Abstract
The present study evaluated the effects of 4 typical subtropical forages on ruminal microbial community composition to formulate a better diet for buffalo. Corn straw silage, elephant grass, cassava residues and sugarcane tail silage were used as substrates for in vitro fermentation. Eight replicates were set up for every substrate, and fermentation was carried out in a 100-mL glass syringe, using buffalo rumen inoculum. Every replicate was anaerobically dispensed with 10 mL of rumen inoculum, 20 mL of McDougall's buffer and 200 mg of dried substrate, and placed in a water bath at 39 °C. Gas production was recorded at 0, 2, 6, 12, 24, 36, 48 and 72 h of incubation. After 24 h, fermentation was ceased for 4 replicates and samples were collected. Volatile fatty acids (VFA) concentrations were measured using gas chromatography. Microbial populations were quantified using quantitative real-time PCR (qRT-PCR), and microbial community was analyzed using high throughput sequencing technology. The results showed, cassava residues as substrate had the highest gas production, acetate, propionate and total VFA concentrations (P < 0.05), and corn straw silage had the lowest acetate:propionate ratio (P < 0.05). The lowest numbers of fungi, Ruminococcus albus and Fibrobacter succinogenes, and the highest number of protozoa were observed with cassava residues (P < 0.05). The least abundances of bacterial phyla Firmicutes, Bacteroidetes and genus Prevotella, and substantially higher abundance of phylum proteobacteria (56%) and genus Succinivibrio (52%) were observed with cassava residues. The most abundances of Methanobrevibacter gottschalkii and Entodinium were observed with cassava residues. Spearman's correlations analysis showed, Succinivibrio had strong positive correlations with propionate, butyrate, Metadinium and M. gottschalkii, indicating fermentation products were related to microbial community. In conclusion, incubation with cassava residues resulted in lower number of fiber degrading microbes but higher protozoal population because of its low fiber contents. The microbial community was highly altered by in vitro incubation with cassava residues, whereas remained similar for the other 3 high fiber containing substrates.
Keywords: Buffalo rumen inoculum, In vitro fermentation, Subtropical forages, Microbial population, Microbial community
1. Introduction
Forages account for 30% to 100% of ruminant rations, which has been reported to influence the microbial community composition differently both in vivo and in vitro (Kong et al., 2010, Martínez et al., 2010). In subtropical areas, a variety of feedstuffs are used for dairy buffalos, mainly including corn straw silage, sugarcane tail silage, cassava residue and elephant grass. Nutritive characterization of forages for the ruminants is important to improve their productivity (Bartocci et al., 2002), as well as to effectively use the forages to make dairy rations based on their fermentation characteristics in the rumen. Although the nutritive values of above mentioned 4 forages have been intensively studied, but their impacts on ruminal fermentation and microbial community are poorly understood, because of constraints on microbial molecular research techniques in the past.
The rumen microbiome is a dynamic system that rapidly changes with diets. The type of forage alters the rumen microbial community composition mainly due to its specific fiber structure, which determines fermentation products and ruminal pH (Fernando et al., 2010, Petri et al., 2012). Changes in microbial community could provide us a clear understanding of interaction between forage and ruminal microbes (Yang et al., 2016). Nowadays with the development of microbial molecular techniques, especially the high throughput sequencing technology, research on ruminal microbial community has become more convenient than before. The in vitro techniques are assumed to be able to adequately mimic fermentation in vivo and are widely used to explore digestibility of different forages and their effects on ruminal fermentation (Kaiser and Weniger, 1994, Mould et al., 2005, Zapletalová et al., 2016). Thus, conducting in vitro experiments to study how the forages influence ruminal fermentation and microbial community is significant, which can clarify the functional differences of different forages in the rumen.
Ménard et al. (2010) and Calabro et al. (2008) reported higher production of gas and total volatile fatty acids (VFA) in buffalo rumen fluid than those of cattle when incubated with the same substrates, but the microbial mechanism behind the difference was not addressed. To the best of our knowledge, the majority of in vitro studies on buffalo rumen inoculum have discussed gas production and digestibility of forages, whereas studies focusing on the rumen microbial community composition and its variation with the variety of feedstuffs are less documented. Therefore, the principal objective of this in vitro study was to investigate differences in fermentation parameters, rumen microbial population and community composition of buffalo rumen inoculum, resulting from incubation with 4 typical subtropical forages.
2. Materials and methods
2.1. Ethical statement
The animals included as donors for rumen inoculum in this study were housed at Buffalo Research Institute, Chinese Academy of Agricultural Sciences, Nanning, Guangxi province, China. All experimental protocols regarding animal handling and treatment were approved by the Animal Care Committee, Guangxi University, under guidance of the International Cooperation Committee of Animal Welfare, China.
2.2. Rumen inoculum donors and their ration
Rumen inoculum in the present study was collected from 3 ruminal-cannulated buffalos (Bubalus bubalis, a hybrid of the Murrah and the local Chinese buffalo). The buffalos were dry pregnant females with similar live weights (around 500 kg). The buffalos received 3 kg/(animal·day) concentrate with ad libitum corn silage and free access of water. Composition of concentrate is as follows (based on dry matter): maize 52%, wheat bran 18.5%, soybean meal 8%, cotton seed meal 15%, stone dust 2%, calcium hydrogen phosphate 1.5%, sodium chloride 2% and premix 1%. The premix contained per kilogram: 119 g of MgSO4·H2O, 2.5 g of FeSO4·7H2O, 0.8 g of CuSO4·5H2O, 3 g of MnSO4·H2O, 5 g of ZnSO4·H2O, 10 mg of Na2SeO3, 40 mg of KI, 30 mg of CoCl2·6H2O, 28.5 g of vitamin A1, 0.44 g of vitamin D, and 16.2 g of vitamin E.
2.3. Substrates and their nutritional composition analysis
Cassava residues (Manihot esculenta), corn straw silage (Zea mays), elephant grass (Pennisetum purpureum) and sugarcane tail silage (Saccharum officinarum) used as substrates for in vitro incubation were from the farm of Buffalo Research Institute, Chinese Academy of Agricultural Sciences, Nanning, Guangxi province, China. These forage samples were dried at 65 °C until their constant weight was achieved, ground with a small shredder and through a 1-mm screen and stored at −20 °C until analysis for chemical composition and in vitro gas production. The nutritional composition of the 4 forages is shown in Table 1. The forage samples were analysed for dry matter (DM) content by oven-drying for 8 h at 105 °C, and crude protein (CP) was calculated as N × 6.25 (AOAC, 1990). Neutral detergent fiber (NDF) and acid detergent fiber (ADF) contents were determined according to the method as described by Van Soest et al. (1991).
Table 1.
