immunomodulatory subtype (IM), a mesenchymal subtype (M), a mesenchymal stem-like subtype (MSL), a luminal androgen receptor subtype (LAR), and an unclassified set that is regarded as unstable (UNS) (Figure 1 and Table 1). The BL1 subtype strongly expresses specific genes that are related to cell proliferation and DNA damage response. It preferentially responds to cisplatin and poly (ADP-ribose) polymerase (PARP) inhibitors. The BL2 subtype is enriched with genes that are associated with growth factor pathways, indicating that growth factor inhibitors may be efficacious for the BL2 subtype. The IM subtype possesses abundant genes that are involved in immune-mediated reactions, and programmed cell death 1/programmed death-ligand 1 (PD1/PDL1) inhibitors are anticipated to be a hopeful therapeutic option for this subtype. Both the M and MSL subtypes specifically express genes that are relevant to cell motility, cellular differentiation, and growth factor pathways, while the MSL subtype expresses lower levels of proliferation genes than those present in the M subtype. The mammalian target of rapamycin (mTOR) inhibitors and epithelial-to-mesenchymal transition (EMT)-targeted agents are candidate drugs for these two subtypes. The LAR subtype is named for the AR enrichment, and anti-androgen treatments (e.g., bicalutamide, an AR antagonist) are undergoing clinical trials [82], [84], [89].
Table 1.
Transcriptomic subtypes of triple-negative breast cancer and potential therapeutic agents
| Subtype | Percentage (%) | Cell line models | Unique pathways | Potential agents |
|---|---|---|---|---|
| BL1 | 17 | HCC2157, HCC1599, HCC1937, HCC1143, HCC3153, HCC38, MDA-MB-468 | Cell cycle DNA damage response Proliferation genes |
PARP inhibitors Cisplatin |
| BL2 | 7 | SUM149PT, CAL-851, HCC70, HCC1806, HDQ-P1 | Cell cycle DNA damage response Growth factor signaling |
mTOR inhibitors Growth factor inhibitors |
| IM | 18 | HCC1187, DU4475 | Immune signaling Cytokine signaling Antigen presentation |
PD1/PDL1 inhibitors |
| M | 24 | BT-549, CAL-51, CAL-120 | Cell motility Cell differentiation Growth factor signaling EMT |
mTOR inhibitors EMT-targeted treatment |
| MSL | 6 | HS578T, MDA-MB-157, SUM159PT, MDA-MB-436, MDA-MB-231 | Cell motility Cell differentiation Growth factor signaling Angiogenesis genes |
PI3K inhibitors Antiangiogenic therapy Src antagonist |
| LAR | 9 | MDA-MB-453, SUM185PE, HCC2185, CAL-148, MFM-223 | Androgen receptor Luminal gene expression |
Antiandrogen therapy PI3K/mTOR inhibitors |
| UNS | 19 | HCC1395, BT20, SW527 | Cell cycle DNA damage response Proliferation genes |
PARP inhibitors Cisplatin |
Note: Data are obtained from [82]. BL, basal-like; IM, immunomodulatory; M, mesenchymal; MSL, mesenchymal stem-like; LAR, luminal androgen receptor; UNS, unstable; PARP, poly(ADP-ribose)polymerase; EMT, epithelial-to-mesenchymal transition; mTOR, mammalian target of rapamycin; PD1, programmed cell death 1; PDL1, programmed death-ligand 1; PI3K, phosphatidylinositide 3-kinase.
Functional characterization of breast cancer through RNAi screening
Genomic analyses have uncovered a rapidly growing number of genetic variants that may participate in cancer initiation and progression. However, two intractable challenges remain. On one hand, genomic analyses fail to distinguish between the “driver” mutations that are critical for pathogenesis and the “passenger” incidents that occur coincidentally. On the other hand, hundreds of unanticipated synthetic lethal (SL) interactions are hidden in cancerous abnormalities. SL interactions refer to gene pair relationships in which the separate inactivation of either gene does not affect the viability of cancer cells, but joint inactivation is lethal [10], [90], [91]. It is an intelligent approach to authenticating the inactivated genes first and then selectively inhibiting their SL partners to efficaciously kill the specific cancer cells. A well-known example is the use of PARP inhibitors in the management of BRCA-mutated breast cancer [92], [93], [94].
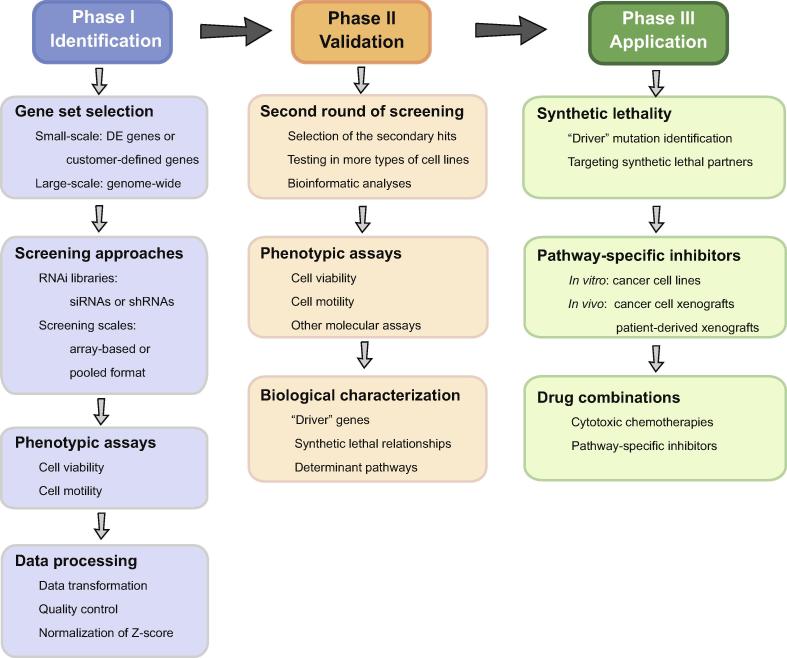
To circumvent the aforementioned limitations of genomic studies, loss-of-function RNAi screening technology has been widely adopted to define the functional genes that are necessary for cancer cells and to disclose SL relationships in exploring novel therapeutic options for cancer treatment [95]. Two types of RNA tools are used for RNAi, i.e., small interfering RNAs (siRNAs) and short hairpin RNAs (shRNAs). siRNAs are applied to achieve transient and short-term gene silencing, whereas vector-based shRNAs enable stable and long-term gene silencing. Both siRNAs and shRNAs can be used in array-based screening or in pooled formats. Figure 2 presents a general flowchart for high-throughput RNAi screening.
Figure 2.