Nutritive profile of substrates used for incubation (on dry matter basis, unless indicated).
| Nutrients | Substrates |
|||
|---|---|---|---|---|
| Cassava residues | Corn straw silage | Elephant grass | Sugarcane tail silage | |
| Dry matter, % | 95.8 | 95.3 | 93.6 | 95.0 |
| Protein, % | 2.26 | 10.8 | 13.7 | 7.68 |
| Neutral detergent fiber, % | 21.5 | 52.4 | 58.0 | 68.3 |
| Acid detergent fiber, % | 15.7 | 30.9 | 32.5 | 37.6 |
2.4. In vitro fermentation and gas production
In vitro fermentation system was set up following the procedure as described by Tang et al. (2008). Equal volumes of rumen inoculum taken from the selected 3 buffalos were mixed together. Rumen contents were strained through 4 layered cheesecloth into a pre-warmed Erlenmeyer flask. All laboratory handling of rumen inoculum was performed under the continuous flow of carbon dioxide (CO2) gas. In vitro fermentation process was carried out in glass syringes (100 mL) fitted with plungers (Tang et al., 2008). Every glass syringe was anaerobically dispensed with fermentation medium comprising 10 mL of buffalo rumen inoculum and 20 mL of McDougall's buffer solution and 200 mg of dried substrate. In total there were 32 glass syringes with 4 substrates and 8 experimental replicates (n = 8). In addition, a similar set of 4 glass syringes containing only fermentation medium was also run to serve as the blank controls to correct gas production resulting from the fermentation of dry matter in the rumen inoculum. Glass syringes containing fermentation medium and substrate were incubated in a shaking water bath at 39 °C and gas production was recorded at 0, 2, 6, 12, 24, 36, 48 and 72 h of incubation.
2.5. Sampling and volatile fatty acid analysis
After 24 h of incubation, fermentation process in 4 of the 8 replicates was ceased by placing glass syringes in an ice-cold water bath, and samples of fermented rumen inoculum were collected. The collected samples were filtered through 4 layered cheesecloth into a 50-mL centrifuge tube. A 2-mL aliquot of the filtrate was immediately subjected to determine the concentrations of VFA using gas chromatography (GC-2010, Shimadzu, Tokyo, Japan), equipped with a flame ionization detector and a capillary column (HP-INNOWAX, 1909N-133, Agilent Technologies, Santa Clara, CA, USA) as described by Zhang et al. (2008). Another 2-mL aliquot of the filtrate was stored at −20 °C for metagenomic DNA extraction.
2.6. DNA extraction and quantitative real-time PCR (qRT-PCR)
DNA was extracted from 2 mL of the preserved fermented rumen inoculum sample, and was further employed to perform qRT-PCR and high throughput sequencing. DNA extraction was performed following the procedure as reported by Rius et al. (2012). Quantitative real-time PCR was performed to quantify the populations of bacteria, methanogen, fungi, protozoa, Ruminococcus albus, Fi. succinogenes, Selenomonas ruminantium, and Prevotella ruminicola, using the method as described by Jiao et al. (2014). Primers used were the same as described by Jiao et al. (2014) (Table 2). Briefly, standard curves were generated by tenfold serial dilutions of plasmid DNA containing the extracts of 16s and 18s rRNA gene inserts from every microbial group and bacterial species. Quantitative real-time PCR assay was performed by using SYBR Green Master Mix (Perfect Real Time Takara, Japan) on a Roche light cycle 480 real time PCR system (Riche, Basel, Switzerland), using 10-μL reaction mixture volume. Each reaction mixture contained 5 μL of Fast SYBR Green Master Mix, 0.5 μL of each primer (20 pmol/μL), 3.5 μL of nuclease-free water and 0.5 μL of DNA template (10 ng/μL). All standard dilutions and samples were assayed in triplicate with amplification carried out according to the following program: 95 °C for 10 min for initial denaturation, then 30 cycles at 95 °C for 20 s, annealing for 1 min at 62 °C, followed by terminal elongation at 72 °C for 5 min. The corresponding qRT-PCR efficiency for every microbial group and bacterial species ranged from 90% to 100% in this study. Total 16S rRNA or 18S rRNA gene copy numbers in samples were determined by relating the threshold cycle values to the standard curves. Copy numbers for the 16S rRNA gene in mL of rumen inoculum were calculated as proposed by Li et al. (2009). Values were converted to log10 for further statistical analysis.
Table 2.
Primers used for quantitative real-time PCR.
| Primer | Sequence (5′ to 3′) | Size, bp | Literature cited |
|---|---|---|---|
| Bacteria-F | CGGCAACGAGCGCAACCC | 146 | Denman and McSweeney (2006) |
| Bacteria-R | CCATTGTAGCACGTGTGTAGCC | ||
| Fungi-F | GAGGAAGTAAAAGTCGTAACAAGGTTTC | 120 | Denman and McSweeney (2006) |
| Fungi-R | CAAATTCACAAAGGGTAGGATGATT | ||
| Protozoa-F | GCTTTCGWTGGTAGTGTATT | 223 | Sylvester et al. (2004) |
| Protozoa-R | CTTGCCCTCYAATCGTWCT | ||
| Methanogen-F | TTCGGTGGATCDCARAGRGC | 140 | Denman et al. (2007) |
| Methanogen-R | GBARGTCGWAWCCGTAGAATCC | ||
| Fibrobacter succinogenes-F | GTTCGGAATTACTGGGCGTAAA | 121 | Denman and McSweeney (2006) |
| Fibrobacter succinogenes-R | CGCCTGCCCCTGAACTATC | ||
| Selenomonas ruminantium-F | CAATAAGCATTCCGCCTGGG | 138 | Stevenson and Weimer (2007) |
| Selenomonas ruminantium-R | TTCACTCAATGTCAAGCCCTGG | ||
| Ruminococcus albus-F | CCCTAAAAGCAGTCTTAGTTCG | 176 | Koike et al. (2001) |
| Ruminococcus albus-R | CCTCCTTGCGGTTAGAACA | ||
| Prevotella ruminicola-F | GAAAGTCGGATTAATGCTCTATGTTG | 74 | Stevenson and Weimer (2007) |
| Prevotella ruminicola-R | CATCCTATAGCGGTAAACCTTTGG |
F = forward; R = reversed.
2.7. High throughput sequencing and bioinformatics analysis
Four DNA samples from every substrate were pooled into one sample to analyze microbial community at 24 h of incubation. Metagenomic DNA samples were sent to the BGI genomic research center in Wuhan, China, for ruminal microbial community composition analysis. High throughput sequencing technique was conducted using illumina Miseq PE 250 platform (Illumina, Santiago, CA, USA). Bacterial and methanogen communities were analyzed using 16s rRNA gene sequencing, and protozoal communities using 18s rRNA gene sequencing (Kittelmann et al., 2013). The primers used for PCR amplifications are shown in Table 3. The sequence data reported in this study have been deposited in the NCBI database (accession No. SRR5483195–SRR5483198). All high throughput sequence data processing including sequence quality control, operational taxonomic unit (OTU) based analysis, taxonomy analysis, and diversity indices calculation were performed using Mothur V 1.31.2 (Patrick and Sarah, 2009). Sequences were grouped into OTU sharing 97% similarity of bacterial, methanogen and protozoal sequences, and put into phylogenetic groups using Basic Local Alignment Search Tool (BLAST). Bacterial 16S rRNA genes were blasted against the Green genes database (V201305; McDonald et al., 2008), methanogen 16S rRNA genes against databases provided by Seedorf et al (2014), and protozoal 18S rRNA genes against databases provided by Kittelmann and Janssen (2011). Bacterial data were summarized at phylum and genus levels, protozoal data at genus level, while methanogen data were summarized using a mixed taxonomic rank scheme (Janssen and Kirs, 2008). Bar charts were drawn using Microsoft Excel 2013, presenting bacterial, methanogen and protozoal abundance for different substrates in buffalo inoculum. Spearman's rank correlation was used to analyze relationship between in vitro ruminal bacterial, methanogen protozoal community and VFA concentrations, regardless of the substrate incubated. Spearman's rank correlations were plotted using the “corrplot” packages of R software (v3.2.3). Microbial taxa, which represented >1% of the total community within every microbial group (bacteria, methanogen, and protozoa), were included in the analysis.
Table 3.
Primers used for microbial community composition analysis.
| Microbes | Primer sequence (5′ to 3′) | Size, bp | Literature cited |
|---|---|---|---|
| Bacteria-F | GGCGVACGGGTGAGTAA | 427 | Hristov et al. (2012) |
| Bacteria-R | CCGCNGCNGCTGGCAC | ||
| Methanogen-F | AGGAATTGGCGGGGGAGCAC | 472 | Jeyanathan et al. (2011) |
| Methanogen-R | GCGGTGTGTGCAAGGAGC | ||
| Protozoa-F | AATTGCAAAGATCTATCCC | 511 | Kittelmann et al. (2013) |
| Protozoa-R | GACTAGGGATTGGAGTGG |
F = forward; R = reversed.
2.8. Statistical analysis
All raw data including rumen fermentation parameters, microbial populations and microbial relative abundance were sorted using Microsoft Excel. Data of ruminal fermentation and microbial population were analyzed as a one-way factorial design using the ANOVA procedure of SAS (2005), according to the following statistical model: Yi = μ + αi + εi, where Yi is the dependent variable, αi is the effect of substrate (i = 1, 4) and εi is the residual error. Differences among means were tested using Duncan's multiple range tests. Statistical significance was considered if P < 0.05.
3. Results
3.1. Effects of substrates on in vitro gas production, fermentation parameters and microbial population
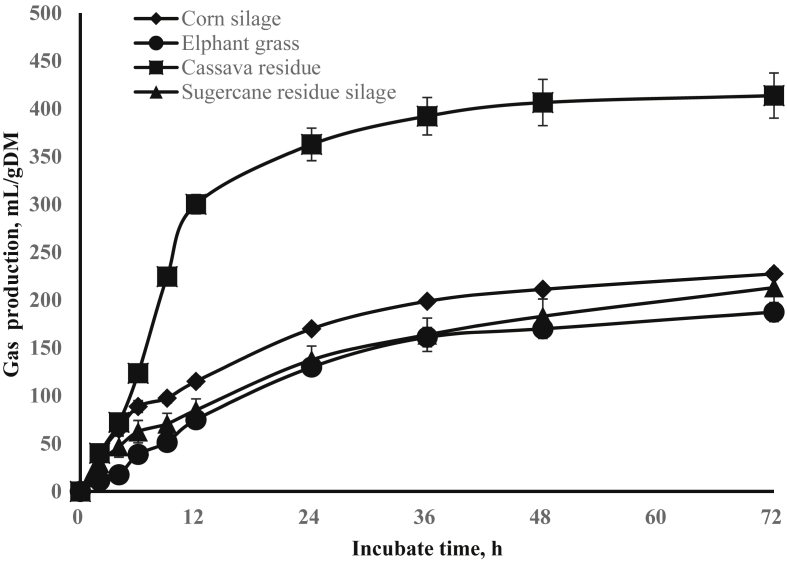
After 72 h of incubation, the highest gas production (GP) was found for cassava residues, followed by corn straw silage (Fig. 1). Elephant grass and sugarcane tail silage produced lower gas than the other 2 substrates (Fig. 1). After 24 h of incubation, the highest GP and concentrations of total VFA, acetate, propionate and butyrate were observed with cassava residues (P < 0.05, Table 4). Corn straw silage had the second highest GP, while total VFA, acetate and propionate concentrations were similar to those observed with elephant grass (P < 0.05). The lowest acetate:propionate ratio (A:P) was also observed with corn straw silage (P < 0.05). Twenty four hour GP was lower with both elephant grass and sugarcane tail silage (P < 0.05), but concentrations of total VFA, acetate and propionate for elephant grass were higher than those found for sugarcane tail silage (P < 0.05). The gene copy numbers of bacteria, methanogens, P. ruminicola, and S. ruminantium after 24 h of incubation were not influenced by the substrate source (P > 0.05, Table 4), while the lowest numbers of fungi, R. albus and Fibrobacter succinogenes and the highest number of protozoa were observed with cassava residues (P < 0.05).
Fig. 1.
The gas production of the 4 different substrates during 72 h incubation process.
Table 4.
Gas production, ruminal fermentation parameters, and microbial gene copy numbers in buffalo inoculum after 24 h of in vitro incubation of 4 different substrates.
| Index | Substrates |
SEM | P-value | |||
|---|---|---|---|---|---|---|
| Cassava residues | Corn straw silage | Elephant grass | Sugarcane tail silage | |||
| Gas production, mL/g | 363a | 170b | 130c | 138c | 3.883 | 0.020 |
| Total volatile fatty acids, mmol/L | 74.3a | 58.8b | 59.5b | 45.4c | 3.786 | <0.010 |
| Acetate, mmol/L | 53.3a | 43.3b | 46.4b | 34.9c | 2.658 | <0.010 |
| Propionate, mmol/L | 11.0a | 10.1a | 9.60a | 6.97b | 0.722 | <0.010 |
| Butyrate, mmol/L | 10.0a | 5.37b | 3.46c | 3.50c | 0.464 | <0.010 |
| Acetate/Propionate | 4.86a | 4.28b | 4.84a | 5.02a | 0.087 | <0.010 |
| Bacteria log10 copies/mL | 9.30 | 9.43 | 9.31 | 9.35 | 0.118 | 0.584 |
| Methanogen log10 copies/mL | 7.42 | 7.09 | 7.23 | 7.14 | 0.182 | 0.192 |
| Fungi log10 copies/mL | 4.35 | 6.03 | 6.71 | 6.16 | 0.263 | <0.010 |
| Protozoa log10 copies/mL | 7.16 | 6.74 | 6.30 | 6.94 | 0.188 | 0.012 |
| Ruminococcus albus log10 copies/mL | 5.45 | 6.21 | 6.43 | 6.16 | 0.137 | <0.010 |
| Fibrobacter succinogenes log10 copies/mL | 5.53 | 6.09 | 6.54 | 6.12 | 0.133 | <0.010 |
| P. ruminicola log10 copies/mL | 7.59 | 7.69 | 7.86 | 7.43 | 0.186 | 0.103 |
| S. ruminantium log10 copies/mL | 8.19 | 8.38 | 8.39 | 8.44 | 0.143 | 0.214 |
a–c Within a row, means with different superscripts differ at P < 0.05.