A general flowchart for high-throughput RNAi screening
High-throughput RNAi screening usually comprises three phases. In phase I, the screening strategies, including gene sets, RNAi libraries, and screening scales, are determined mainly depending on the researchers’ purposes. The results of phenotypic assays are evaluated and normalized for the selection of effective hits. In phase II, the primary hits are validated by a second round of screening to confirm the “driver” genes, uncover the hidden synthetic lethal relationships, and disclose the critical signaling pathways. In phase III, targeted agents are tested both in vitro and in vivo, alone or in combination with other approved therapies. DE, differentially expressed; siRNA, small interfering RNA; shRNA, short hairpin RNA.
In RNAi screening, a small-scale array can be used to selectively suppress the up-regulated genes that have been detected in previous transcriptomic analyses or the genes that are differentially expressed among different cancer subtypes [96], [97], [98], [99], [100], [101], [102]. Bauer et al. have made the first attempt to perform a vector-based shRNA screening targeting 428 genes that are derived from the overlay of a pool of abnormal transcripts in breast cancer and the druggable gene list. They find that inhibiting both PPMID and SP1 significantly reduces the viability of two TNBC cell lines and increases their sensitivity to paclitaxel. When combined with paclitaxel, both CCT007093 and mithramycin, the respective chemical inhibitors of protein phosphatase Mg2+/Mn2+ dependent 1D (PPMID) and specificity protein 1 (SP1), suppress the growth of the paclitaxel-resistant TNBC cells [96]. In the same year, Kourtidis et al. have carried out a shRNA screen targeting 150 genes that are co-overexpressed with HER2 based on previous meta-analyses and discovered that both NR1D1 and PBP are novel survival factors essential for HER2+ breast cancer cells [97]. These two independent studies focus on two different types of breast cancers respectively, and uncover the distinct determinant genes between TNBC and HER2+ breast cancer. Subsequently, Marotta et al. further expand the number of breast cancer candidate genes and find that the IL-6/JAK2/Stat3 axis is significantly activated in CD44+CD24− breast cancer cells [99]. In addition, two other groups perform siRNA screening by selectively focusing on the genes that are enriched in the aberrantly amplified regions in breast cancer, and identify several candidate oncogenic driver genes, such as RAD21, EIF3H, CHRAC1, TANC2, and GNAS [101], [102].
The development of large-scale RNAi libraries has enabled non-biased genome-wide loss-of-function screening [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115]. Using high-throughput siRNA screening targeting the kinome, Brough et al. have defined a set of pharmacologically tractable genes in 34 breast cancer cell lines and uncovered the SL interactions between PTEN and TTK genes [103]. They further investigate the dependencies of kinase genes in ten cancers and utilize the resultant screening data to predict the drug sensitivity of the designated tumor cell lines by integrating with other molecular profiling datasets. They find that both ERBB3 and CCND1 are frequently amplified in breast cancer, whereas some skeletal system morphogenesis-related genes, such as PDGFRA, ACVR2B, TGFBR2, DLG1, FGFR1, and FGFR2, are highly-expressed in osteosarcoma [113]. In addition, using a pool of siRNAs targeting 17,378 genes, Petrocca et al. confirm that 154 genes are relevant to poor prognosis in breast cancer [107]. Marcotte et al. have conducted a genome-wide pooled screening containing 78,432 shRNAs of 16,056 unique genes in 72 cell lines for breast, pancreatic, and ovarian cancer. They discover that 297 genes are generally essential across all the cell lines examined [104]. Their further study on 77 breast cancer cell lines reveals that BRD4 is a putative targeted option for luminal breast cancer and PIK3CA mutations probably determine the resistance to bromodomain and extra-terminal domain (BET)-inhibitors [112]. Moreover, by performing deep RNAi screening in 398 cancer cell lines, a recent study has identified a wide variety of cancer genes and constructed interaction networks among protein complexes and signaling pathways [95]. Taken together, these studies indicate that RNAi screening is a direct and impactful approach to identifying key determinants and informing novel therapeutic agents and drug combination strategies in breast cancer.
miRNA signatures for TNBC
The majority of human genome, approximately 98%, is transcribed into ncRNAs [53], which consist of housekeeping ncRNAs and regulatory ncRNAs. The former includes rRNA, tRNA, small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and guide RNA (gRNA), whereas the latter includes miRNA, siRNA, piwi-interacting RNA (piRNA), and long ncRNA (lncRNA) [53], [71], [116]. miRNAs are well known for their involvement in various biological processes [117], [118], and a large number of miRNAs are deregulated in breast cancer [119], [120], [121], [122], [123], [124]. Using miRNA profiling in 31 primary TNBC cases and 13 lymph node metastatic samples in comparison with those from 23 matched normal counterparts, Avery-Kiejda et al. have identified 27 miRNAs related to the metastatic capabilities of TNBC cells [125]. Additionally, Koduru et al. have compared the publicly available small RNA sequencing data derived from 24 TNBC cases with those from 14 adjacent normal tissue samples and find that 55 aberrantly expressed miRNAs participate in the TGF-β signaling pathway [126].
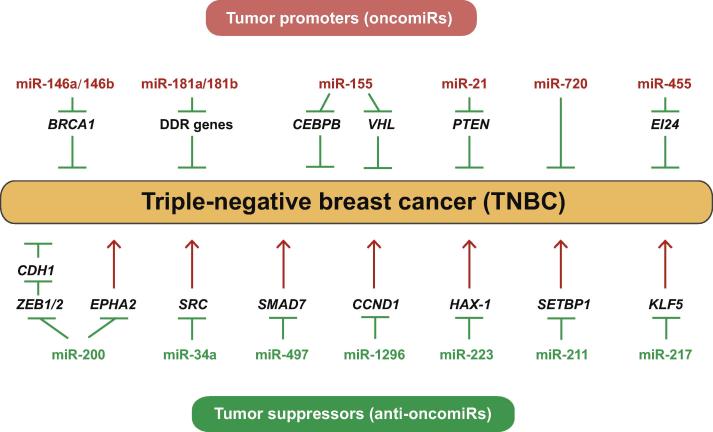
The expression of some miRNAs is up-regulated in TNBC and these miRNAs may function as tumor promoters to increase the proliferation and/or invasion of TNBC cells. This type of miRNAs is thus termed as oncomiRs, which include miR-146a/146b [127], miR-181a/181b [128], [129], miR-155 [130], [131], miR-21 [132], [133], miR-720 [134], and miR-455 [135]. In contrast, the expression of some other miRNAs is decreased in TNBC and may function as tumor suppressors to inhibit cancer cell growth, induce apoptosis, and ameliorate metastasis. These miRNAs are termed as anti-oncomiRs, which include the miR-200 family [136], [137], [138], miR-34a [139], miR-497 [140], miR-1296 [141], miR-223 [142], miR-211 [143], and miR-217 [144]. These functional miRNAs and their corresponding targets are shown in Figure 3. Studies of the systemic delivery of miRNA mimics or inhibitors via nanotechnologies are ongoing and hold great promise for cancer management [145], [146], [147], [148], [149], [150], [151]. For example, RNA nanoparticles with an 8-nt sequence that is complementary to the seed region of miR-21 inhibit TNBC tumors in mouse models without affecting other healthy organs [149].