3.2. Effects of substrates on ruminal bacterial community
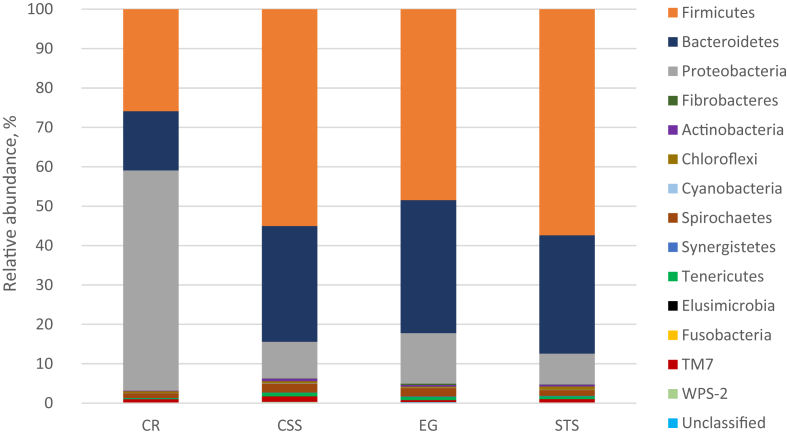
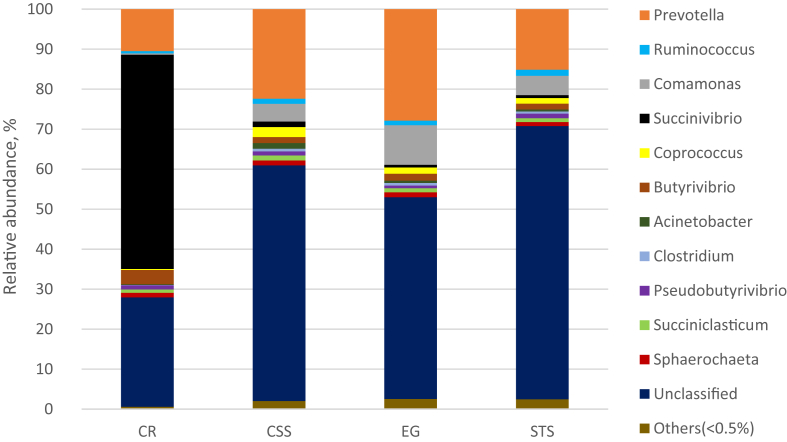
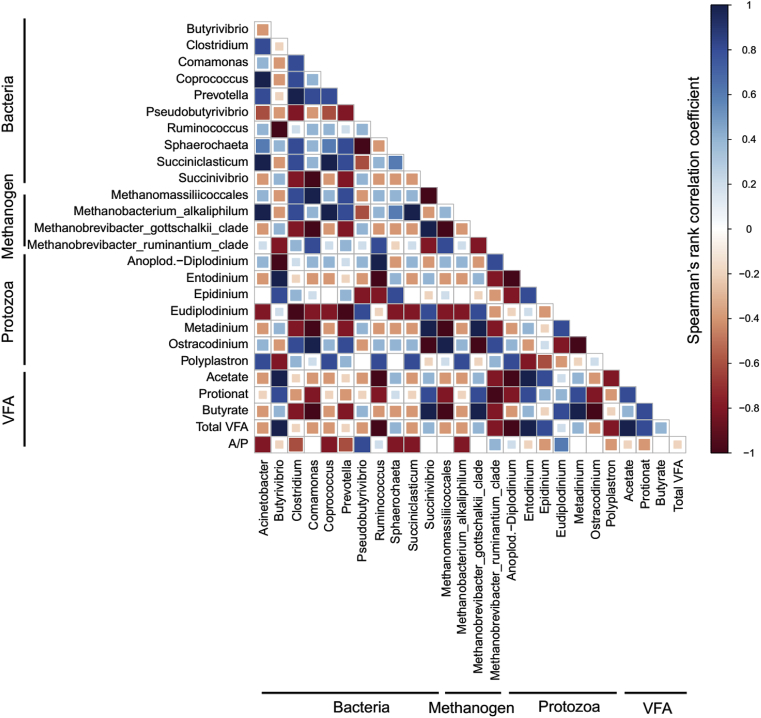
After 24 h of incubation, the lowest bacterial Chao1 and Shannon diversity index were observed for cassava residues (Table 5). Bacterial community composition analysis showed, Firmicutes (>45%) and Bacteroidetes (>29%) were the dominant bacterial phyla with corn straw silage, elephant grass and sugarcane tail silage (Fig. 2, Table A.1). The abundance of phylum Proteobacteria was substantially increased (55.9%) with cassava residues, while the abundance of Proteobacteria was not that much higher for other three substrates (<13%). At genus level, Prevotella was the most dominant, irrespective of the substrate incubated; but the lowest abundance of Prevotella (10.3%) was observed with cassava residues (Fig. 3, Table A.1). Moreover, the abundances of Succinivibrio and Butyrivibrio were higher with cassava residues as compared to those observed for the other three substrates; especially, Succinivibrio accounted for 52.5% of total bacterial abundance, while less than 1.5% with the other three substrates. Spearman's correlation analysis demonstrated that Succinivibrio had strong positive correlations with propionate, butyrate, Metadinium and Methanobrevibacter gottschalkii, and negative correlations with Ostracodinium and Methanomassiliicoccales (Fig. 4).
Table 5.
Rumen microbial alpha diversity statistics after 24 h of in vitro incubation of 4 different substrates.
| Microbes | Index | Substrate |
|||
|---|---|---|---|---|---|
| Cassava residues | Corn straw silage | Elephant grass | Sugarcane tail silage | ||
| Bacteria | Chao1 | 833 | 892 | 918 | 986 |
| Shannon | 3.52 | 5.75 | 5.58 | 5.86 | |
| Simpson | 0.24 | 0.01 | 0.01 | 0.01 | |
| Methanogens | Chao1 | 16.0 | 15.0 | 16.0 | 17.0 |
| Shannon | 0.89 | 1.05 | 1.29 | 1.17 | |
| Simpson | 0.56 | 0.48 | 0.41 | 0.43 | |
| Protozoa | Chao1 | 318 | 354 | 332 | 345 |
| Shannon | 3.34 | 3.33 | 3.42 | 3.37 | |
| Simpson | 0.09 | 0.09 | 0.07 | 0.09 | |
Fig. 2.