Figure 3.
Functional miRNAs involved in TNBC
Many miRNAs are differentially expressed in TNBC. They may act as tumor promoters (oncomiRs) or tumor suppressors (anti-oncomiRs) to regulate the capacities of proliferation and/or invasion of TNBC by suppressing their corresponding targets. DDR, DNA damage response.
According to the critical roles of miRNAs in cell function and fate determination, a “Helm” model has been proposed to describe miRNAs as functional signatures for precisely characterizing cell identities in temporal-spatial specific status that primarily depends on the abundances of different miRNAs and the balance between these miRNAs and their corresponding targets [152]. In the case of breast cancer, comprehensive analyses of miRNA profiles combined with the mRNA expression patterns facilitate the illumination of the balance of miRNA-target pairs. We may be able to reclassify the breast cancer subtypes and clarify the unique biological capabilities of the selected cancer cells based on the dominant functional miRNAs.
Conclusion and perspectives
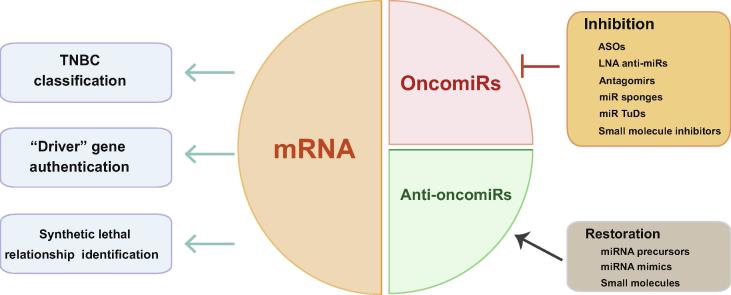
Transcriptomic analyses have provided massive amounts of information on the gene expression patterns in breast cancer. For clinical applications, the mRNA expression profiles can be employed to classify TNBC into unique molecular subtypes and to propose reliable therapeutic targets. Loss-of-function RNAi screening can be performed to discover the driver mutations and the SL partners of these inactivated genes for exploring novel targeted options. Moreover, an increasing number of miRNA are detected to be differentially expressed in breast cancer, many of which play critical roles as tumor promoters (oncomiRs) or tumor suppressors (anti-oncomiRs). The applications of transcriptomics in breast cancer are summarized in Figure 4.
Figure 4.
Applications of mRNA and miRNA indexes in breast cancer
Transcriptomic analyses reveal the expression patterns of both mRNAs and miRNAs. TNBC has been classified into different subtypes according to the cluster analysis of the distinct mRNA expression profiles. Additionally, high-throughput RNAi screening is widely applied to authenticate the “driver” inactivated genes and identify the hidden synthetic lethal relationships. Transcriptomic analyses also facilitate the discovery and validation of breast cancer-associated miRNAs. Therapeutic strategies, based on the inhibition and restoration of deregulated miRNAs, are now undergoing trials and hold great promise for breast cancer treatment. ASO, antisense oligonucleotide; LNA, locked nucleic acid; TuD, tough decoy.
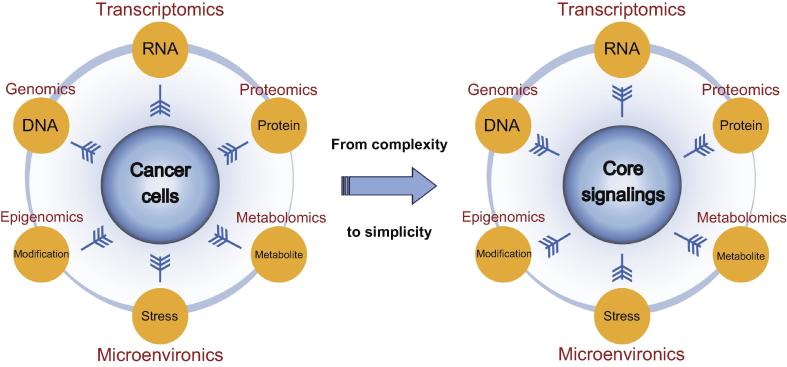
With the development of high-throughput sequencing technologies and computational analysis tools, it has become much easier to obtain and decipher enormous datasets that are relevant to different biological layers of human cancers besides genomic and transcriptomic studies. Epigenomic studies reveal the architecture of epigenetic alterations in human genes, including DNA methylation and chromatin modifications [153], [154], [155], [156], [157]. Proteomic [158], [159], [160], [161], [162], [163] and metabolomic [164], [165], [166] studies are also useful for elucidating additional faces of cancer biology. Novel strategies for integrating the genomic, transcriptomic, epigenomic, proteomic, and metabolomic data are demanded for a holistic understanding of tumor evolution and development [167], [168], [169], [170], [171]. In addition, cancer cells are not isolated entities; rather, they communicate with other stromal cells and adjacent tumor sub-clones. The influence of the tumor microenvironment has come into public notice in recent years [172]. It is important and necessary to take the interplay between cancer cells and their microenvironment into account to understand the effects of external stresses on cancer initiation and progression, as illustrated in Figure 5.
Figure 5.
Integration of multi-omic data to converge at the core signaling transduction pathways in cancer management
Comprehensive analyses of genome, transcriptome, proteome, epigenome, metabolome, and tumor microenvironment delineate the diverse aspects of cancer biology. Integration of these multi-omic data into the core intracellular signaling transduction pathways helps to provide accurate guidelines for cancer diagnosis and treatment.
The generation of multi-omic data has become an addictive routine for cancer studies. However, an intractable puzzle arises, that is, it is becoming increasingly intricate to assimilate the rapidly growing number of “big data”, as mentioned by Dr. Weinberg [173], [174], [175], [176]. Intelligent utilization and management of these data require massive computational resources and accurate statistical methodologies to unearth the hidden links among different subcomponents [174]. Although several data integration algorithms and a panel of software tools have been developed [177], there is still a lack of an impactful paradigm to solve Weinberg’s puzzle of how to effectively integrate multifarious information on cancer biology [173]. Multi-omic data display the myriad layers of cancer biology in detail, but the endless complexity seems to confuse our vision of the nature and the “Achilles’ heel” of cancers. Moreover, extreme inter- and intra-tumor heterogeneities originate from the rapid evolution of tumor cells. They may lead to a concept of personalized medicine for cancer therapy, which is based on every individual difference, rather than a concept that tumors are substantially a class of diseases that can be divided into several well-defined categories. We may return to simplicity and attempt to solve the complicated problems by identifying the common master regulators that correspond to the key hallmarks of cancers (e.g., the capabilities of proliferation, metastasis, immune evasion, and energy metabolism) in different malignant cells.
We suppose that the core signaling networks that are derived from hundreds of unique cancer-related signaling transduction pathways may be the appropriate candidates to unravel Weinberg’s puzzle. These networks reflect the cell identities, including the varieties of biological capabilities and the temporal–spatial status in cell differentiation, and may be used as functional classification criteria for the characterization of different subtypes of human cancers. Accordingly, pathway-targeted drugs and therapeutic strategies may be precisely designed to redress the deregulated networks. The integration of multi-omic data into the core signaling transduction channels that determine the development of cancers may be a potential direction for us to go out from Weinberg’s puzzle of the biological “big data” and helps to provide accurate guidelines for diagnosis and management for cancer patients (Figure 5).
Breast cancer is well-classified using both cellular and molecular features. Titanic efforts have been made to acquire a great deal of multi-omic data that display diverse biological signatures of the development of breast cancer, particularly in TNBC, which has not been well-characterized and for which no targeted drugs are yet available. It is high time for us to take advantage of these rich resources to identify the core signaling networks in TNBC and we are filled with hope that this most difficult-to-treat cancer will be precisely targeted in the near future.
Competing interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31230042, 31671349, and 31700712).
Handled by Mofang Liu
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Marrone M., Schilsky R.L., Liu G., Khoury M.J., Freedman A.N. Opportunities for translational epidemiology: the important role of observational studies to advance precision oncology. Cancer Epidemiol Biomarkers Prev. 2015;24:484–489. doi: 10.1158/1055-9965.EPI-14-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu K.H., Snyder M. Omics profiling in precision oncology. Mol Cell Proteomics. 2016;15:2525–2536. doi: 10.1074/mcp.O116.059253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed S., Zhou Z., Zhou J., Chen S.Q. Pharmacogenomics of drug metabolizing enzymes and transporters: relevance to precision medicine. Genomics Proteomics Bioinformatics. 2016;14:298–313. doi: 10.1016/j.gpb.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins F.S., Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter D.J. Uncertainty in the era of precision medicine. N Engl J Med. 2016;375:711–713. doi: 10.1056/NEJMp1608282. [DOI] [PubMed] [Google Scholar]
- 6.Cohen R.L., Settleman J. From cancer genomics to precision oncology—tissue's still an issue. Cell. 2014;157:1509–1514. doi: 10.1016/j.cell.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 7.Arnedos M., Vicier C., Loi S., Lefebvre C., Michiels S., Bonnefoi H. Precision medicine for metastatic breast cancer—limitations and solutions. Nat Rev Clin Oncol. 2015;12:693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 8.Biankin A.V., Piantadosi S., Hollingsworth S.J. Patient-centric trials for therapeutic development in precision oncology. Nature. 2015;526:361–370. doi: 10.1038/nature15819. [DOI] [PubMed] [Google Scholar]
- 9.Roychowdhury S., Chinnaiyan A.M. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin. 2016;66:75–88. doi: 10.3322/caac.21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senft D., Leiserson M.D.M., Ruppin E., Ronai Z.A. Precision oncology: the road ahead. Trends Mol Med. 2017;23:874–898. doi: 10.1016/j.molmed.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G., Shi T., Shi L. Characterizing and annotating the genome using RNA-seq data. Sci China Life Sci. 2017;60:116–125. doi: 10.1007/s11427-015-0349-4. [DOI] [PubMed] [Google Scholar]
- 12.Sestak I., Cuzick J. Update on breast cancer risk prediction and prevention. Curr Opin Obstet Gynecol. 2015;27:92–97. doi: 10.1097/GCO.0000000000000153. [DOI] [PubMed] [Google Scholar]
- 13.Ellis A.J., Hendrick V.M., Williams R., Komm B.S. Selective estrogen receptor modulators in clinical practice: a safety overview. Expert Opin Drug Saf. 2015;14:921–934. doi: 10.1517/14740338.2015.1014799. [DOI] [PubMed] [Google Scholar]
- 14.Lumachi F., Santeufemia D.A., Basso S.M. Current medical treatment of estrogen receptor-positive breast cancer. World J Biol Chem. 2015;6:231–239. doi: 10.4331/wjbc.v6.i3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gradishar W.J. HER2 therapy — an abundance of riches. N Engl J Med. 2012;366:176–178. doi: 10.1056/NEJMe1113641. [DOI] [PubMed] [Google Scholar]
- 16.Figueroa-Magalhães M.C., Jelovac D., Connolly R., Wolff A.C. Treatment of HER2-positive breast cancer. Breast. 2014;23:128–136. doi: 10.1016/j.breast.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 18.Hurvitz S., Mead M. Triple-negative breast cancer: advancements in characterization and treatment approach. Curr Opin Obstet Gynecol. 2016;28:59–69. doi: 10.1097/GCO.0000000000000239. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann B.D., Pietenpol J.A. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirshfield K.M., Ganesan S. Triple-negative breast cancer: molecular subtypes and targeted therapy. Curr Opin Obstet Gynecol. 2014;26:34–40. doi: 10.1097/GCO.0000000000000038. [DOI] [PubMed] [Google Scholar]
- 21.Judes G., Rifaï K., Daures M., Dubois L., Bignon Y.J., Penault-Llorca F. High-throughput «Omics» technologies: new tools for the study of triple-negative breast cancer. Cancer Lett. 2016;382:77–85. doi: 10.1016/j.canlet.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Jia L.Y., Shanmugam M.K., Sethi G., Bishayee A. Potential role of targeted therapies in the treatment of triple-negative breast cancer. Anticancer Drugs. 2016;27:147–155. doi: 10.1097/CAD.0000000000000328. [DOI] [PubMed] [Google Scholar]
- 23.Ding L., Ellis M.J., Li S., Larson D.E., Chen K., Wallis J.W. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerji S., Cibulskis K., Rangel-Escareno C., Brown K.K., Carter S.L., Frederick A.M. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popova T., Manié E., Rieunier G., Caux-Moncoutier V., Tirapo C., Dubois T. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 27.Shah S.P., Roth A., Goya R., Oloumi A., Ha G., Zhao Y. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J.C., Alvarez M.J., Talos F., Dhruv H., Rieckhof G.E., Iyer A. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell. 2014;159:402–414. doi: 10.1016/j.cell.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foedermayr M., Sebesta M., Rudas M., Berghoff A.S., Promberger R., Preusser M. BRCA-1 methylation and TP53 mutation in triple-negative breast cancer patients without pathological complete response to taxane-based neoadjuvant chemotherapy. Cancer Chemother Pharmacol. 2014;73:771–778. doi: 10.1007/s00280-014-2404-1. [DOI] [PubMed] [Google Scholar]