The taxonomic composition of bacterial phyla and their relative abundance in buffalo inoculum after 24 h of in vitro incubation of different substrates. CR = cassava residues; CSS = corn straw silage; EG = elephant grass; STS = sugarcane tail silage.
Fig. 3.
The taxonomic composition of bacterial genera and their relative abundance in buffalo inoculum after 24 h of in vitro incubation of different substrates. CR = cassava residues; CSS = corn straw silage; EG = elephant grass; STS = sugarcane tail silage. The genera of which abundance was less than 0.5% in all samples were classified into “others”.
Fig. 4.
Spearman's rank correlation matrix of the dominant microbes and volatile fatty acids (VFA) across domains in analyzed buffalo inoculum samples after 24 of in vitro incubation, regardless of the substrate source incubated. Microbial populations representing at least 1% of the bacterial, methanogenic and protozoa communities were selected to perform the analysis. Strong correlations are indicated by large squares, whereas weak correlations are indicated by small squares. Color denotes the nature of the correlation: 1 (dark blue) indicating perfect positive correlation; −1 (dark red) indicating perfect negative correlation between 2 microbial populations.
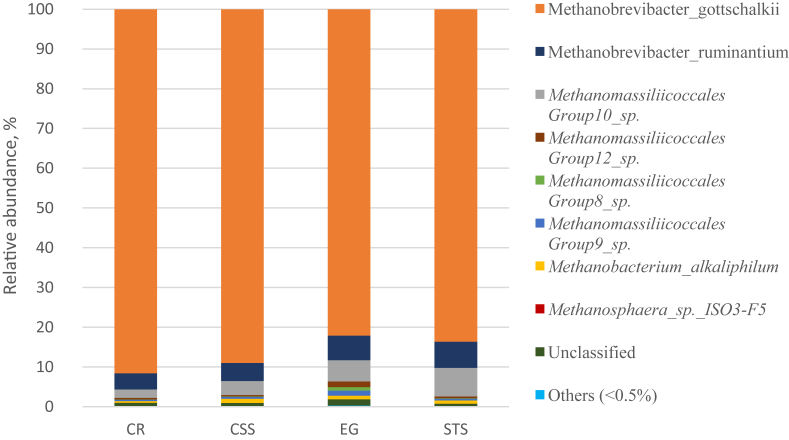
3.3. Effects of substrates on ruminal methanogen community
No significant differences in methanogen diversity indices were observed for the 4 substrates (Table 5). The differences existed in the abundances of methanogen at genus level, but not as obvious as those observed in bacterial community. Generally, Methanobrevibacter was the dominant genus and M. gottschalkii was the dominant species irrespective of the substrate incubated (Fig. 5, Table A.2), and the highest abundance of M. gottschalkii (91.6%) was observed with cassava residues. Spearman's correlation analysis demonstrated that M. gottschalkii had strong positive correlations with butyrate, Succinivibrio and Metadinium, and negative correlations with Comamonas and Methanomassiliicoccales (Fig. 4).
Fig. 5.
The taxonomic composition of methanogen species and their relative abundance in buffalo inoculum after 24 h of in vitro incubation of different substrates. CR = cassava residues; CSS = corn straw silage; EG = elephant grass; STS = sugarcane tail silage. The species of which abundance was less than 0.5% in all samples were classified into “others”.
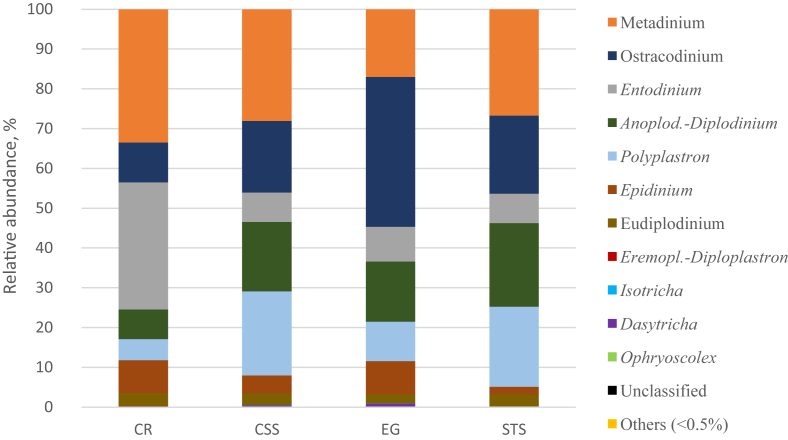
3.4. Effects of substrates on ruminal protozoal community
No significant differences in protozoal diversity indices were observed for the 4 substrates (Table 5). Protozoal community, which appeared with prominent variations with the 4 different substrates, was dominated by Entodinium (32.0%) with cassava residues, Ostracodinium (37.6%) with elephant grass, and Metadinium with both corn straw silage and sugarcane tail silage (Fig. 6, Table A.3). In addition, the abundance of Polyplastron with both elephant grass and cassava residues was lower than those with the other 2 substrates (Fig. 6, Table A.3). Spearman's correlations analysis showed that Metadinium had strong positive correlations with butyrate, Succinivibrio and M. gottschalkii, and negative correlations with Comamonas and Methanomassiliicoccales. Entodinium had strong positive correlations with acetate, total VFA and Butyrivibrio, and negative correlations with Ruminococcus and Anoplodinium diplodinium (Fig. 4).
Fig. 6.
The taxonomic composition of protozoal genera and their relative abundance in buffalo inoculum after 24 h of in vitro incubation of different substrates. CR = cassava residues; CSS = corn straw silage; EG = elephant grass; STS = sugarcane tail silage. The genera of which abundance was less than 0.5% in all samples were classified into “others”.
4. Discussion
In the present study, the in vitro gas production and total VFA recorded were highest for cassava residues, followed by corn straw silage. This might be because cassava residues had the lowest and corn straw silage had the second lowest ADF contents among the 4 substrates. Smith et al. (1988) also reported that cassava residues were rich in non-forage fiber and soluble carbohydrates and its rumen degradability value can reach 88.5% in 12 h. In addition, we also found that sugarcane tail silage, which was with the highest ADF content among the 4 substrates, produced lower gas at 72 h and the lowest concentrations of total VFA at 24 h of incubation. This further affirmed that ADF content was the key factor influencing in vitro degradability of substrates. We found, the populations of fiber degrading microbes, such as bacteria, fungi, R. albus and F. succinogenes, were decreased by in vitro incubation with cassava residues even though in vitro GP and VFA concentration were higher. These microbes, especially fungi which was responsible for fiber degradation in the rumen, was decreased by incubation of substrates with low fiber contents. This result was consistent with the findings of Saro et al. (2014), who reported that populations of ruminal F. succinogenes, Ruminococcus flavefaciens and fungi were increased in the sheep fed high NDF grass hay than in the sheep fed low NDF alfalfa hay. This result was also similar to the findings of Huws et al. (2010) who reported greater abundance of cellulolytic bacteria in the rumen of steers fed high NDF grass silage than in the rumen of steers fed low NDF red clover silage. Likewise, this study confirmed populations of fiber degrading microbes were increased by incubation with high fiber feedstuffs, similar to in vivo environment.