- 33.Burstein M.D., Tsimelzon A., Poage G.M., Covington K.R., Contreras A., Fuqua S.A. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M.J. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao R., Davis A., McDonald T.O., Sei E., Shi X., Wang Y. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet. 2016;48:1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira B., Chin S.F., Rueda O.M., Vollan H.K., Provenzano E., Bardwell H.A. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nature Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yates L.R., Knappskog S., Wedge D., Farmery J.H.R., Gonzalez S., Martincorena I. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32:169–184. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michailidou K., Lindström S., Dennis J., Beesley J., Hui S., Kar S. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin L.A., Ribas R., Simigdala N., Schuster E., Pancholi S., Tenev T. Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat Commun. 2017;8:1865. doi: 10.1038/s41467-017-01864-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polak P., Kim J., Braunstein L.Z., Karlic R., Haradhavala N.J., Tiao G. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rheinbay E., Parasuraman P., Grimsby J., Tiao G., Engreitz J.M., Kim J. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamel H.F.M., Al-Amodi H.S.A.B. Exploitation of gene expression and cancer biomarkers in paving the path to era of personalized medicine. Genomics Proteomics Bioinformatics. 2017;15:220–235. doi: 10.1016/j.gpb.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGee S.R., Tibiche C., Trifiro M., Wang E. Network analysis reveals a signaling regulatory loop in PIK3CA-mutated breast cancer predicting survival outcome. Genomics Proteomics Bioinformatics. 2017;15:121–129. doi: 10.1016/j.gpb.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haiman C.A., Chen G.K., Vachon C.M., Canzian F., Dunning A., Millikan R.C. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siddiq A., Couch F.J., Chen G.K. Lindstro¨m S, Eccles D, Millikan RC. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghoussaini M., Fletcher O., Michailidou K., Turnbull C., Schmidt M.K., Dicks E. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet. 2012;44:312–318. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Walavalkar N.M., Dozmorov M.G., Rich S.S., Civelek M., Guertin M.J. Identification of breast cancer associated variants that modulate transcription factor binding. PLoS Genet. 2017;13:e1006761. doi: 10.1371/journal.pgen.1006761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Y., Ye P., Long X. Differential expression profiles of the transcriptome in breast cancer cell lines revealed by next generation sequencing. Cell Physiol Biochem. 2017;44:804–816. doi: 10.1159/000485344. [DOI] [PubMed] [Google Scholar]
- 53.Casamassimi A., Federico A., Rienzo M., Esposito S., Ciccodicola A. Transcriptome profiling in human diseases: new advances and perspectives. Int J Mol Sci. 2017;18:1652. doi: 10.3390/ijms18081652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C.G., Calin G.A., Volinia S., Croce C.M. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3:563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 55.Yin J.Q., Zhao R.C., Morris K.V. Profiling microRNA expression with microarrays. Trends Biotechnol. 2008;26:70–76. doi: 10.1016/j.tibtech.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y., Gelfond J.A., McManus L.M., Shireman P.K. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics. 2009;10:407. doi: 10.1186/1471-2164-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sørlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Z., Fan C., Oh D.S., Marron J.S., He X., Qaqish B.F. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu X., Stern H.M., Ge L., O'Brien C., Haydu L., Honchell C.D. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7:511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 61.Hollestelle A., Nagel J.H., Smid M., Lam S., Elstrodt F., Wasielewski M. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat. 2010;121:53–64. doi: 10.1007/s10549-009-0460-8. [DOI] [PubMed] [Google Scholar]
- 62.Castaneda C.A., Andrés E., Barcena C., Gómez H.L., Cortés-Funés H., Ciruelos E. Behaviour of breast cancer molecular subtypes through tumour progression. Clin Transl Oncol. 2012;14:481–485. doi: 10.1007/s12094-012-0827-x. [DOI] [PubMed] [Google Scholar]
- 63.Engstrøm M.J., Opdahl S., Hagen A.I., Romundstad P.R., Akslen L.A., Haugen O.A. Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients. Breast Cancer Res Treat. 2013;140:463–473. doi: 10.1007/s10549-013-2647-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kimbung S., Kovács A., Danielsson A., Bendahl P.O., Lövgren K., Stolt M.F. Contrasting breast cancer molecular subtypes across serial tumor progression stages: biological and prognostic implications. Oncotarget. 2015;6:33306–33318. doi: 10.18632/oncotarget.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen C., Li Z., Yang Y., Xiang T., Song W., Liu S. Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer. Cancer Biol Ther. 2015;16:856–865. doi: 10.1080/15384047.2015.1040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karagoz K., Sinha R., Arga K.Y. Triple negative breast cancer: a multi-omics network discovery strategy for candidate targets and driving pathways. OMICS. 2015;19:115–130. doi: 10.1089/omi.2014.0135. [DOI] [PubMed] [Google Scholar]
- 67.Jiang Y.Z., Liu Y.R., Xu X.E., Jin X., Hu X., Yu K.D. Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer Res. 2016;76:2105–2114. doi: 10.1158/0008-5472.CAN-15-3284. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y.R., Jiang Y.Z., Xu X.E., Hu X., Yu K.D., Shao Z.M. Comprehensive transcriptome profiling reveals multigene signatures in triple-negative breast cancer. Clin Cancer Res. 2016;22:1653–1662. doi: 10.1158/1078-0432.CCR-15-1555. [DOI] [PubMed] [Google Scholar]
- 69.Peng C., Ma W., Xia W., Zheng W. Integrated analysis of differentially expressed genes and pathways in triple-negative breast cancer. Mol Med Rep. 2017;15:1087–1094. doi: 10.3892/mmr.2017.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costa V., Angelini C., De Feis I., Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. J Biomed Biotechnol. 2010;2010:853916. doi: 10.1155/2010/853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Metzker M.L. Sequencing technologies–the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 73.Costa V., Aprile M., Esposito R., Ciccodicola A. RNA-Seq and human complex diseases: recent accomplishments and future perspectives. Eur J Hum Genet. 2013;21:134–142. doi: 10.1038/ejhg.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 75.Edgren H., Murumagi A., Kangaspeska S., Nicorici D., Hongisto V., Kleivi K. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12:R6. doi: 10.1186/gb-2011-12-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ha K.C., Lalonde E., Li L., Cavallone L., Natrajan R., Lambros M.B. Identification of gene fusion transcripts by transcriptome sequencing in BRCA1-mutated breast cancers and cell lines. BMC Med Genomics. 2011;4:75. doi: 10.1186/1755-8794-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim J., Kim S., Ko S., In Y.H., Moon H.G., Ahn S.K. Recurrent fusion transcripts detected by whole-transcriptome sequencing of 120 primary breast cancer samples. Genes Chromosomes Cancer. 2015;54:681–691. doi: 10.1002/gcc.22279. [DOI] [PubMed] [Google Scholar]
- 78.Kumar-Sinha C., Kalyana-Sundaram S., Chinnaiyan A.M. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129. doi: 10.1186/s13073-015-0252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Veeraraghavan J., Ma J., Hu Y., Wang X.S. Recurrent and pathological gene fusions in breast cancer: current advances in genomic discovery and clinical implications. Breast Cancer Res Treat. 2016;158:219–232. doi: 10.1007/s10549-016-3876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buermans H.P., Ariyurek Y., van Ommen G., den Dunnen J.T., 't Hoen P.A. New methods for next generation sequencing based microRNA expression profiling. BMC Genomics. 2010;11:716. doi: 10.1186/1471-2164-11-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pritchard C.C., Cheng H.H., Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eswaran J., Cyanam D., Mudvari P., Reddy S.D.N., Pakala S.B., Nair S.S. Transcriptomic landscape of breast cancers through mRNA sequencing. Sci Rep. 2012;2:264. doi: 10.1038/srep00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abramson V.G., Lehmann B.D., Ballinger T.J., Pietenpol J.A. Subtyping of triple-negative breast cancer: implications for therapy. Cancer. 2015;121:8–16. doi: 10.1002/cncr.28914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Le Du F., Eckhardt B.L., Lim B., Litton J.K., Moulder S., Meric-Bernstam F. Is the future of personalized therapy in triple-negative breast cancer based on molecular subtype? Oncotarget. 2015;6:12890–12908. doi: 10.18632/oncotarget.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kalimutho M., Parsons K., Mittal D., López J.A., Srihari S., Khanna K.K. Targeted therapies for triple-negative breast cancer: combating a stubborn disease. Trends Pharmacol Sci. 2015;36:822–846. doi: 10.1016/j.tips.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 87.Liu Y.R., Jiang Y.Z., Xu X.E., Yu K.D., Jin X., Hu X. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res. 2016;18:33. doi: 10.1186/s13058-016-0690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andreopoulou E., Kelly C.M., McDaid H.M. Therapeutic advances and new directions for triple-negative breast cancer. Breast Care (Basel) 2017;12:21–28. doi: 10.1159/000455821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mayer I.A., Abramson V.G., Lehmann B.D., Pietenpol J.A. New strategies for triple-negative breast cancer-deciphering the heterogeneity. Clin Cancer Res. 2014;20:782–790. doi: 10.1158/1078-0432.CCR-13-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McLornan D.P., List A., Mufti G.J. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med. 2014;371:1725–1735. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
- 91.Jerby-Arnon L., Pfetzer N., Waldman Y.Y., McGarry L., James D., Shanks E. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell. 2014;158:1199–1209. doi: 10.1016/j.cell.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 92.Rios J., Puhalla S. PARP inhibitors in breast cancer: BRCA and beyond. Oncology (Williston Park) 2011;25:1014–1025. [PubMed] [Google Scholar]
- 93.Arun B., Akar U., Gutierrez-Barrera A.M., Hortobagyi G.N., Ozpolat B. The PARP inhibitor AZD2281 (Olaparib) induces autophagy/mitophagy in BRCA1 and BRCA2 mutant breast cancer cells. Int J Oncol. 2015;47:262–268. doi: 10.3892/ijo.2015.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Livraghi L., Garber J.E. PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Med. 2015;13:188. doi: 10.1186/s12916-015-0425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McDonald E.R., 3rd, de Weck A., Schlabach M.R., Billy E., Mavrakis K.J., Hoffman G.R. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell. 2017;170:577–592. doi: 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 96.Bauer J.A., Ye F., Marshall C.B., Lehmann B.D., Pendleton C.S., Shyr Y. RNA interference (RNAi) screening approach identifies agents that enhance paclitaxel activity in breast cancer cells. Breast Cancer Res. 2010;12:R41. doi: 10.1186/bcr2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kourtidis A., Jain R., Carkner R.D., Eifert C., Brosnan M.J., Conklin D.S. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res. 2010;70:1783–1792. doi: 10.1158/0008-5472.CAN-09-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boimel P.J., Cruz C., Segall J.E. A functional in vivo screen for regulators of tumor progression identifies HOXB2 as a regulator of tumor growth in breast cancer. Genomics. 2011;98:164–172. doi: 10.1016/j.ygeno.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marotta L.L., Almendro V., Marusyk A., Shipitsin M., Schemme J., Walker S.R. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24- stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boyer A.P., Collier T.S., Vidavsky I., Bose R. Quantitative proteomics with siRNA screening identifies novel mechanisms of Trastuzumab resistance in HER2 amplified breast cancers. Mol Cell Proteomics. 2013;12:180–193. doi: 10.1074/mcp.M112.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mahmood S.F., Gruel N., Chapeaublanc E., Lescure A., Jones T., Reyal F. A siRNA screen identifies RAD21, EIF3H, CHRAC1 and TANC2 as driver genes within the 8q23, 8q24.3 and 17q23 amplicons in breast cancer with effects on cell growth, survival and transformation. Carcinogenesis. 2014;35:670–682. doi: 10.1093/carcin/bgt351. [DOI] [PubMed] [Google Scholar]
- 102.Garcia-Murillas I., Sharpe R., Pearson A., Campbell J., Natrajan R., Ashworth A. An siRNA screen identifies the GNAS locus as a driver in 20q amplified breast cancer. Oncogene. 2014;33:2478–2486. doi: 10.1038/onc.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brough R., Frankum J.R., Sims D., Mackay A., Mendes-Pereira A.M., Bajrami I. Functional viability profiles of breast cancer. Cancer Discov. 2011;1:260–273. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marcotte R., Brown K.R., Suarez F., Sayad A., Karamboulas K., Krzyzanowski P.M. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Giamas G., Filipović A., Jacob J., Messier W., Zhang H., Yang D. Kinome screening for regulators of the estrogen receptor identifies LMTK3 as a new therapeutic target in breast cancer. Nat Med. 2011;17:715–719. doi: 10.1038/nm.2351. [DOI] [PubMed] [Google Scholar]
- 106.Hu K., Law J.H., Fotovati A., Dunn S.E. Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells. Breast Cancer Res. 2012;14:R22. doi: 10.1186/bcr3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Petrocca F., Altschuler G., Tan S.M., Mendillo M.L., Yan H., Jerry D.J. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24:182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garimella S.V., Gehlhaus K., Dine J.L., Pitt J.J., Grandin M., Chakka S. Identification of novel molecular regulators of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in breast cancer cells by RNAi screening. Breast Cancer Res. 2014;16:R41. doi: 10.1186/bcr3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Deng T., Liu J.C., Chung P.E., Uehling D., Aman A., Joseph B. shRNA kinome screen identifies TBK1 as a therapeutic target for HER2+ breast cancer. Cancer Res. 2014;74:2119–2130. doi: 10.1158/0008-5472.CAN-13-2138. [DOI] [PubMed] [Google Scholar]