It has been reported that ruminal microbial community can be influenced by in vitro incubation with different substrates (Lengowski et al., 2016). This can be further evidenced in this study where Shannon diversity index of ruminal bacteria was lower for low NDF containing substrate such as cassava residues, while higher for the other 3 high NDF containing substrates. Grilli et al. (2016) also found that goat rumen bacterial Shannon diversity index was increased when fed 100% alfalfa hay as compared when fed 60% alfalfa hay. This indicated, the fiber contents of diet can increase the richness of species in the rumen liquor, which was consistent with results of the present study. Henderson et al. (2015) reported that ruminants share a core ruminal microbiome which is mainly composed of Firmicutes, Bacteroidetes and Proteobacteria regardless of the ruminant species; this was supported by the results of current study. Bacterial community in buffalo inoculum was dominated by Firmicutes, followed by Bacteroidetes after in vitro incubation with the 3 high fiber containing substrates. However, the abundances of phylum Proteobacteria as well as genus Succinivibrio, which is typically a group of Gram-negative and strictly anaerobic bacteria belonging to phylum Proteobacteria, were greatly increased; and the abundances of genus Prevotella and Ruminococcus were decreased by incubation with cassava residues, indicating special effects of cassava residues on ruminal bacterial community were due to its different nutrients composition. Thus, concentrations of total VFA and acetate after incubation of cassava residues were higher than those of other substrates, was probably due to the increased abundance of Succinivibrio, because members of family Succinivibrio can effectively ferment carbohydrates to succinate and acetate (Santos and Thompson, 2014). In addition, spearman's correlation analysis also showed that Succinivibrio had strong positive correlations with propionate and butyrate, indicating Succinivibrio was a main contributor for higher concentrations of butyrate after incubation with cassava residues. Interestingly, higher abundance of Succinivibrio, which was the reason of low methane production from hindgut of Tammar wallabies (Pope, 2011), was also observed in the inoculum incubated with cassava residues. This was also consistent with the conclusion that the ruminal methane production is always lower by feeding low fiber feedstuffs to ruminants (Gemeda and Hassen, 2015).
Methanogens in the rumen are the only microbes responsible for methane production. Though, there were no differences in methanogen populations resulting with different substrates, but the methanogen community was different for the different substrates. In particular, elephant grass and sugarcane tail silage, which are 2 high ADF substrates, caused low abundance of M. gottschalkii as compared with other 2 substrates, indicating that ADF rather than NDF probably inhibited the growth of M. gottschalkii, but this prediction needs further exploration. McCabe et al. (2015) reported that a strong inverse relationship existed between Succinivibrionaceae and M. gottschalkii in the rumen of cattle in vivo. In contrast, the present in vitro study showed the relationship was positive. The reason for inconsistency might be the difference between in vitro and in vivo environments.
The role of ruminal protozoa is to engulf ruminal bacteria and starch granules to keep ruminal pH steady (Newbold et al., 2015). It has been reported that if there is a readily available carbohydrate source, the protozoa can grow constantly and represent a larger population in the rumen (Veira, 1986). This conclusion was consistent with the results of the present study, where it was found that protozoal population was greatly increased upon incubation of cassava residues which was a high starch containing substrate. The ruminal protozoal community appeared greater variation after incubation with different substrates compared with that of bacteria and methanogens. Entodinium was the dominant genus (around 31%) on incubation of cassava residues, however its abundance was much lower (<8.8%) in the other 3 high fiber containing substrates. This was consistent with the findings of Coleman (1992), who reported that Entodinium species had the highest starch uptake rates as compared with other protozoal species. The abundance of Metadinium was lower and the abundance of Ostracodinium was higher with elephant grass compared with that of other substrates, suggesting there were some peculiar characteristics displayed by elephant grass compared with sugarcane tail silage and corn straw silage, even though they had similar NDF contents.
5. Conclusion
The 24-h in vitro gas and total VFA production were higher for cassava residues, indicating its higher digestibility and nutritional value compared with the 3 other forages. Fungal and some cellulolytic bacterial populations were reduced, and protozoal population was increased due to low NDF and high starch contents of cassava residues. Furthermore, the abundance of Succinivibrio, M. gottschalkii and Entodinium were higher for cassava residue compared with the other 3 forages, probably due to higher degradation rate of cassava residues. Similar microbial populations, bacterial and methanogen communities were observed for corn straw silage, elephant grass and sugarcane tail silage, but different protozoal community was observed for elephant grass.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Appendices
Table A.1.
Bacterial abundance in buffalo inoculum at the phylum and genus level after 24 h in vitro incubation.
| Taxon | Substrate |
|||
|---|---|---|---|---|
| Cassava residues | Corn straw silage | Elephant grass | Sugarcane tail silage | |
| Phylum | ||||
| Actinobacteria | 0.170 | 0.650 | 0.420 | 0.460 |
| Bacteroidetes | 15.02 | 29.36 | 33.76 | 30.03 |
| Chloroflexi | 0.450 | 0.480 | 0.330 | 0.850 |
| Firmicutes | 25.90 | 55.05 | 48.45 | 57.38 |
| Proteobacteria | 55.91 | 9.300 | 12.84 | 7.820 |
| Spirochetes | 1.230 | 2.250 | 2.120 | 1.470 |
| TM7 | 0.840 | 1.420 | 0.500 | 0.890 |
| Tenericutes | 0.280 | 0.880 | 0.830 | 0.720 |
| Genus | ||||
| Acinetobacter | 0.168 | 1.337 | 0.551 | 0.477 |
| Butyrivibrio | 3.474 | 1.433 | 1.653 | 1.388 |
| Clostridium | 0.123 | 0.634 | 0.656 | 0.511 |
| Comamonas | 0.290 | 4.203 | 9.509 | 4.581 |
| Coprococcus | 0.257 | 2.356 | 1.535 | 1.337 |
| Prevotella | 10.29 | 21.25 | 26.85 | 14.38 |
| Pseudobutyrivibrio | 1.005 | 0.965 | 0.616 | 1.081 |
| Ruminococcus | 0.547 | 1.213 | 1.128 | 1.473 |
| Sphaerochaeta | 1.139 | 1.143 | 1.207 | 0.971 |
| Succiniclasticum | 0.793 | 1.213 | 0.984 | 0.903 |
| Succinivibrio | 52.46 | 1.309 | 0.630 | 0.707 |
| Unclassified | 26.81 | 55.90 | 48.57 | 64.96 |
| Others (<0.5%) | 0.559 | 1.915 | 2.453 | 2.342 |
Table A.2.