- 110.Bhola N.E., Jansen V.M., Bafna S., Giltnane J.M., Balko J.M., Estrada M.V. Kinome-wide functional screen identifies role of PLK1 in hormone-independent, ER-positive breast cancer. Cancer Res. 2015;75:405–414. doi: 10.1158/0008-5472.CAN-14-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Roosmalen W., Le Dévédec S.E., Golani O., Smid M., Pulyakhina I., Timmermans A.M. Tumor cell migration screen identifies SRPK1 as breast cancer metastasis determinant. J Clin Invest. 2015;125:1648–1664. doi: 10.1172/JCI74440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marcotte R., Sayad A., Brown K.R., Sanchez-Garcia F., Reimand J., Haider M. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Campbell J., Ryan C.J., Brough R., Bajrami I., Pemberton H.N., Chong I.Y. Large-scale profiling of kinase dependencies in cancer cell lines. Cell Rep. 2016;14:2490–2501. doi: 10.1016/j.celrep.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Horiuchi D., Camarda R., Zhou A.Y., Yau C., Momcilovic O., Balakrishnan S. PIM1 kinase inhibition as a targeted therapy against triple-negative breast tumors with elevated MYC expression. Nat Med. 2016;22:1321–1329. doi: 10.1038/nm.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Workenhe S.T., Ketela T., Moffat J., Cuddington B.P., Mossman K.L. Genome-wide lentiviral shRNA screen identifies serine/arginine-rich splicing factor 2 as a determinant of oncolytic virus activity in breast cancer cells. Oncogene. 2016;35:2465–2474. doi: 10.1038/onc.2015.303. [DOI] [PubMed] [Google Scholar]
- 116.Carninci P., Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev. 2007;17:139–144. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 117.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 118.Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 119.Dvinge H., Git A., Graf S., Salmon-Divon M., Curtis C., Sottoriva A. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 120.Riaz M., van Jaarsveld M.T., Hollestelle A., Prager-van der Smissen W.J., Heine A.A., Boersma A.W. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 2013;15:R33. doi: 10.1186/bcr3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gyparaki M.T., Basdra E.K., Papavassiliou A.G. MicroRNAs as regulatory elements in triple negative breast cancer. Cancer Lett. 2014;354:1–4. doi: 10.1016/j.canlet.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 122.Sui X., Wang X., Han W., Li D., Xu Y., Lou F. MicroRNAs-mediated cell fate in triple negative breast cancers. Cancer Lett. 2015;361:8–12. doi: 10.1016/j.canlet.2015.02.048. [DOI] [PubMed] [Google Scholar]
- 123.Mathe A., Scott R.J., Avery-Kiejda K.A. MiRNAs and other epigenetic changes as biomarkers in triple negative breast cancer. Int J Mol Sci. 2015;16:28347–28376. doi: 10.3390/ijms161226090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bertoli G., Cava C., Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Avery-Kiejda K.A., Braye S.G., Mathe A., Forbes J.F., Scott R.J. Decreased expression of key tumour suppressor microRNAs is associated with lymph node metastases in triple negative breast cancer. BMC Cancer. 2014;14:51. doi: 10.1186/1471-2407-14-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Koduru S.V., Tiwari A.K., Leberfinger A., Hazard S.W., Kawasawa Y.I., Mahajan M. A comprehensive NGS data analysis of differentially regulated miRNAs, piRNAs, lncRNAs and sn/snoRNAs in triple negative breast cancer. J Cancer. 2017;8:578–596. doi: 10.7150/jca.17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Garcia A.I., Buisson M., Bertrand P., Rimokh R., Rouleau E., Lopez B.S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3:279–290. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Taylor M.A., Sossey-Alaoui K., Thompson C.L., Danielpour D., Schiemann W.P. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123:150–163. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bisso A., Faleschini M., Zampa F., Capaci V., De Santa J., Santarpia L. Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle. 2013;12:1679–1687. doi: 10.4161/cc.24757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Johansson J., Berg T., Kurzejamska E., Pang M.F., Tabor V., Jansson M. MiR-155-mediated loss of C/EBPβ shifts the TGF-β response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene. 2013;32:5614–5624. doi: 10.1038/onc.2013.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kong W., He L., Richards E.J., Challa S., Xu C.X., Permuth-Wey J. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.MacKenzie T.A., Schwartz G.N., Calderone H.M., Graveel C.R., Winn M.E., Hostetter G. Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am J Pathol. 2014;184:3217–3225. doi: 10.1016/j.ajpath.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fang H., Xie J., Zhang M., Zhao Z., Wan Y., Yao Y. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am J Transl Res. 2017;9:953–961. [PMC free article] [PubMed] [Google Scholar]
- 134.Das S.G., Romagnoli M., Mineva N.D., Barillé-Nion S., Jézéquel P., Campone M. miR-720 is a downstream target of an ADAM8-induced ERK signaling cascade that promotes the migratory and invasive phenotype of triple-negative breast cancer cells. Breast Cancer Res. 2016;18:40. doi: 10.1186/s13058-016-0699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Z., Meng Q., Pan A., Wu X., Cui J., Wang Y. MicroRNA-455-3p promotes invasion and migration in triple negative breast cancer by targeting tumor suppressor EI24. Oncotarget. 2017;8:19455–19466. doi: 10.18632/oncotarget.14307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Truong H.H., Xiong J., Ghotra V.P., Nirmala E., Haazen L., Le Dévédec S.E. β1 integrin inhibition elicits a prometastatic switch through the TGFβ-miR-200-ZEB network in E-cadherin-positive triple-negative breast cancer. Sci Signal. 2014;7:ra15. doi: 10.1126/scisignal.2004751. [DOI] [PubMed] [Google Scholar]
- 137.Tsouk E., Wang J., Frigo D.E., Aydoğdu E., Williams C. miR-200a inhibits migration of triple-negative breast cancer cells through direct repression of the EPHA2 oncogene. Carcinogenesis. 2015;36:1051–1060. doi: 10.1093/carcin/bgv087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.D'Ippolito E., Plantamura I., Bongiovanni L., Casalini P., Baroni S., Piovan C. miR-9 and miR-200 regulate PDGFRβ-mediated endothelial differentiation of tumor cells in triple-negative breast cancer. Cancer Res. 2016;76:5562–5572. doi: 10.1158/0008-5472.CAN-16-0140. [DOI] [PubMed] [Google Scholar]
- 139.Adams B.D., Wali V.B., Cheng C.J., Inukai S., Booth C.J., Agarwal S. miR-34a silences c-SRC to attenuate tumor growth in triple-negative breast cancer. Cancer Res. 2016;76:927–939. doi: 10.1158/0008-5472.CAN-15-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu J., Zhou Y., Shi Z., Hu Y., Meng T., Zhang X. microRNA-497 modulates breast cancer cell proliferation, invasion, and survival by targeting SMAD7. DNA Cell Biol. 2016;35:521–529. doi: 10.1089/dna.2016.3282. [DOI] [PubMed] [Google Scholar]