Methanogen abundance of Buffalo and Jersey cow's rumen at genus level after 24 h in vitro fermentation with 4 different forages (percentage of bacteria in total bacteria > 0.5%).
| Species | Substrate |
|||
|---|---|---|---|---|
| Cassava residues | Corn straw silage | Elephant grass | Sugarcane tail Silage | |
| Methanomassiliicoccales Group10_sp. | 2.148 | 3.506 | 5.368 | 7.179 |
| Methanomassiliicoccales Group12_sp. | 0.421 | 0.347 | 1.420 | 0.466 |
| Methanomassiliicoccales Group8_sp. | 0.051 | 0.142 | 0.850 | 0.129 |
| Methanomassiliicoccales Group9_sp. | 0.340 | 0.456 | 1.298 | 0.406 |
| Methanobacterium alkaliphilum | 0.442 | 1.031 | 0.919 | 0.833 |
| Methanobrevibacter gottschalkii | 91.62 | 89.02 | 82.12 | 83.65 |
| Methanobrevibacter ruminantium | 4.011 | 4.537 | 6.165 | 6.570 |
| Methanosphaera_sp._ISO3_F5 | 0.006 | 0.000 | 0.000 | 0.004 |
| Unclassified | 0.828 | 0.808 | 1.557 | 0.678 |
| Others (<0.5%) | 0.137 | 0.152 | 0.304 | 0.086 |
Table A.3.
Protozoal abundance of buffalo and Jersey cow's rumen at genus level after 24 h in vitro fermentation with 4 different forages (percentage of bacteria in total bacteria > 0.5%).
| Genus | Substrate |
|||
|---|---|---|---|---|
| Cassava residues | Corn straw silage | Elephant grass | Sugarcane tail silage | |
| Anoplodinium diplodinium | 7.450 | 17.46 | 15.11 | 21.04 |
| Entodinium | 31.99 | 7.400 | 8.750 | 7.360 |
| Epidinium | 8.270 | 4.460 | 8.310 | 1.910 |
| Diploplastron | 0.050 | 0.020 | 0.010 | 0.010 |
| Eudiplodinium | 3.220 | 2.980 | 2.310 | 3.140 |
| Isotricha | 0.020 | 0.040 | 0.060 | 0.020 |
| Metadinium | 33.46 | 28.06 | 17.03 | 26.76 |
| Ophryoscolex | 0.000 | 0.000 | 0.000 | 0.000 |
| Ostracodinium | 10.01 | 18.01 | 37.63 | 19.61 |
| Polyplastron | 5.310 | 21.11 | 9.940 | 20.10 |
References
- AOAC . 15th ed. AOAC International; Arlington: 1990. Official methods of analysis; p. 1117p. [Google Scholar]
- Bartocci S., Tripaldi C., Terramoccia S. Characteristics of foodstuffs and diets, and the quanti-qualitative milk parameters of Mediterranean buffaloes bred in Italy using the intensive system. Livest Prod Sci. 2002;77:45–48. [Google Scholar]
- Calabro S., Moniello G., Piccolo V., Bovera F., Infascelli F., Tudisco R. Rumen fermentation and degradability in buffalo and cattle using the in vitro gas production technique. J Anim Physiol Anim Nutr. 2008;92:356–362. doi: 10.1111/j.1439-0396.2007.00799.x. [DOI] [PubMed] [Google Scholar]
- Coleman G.S. The rate of uptake and metabolism of starch grains and cellulose particles by Entodinium species, Eudiplodinium maggii, some other entodiniomorphid protozoa and natural protozoal populations taken from the ovine rumen. J Appl Bacteriol. 1992;73:507–513. doi: 10.1111/j.1365-2672.1992.tb05013.x. [DOI] [PubMed] [Google Scholar]
- Denman S.E., McSweeney C.S. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol. 2006;58:572–582. doi: 10.1111/j.1574-6941.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- Denman S.E., Tomkins N.W., McSweeney C.S. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol. 2007;62:313–322. doi: 10.1111/j.1574-6941.2007.00394.x. [DOI] [PubMed] [Google Scholar]
- Fernando S.C., Purvis H.T., Najar F.Z., Sukharnikov L.O., Krehbiel C.R., Nagaraja T.G. Rumen microbial population dynamics during adaptation to a high grain diet. Appl Environ Microbiol. 2010;76:7482–7490. doi: 10.1128/AEM.00388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemeda B.S., Hassen A. Methane production of two roughage and total mixed ration as influenced by cellulase and xylanase enzyme addition. Sci Agric. 2015;72:11–19. [Google Scholar]
- Grilli D.J., Fliegerová K., Kopečný J., Lama S.P., Egea V., Sohaefer N. Analysis of the rumen bacterial diversity of goats during shift from forage to concentrate diet. Anaerobe. 2016;42:17–26. doi: 10.1016/j.anaerobe.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Henderson G., Cox F., Ganesh S., Jonker A., Young W., Global Rumen Census Collaborators. Janssen P.H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristov A.N., Lee C., Hristova R., Huhtanen P., Firkins J.L. A meta-analysis of variability in continuous-culture ruminal fermentation and digestibility data. J Dairy Sci. 2012;95:5299–5307. doi: 10.3168/jds.2012-5533. [DOI] [PubMed] [Google Scholar]
- Huws S.A., Lee M.R.F., Muetzel S.M., Scott M.B., Wallace R.J., Scollan N.D. Forage type and fish oil cause shifts in rumen bacterial diversity. FEMS Microbiol Ecol. 2010;73:396–407. doi: 10.1111/j.1574-6941.2010.00892.x. [DOI] [PubMed] [Google Scholar]
- Janssen P.H., Kirs M. Structure of the archaeal community of the rumen. Appl Environ Microbiol. 2008;74:3619–3625. doi: 10.1128/AEM.02812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyanathan J., Kirs M., Ronimus R.S., Hoskin S.O., Janssen H.P. Methanogen community structure in the rumens of farmed sheep, cattle and red deer fed different diets. FEMS Microbiol Ecol. 2011;76:311–326. doi: 10.1111/j.1574-6941.2011.01056.x. [DOI] [PubMed] [Google Scholar]
- Jiao J., Lu Q., Tan Z., Guan L., Zhou C., Tang S. In vitro evaluation of effects of gut region and fibre structure on the intestinal dominant bacterial diversity and functional bacterial species. Anaerobe. 2014;28:168–177. doi: 10.1016/j.anaerobe.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Kaiser D., Weniger J.H. In vivo and in vitro investigations for nutrient digestibility and heat production of ruminants under heat stress and different nutritional levels. V. Comparison of in vivo and in vitro investigations with respect to energy metabolism and energy content of rations. Arch Tierz. 1994;37:535–545. [Google Scholar]
- Kittelmann S., Seedorf H., Walters W.A., Clemente J.C., Knight R., Gordon J.I. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One. 2013;8 doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelmann S., Janssen P.H. Characterization of rumen ciliate community composition in domestic sheep, deer, and cattle, feeding on varying diets, by means of PCR-DGGE and clone libraries. FEMS Microbiol Ecol. 2011;75:468–481. doi: 10.1111/j.1574-6941.2010.01022.x. [DOI] [PubMed] [Google Scholar]
- Kong Y., Teather R., Forster R. Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol Ecol. 2010;74:612–622. doi: 10.1111/j.1574-6941.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- Koike S., Kobayashi Y. Development and use of competitive PCR assays for the rumen bacteria: Fibrobacter succinogenes, Ruminococcus albus and Ruminococcus flavefaciens. FEMS Microbiol Lett. 2001;13:361–366. doi: 10.1111/j.1574-6968.2001.tb10911.x. [DOI] [PubMed] [Google Scholar]
- Li M., Penner G.B., Hernandez-Sanabria E., Oba M., Guan L.L. Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. J Appl Microbiol. 2009;107:1924–1934. doi: 10.1111/j.1365-2672.2009.04376.x. [DOI] [PubMed] [Google Scholar]
- Lengowski M.B., Zuber K.H., Witzig M., Möhring J., Boguhn J., Rodehutscord M. Changes in rumen microbial community composition during adaption to an in vitro system and the impact of different forages. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0150115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez M.E., Ranilla M.J., Tejido M.L., Saro C., Carro M.D. Comparison of fermentation of diets of variable composition and microbial populations in the rumen of sheep and Rusitec fermenters. II. Protozoa population and diversity of bacterial communities. J Dairy Sci. 2010;93:3699–3712. doi: 10.3168/jds.2009-2934. [DOI] [PubMed] [Google Scholar]
- McCabe M., Cormican S.P., Keogh K., O'Connor A., O'Hara E., Palladino R.A. Illumina MiSeq phylogenetic amplicon sequencing shows a large reduction of an uncharacterized succinivibrionaceae and an increase of the methanobrevibacter gottschalkii clade in feed restricted cattle. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald I.R., Bodrossy L., Chen Y., Murrell J.C. Molecular ecology techniques for the study of aerobic methanotrophs. Appl Environ Microbiol. 2008;74:1305–1315. doi: 10.1128/AEM.02233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard O., Ahmad S., Rousseau F., Briard-Bion V., Gaucheron F., Lopez C. Buffalo vs. cow milk fat globules: size distribution, zeta-potential, compositions in total fatty acids and in polar lipids from the milk fat globule membrane. Food Chem. 2010;120:544–551. [Google Scholar]
- Mould F.L., Kleim K.E., Morgan R., Mauricio R.M. In vitro microbial inoculum: a review of its function and properties. Anim Feed Sci Technol. 2005;123–124:31–50. [Google Scholar]
- Newbold C.J., de la Fuente G., Belanche A., Ramos-Morales E., McEwan N.R. The role of ciliate protozoa in the rumen. Front Microbiol. 2015;26:6–13. doi: 10.3389/fmicb.2015.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D.S., Sarah L.W. Introducing mothur: open-source, platform- independent, and community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R.M., Forster R.J., Yang W., McKinnon J.J., McAllister T.A. Characterization of rumen bacterial diversity and fermentation parameters in concentrate fed cattle with and without forage. J Appl Microbiol. 2012;112:1152–1162. doi: 10.1111/j.1365-2672.2012.05295.x. [DOI] [PubMed] [Google Scholar]
- Pope P.B. Isolation of Succinivibrionaceae implicated in low methane emissions from tammar wallabies. Science. 2011;333:646. doi: 10.1126/science.1205760. [DOI] [PubMed] [Google Scholar]
- Rius A.G., Kittelmann S., Macdonald K.A., Waghorn G.C., Janssen P.H. Nitrogen metabolism and rumen microbial enumeration in lactating cows with divergent residual feed intake fed high-digestibility pasture. J Dairy Sci. 2012;95:5024–5034. doi: 10.3168/jds.2012-5392. [DOI] [PubMed] [Google Scholar]
- Saro C., Ranilla M.J., Tejido M.L., Carro M.D. Influence of forage type in the diet of sheep on rumen microbiota and fermentation characteristics. Livest Sci. 2014;160:52–59. [Google Scholar]
- Santos E., Thompson F. The family succinivibrionaceae. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.H., Stackebrandt E., editors. The prokaryotes. Springer; Heidelberg Berlin: 2014. pp. 639–648. [Google Scholar]
- Seedorf H., Kittelmann S., Henderson G., Janssen P.H. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. Peer J. 2014;2:e494. doi: 10.7717/peerj.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest P.J., Robertson J.B., Lewis B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- Smith O.B., Idowu O.A., Asaolu V.O., Odunlami M.O. Comparative rumen degradability of forages, browses, crop residues and by-products. In: Wilson R.T., Azeb M., editors. African small ruminant research and development. ILCA; Addis Ababa, Ethiopia: 1988. pp. 204–208. [Google Scholar]
- Stevenson D.M., Weimer P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol. 2007;75:165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- Sylvester J.T., Karnati S.K., Yu Z., Morrison M., Firkins J.L. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J Nutr. 2004;134:3378–3384. doi: 10.1093/jn/134.12.3378. [DOI] [PubMed] [Google Scholar]
- Tang S.X., Tayo G.O., Tan Z.L., Sun Z.H., Shen L.X. Effects of yeast culture and fibrolytic enzyme supplementation on in vitro fermentation characteristics of low-quality cereal straws. J Anim Sci. 2008;86:1164–1172. doi: 10.2527/jas.2007-0438. [DOI] [PubMed] [Google Scholar]
- Veira D.M. The role of ciliate protozoa in nutrition of the ruminant. J Anim Sci. 1986;63:1547–1560. doi: 10.2527/jas1986.6351547x. [DOI] [PubMed] [Google Scholar]
- Yang C.T., Bing-wen S.I., Diao Q.Y., Jin H., Zeng S.Q., Tu Y. Rumen fermentation and bacterial communities in weaned Chahaer lambs on diets with different protein levels. J Integrat Agri. 2016;15:1564–1574. [Google Scholar]
- Zapletalová M., Kasparovská J., Krízova L., Kašparovský T., Šer O., Lochman Bacterial community dynamics in a rumen fluid bioreactor during in-vitro cultivation. J Biotechnol. 2016;234:43–49. doi: 10.1016/j.jbiotec.2016.07.013. [DOI] [PubMed] [Google Scholar]
- Zhang C.M., Guo Y.Q., Yuan Z.P., Wu Y.M., Wang J.K., Liu J.X. Effect of octadeca carbon fatty acids on microbial fermentation methanogenesis and microbial flora in vitro. Anim Feed Sci Technol. 2008;146:259–269. [Google Scholar]