- 141.Phan B., Majid S., Ursu S., de Semir D., Nosrati M., Bezrookove V. Tumor suppressor role of microRNA-1296 in triple-negative breast cancer. Oncotarget. 2016;7:19519–19530. doi: 10.18632/oncotarget.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sun X., Li Y., Zheng M., Zuo W., Zheng W. MicroRNA-223 increases the sensitivity of triple-negative breast cancer stem cells to trail-induced apoptosis by targeting HAX-1. PLoS One. 2016;11:e0162754. doi: 10.1371/journal.pone.0162754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen L.L., Zhang Z.J., Yi Z.B., Li J.J. MicroRNA-211-5p suppresses tumour cell proliferation, invasion, migration and metastasis in triple-negative breast cancer by directly targeting SETBP1. Br J Cancer. 2017;117:78–88. doi: 10.1038/bjc.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou W., Song F., Wu Q., Liu R., Wang L., Liu C. miR-217 inhibits triple-negative breast cancer cell growth, migration, and invasion through targeting KLF5. PLoS One. 2017;12:e0176395. doi: 10.1371/journal.pone.0176395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kota J., Chivukula R.R., O'Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ling H., Fabbri M., Calin G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li Z., Rana T.M. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 148.Cheng C.J., Bahal R., Babar I.A., Pincus Z., Barrera F., Liu C. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shu D., Li H., Shu Y., Xiong G., Carson W.E., 3rd, Haque F. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Beavers K.R., Werfel T.A., Shen T., Kavanaugh T.E., Kilchrist K.V., Mares J.W. Porous silicon and polymer nanocomposites for delivery of peptide nucleic acids as anti-microRNA therapies. Adv Mater. 2016;28:7984–7992. doi: 10.1002/adma.201601646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 152.Xie S.J., Zhang Y., Qu L.H., Xu H. A helm model for microRNA regulation in cell fate decision and conversion. Sci China Life Sci. 2013;56:897–906. doi: 10.1007/s11427-013-4547-4. [DOI] [PubMed] [Google Scholar]
- 153.Fang F., Turcan S., Rimner A., Kaufman A., Giri D., Morris L.G. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3:75ra25. doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Stirzaker C., Zotenko E., Song J.Z., Qu W., Nair S.S., Locke W.J. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat Commun. 2015;6:5899. doi: 10.1038/ncomms6899. [DOI] [PubMed] [Google Scholar]
- 155.Jones P.A., Issa J.P., Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 156.Zhao Q.Y., Lei P.J., Zhang X.R., Zheng J.Y., Wang H.Y., Zhao J. Global histone modification profiling reveals the epigenomic dynamics during malignant transformation in a four-stage breast cancer model. Clin Epigenetics. 2016;8:34. doi: 10.1186/s13148-016-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fleischer T., Tekpli X., Mathelier A., Wang S., Nebdal D., Dhakal H.P. DNA methylation at enhancers identifies distinct breast cancer lineages. Nat Commun. 2017;8:1379. doi: 10.1038/s41467-017-00510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Geiger T., Madden S.F., Gallagher W.M., Cox J., Mann M. Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 2012;72:2428–2439. doi: 10.1158/0008-5472.CAN-11-3711. [DOI] [PubMed] [Google Scholar]
- 159.Muñiz Lino M.A., Palacios-Rodríguez Y., Rodríguez-Cuevas S., Bautista-Piña V., Marchat L.A., Ruíz-García E. Comparative proteomic profiling of triple-negative breast cancer reveals that up-regulation of RhoGDI-2 is associated to the inhibition of caspase 3 and caspase 9. J Proteomics. 2014;111:198–211. doi: 10.1016/j.jprot.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 160.Lawrence R.T., Perez E.M., Hernandez D., Miller C.P., Haas K.M., Irie H.Y. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11:630–644. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tyanova S., Albrechtsen R., Kronqvist P., Cox J., Mann M., Geiger T. Proteomic maps of breast cancer subtypes. Nat Commun. 2016;7:10259. doi: 10.1038/ncomms10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang Y., Zhu H. Protein array-based approaches for biomarker discovery in cancer. Genomics Proteomics Bioinformatics. 2017;15:73–81. doi: 10.1016/j.gpb.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li X., Wang W., Chen J. Recent progress in mass spectrometry proteomics for biomedical research. Sci China Life Sci. 2017;60:1093–1113. doi: 10.1007/s11427-017-9175-2. [DOI] [PubMed] [Google Scholar]
- 164.Denkert C., Bucher E., Hilvo M., Salek R., Orešic M., Griffin J. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Med. 2012;4:37. doi: 10.1186/gm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mishra P., Ambs S. Metabolic signatures of human breast cancer. Mol Cell Oncol. 2015;2:e992217. doi: 10.4161/23723556.2014.992217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huang S., Chong N., Lewis N.E., Jia W., Xie G., Garmire L.X. Novel personalized pathway-based metabolomics models reveal key metabolic pathways for breast cancer diagnosis. Genome Med. 2016;8:34. doi: 10.1186/s13073-016-0289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shen H., Laird P.W. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Clare S.E., Shaw P.L. “Big Data” for breast cancer: where to look and what you will find. NPJ Breast Cancer. 2016;2:16031. doi: 10.1038/npjbcancer.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sandhu C., Qureshi A., Emili A. Panomics for precision medicine. Trends Mol Med. 2018;24:85–101. doi: 10.1016/j.molmed.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Letai A. Functional precision cancer medicine—moving beyond pure genomics. Nat Med. 2017;23:1028–1035. doi: 10.1038/nm.4389. [DOI] [PubMed] [Google Scholar]
- 171.Wang E., Cho W.C.S., Wong S.C.C., Liu S. Disease biomarkers for precision medicine: challenges and future opportunities. Genomics Proteomics Bioinformatics. 2017;15:57–58. doi: 10.1016/j.gpb.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Soysal S.D., Tzankov A., Muenst S.E. Role of the tumor microenvironment in breast cancer. Pathobiology. 2015;82:142–152. doi: 10.1159/000430499. [DOI] [PubMed] [Google Scholar]
- 173.Weinberg R.A. Coming full circle—from endless complexity to simplicity and back again. Cell. 2014;157:267–271. doi: 10.1016/j.cell.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 174.Alyass A., Turcotte M., Meyre D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med Genomics. 2015;8:33. doi: 10.1186/s12920-015-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Elefsinioti A., Bellaire T., Wang A., Quast K., Seidel H., Braxenthaler M. Key factors for successful data integration in biomarker research. Nat Rev Drug Discov. 2016;15:369–370. doi: 10.1038/nrd.2016.74. [DOI] [PubMed] [Google Scholar]
- 176.McCue M.E., McCoy A.M. The scope of big data in one medicine: unprecedented opportunities and challenges. Front Vet Sci. 2017;4:194. doi: 10.3389/fvets.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang S., Chaudhary K., Garmire L.X. More is better: recent progress in multi-omics data integration methods. Front Genet. 2017;8:84. doi: 10.3389/fgene.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]