Abstract
Protein–RNA interaction networks are essential to understand gene regulation control. Identifying binding sites of RNA-binding proteins (RBPs) by the UV-crosslinking and immunoprecipitation (CLIP) represents one of the most powerful methods to map protein–RNA interactions in vivo. However, the traditional CLIP protocol is technically challenging, which requires radioactive labeling and suffers from material loss during PAGE-membrane transfer procedures. Here we introduce a super-efficient CLIP method (GoldCLIP) that omits all gel purification steps. This nonisotopic method allows us to perform highly reproducible CLIP experiments with polypyrimidine tract-binding protein (PTB), a classical RBP in human cell lines. In principle, our method guarantees sequencing library constructions, providing the protein of interest can be successfully crosslinked to RNAs in living cells. GoldCLIP is readily applicable to diverse proteins to uncover their endogenous RNA targets.
Keywords: RNA binding protein, UV crosslinking, CLIP, HaloTag, PTB
Introduction
RNAs, both coding and non-coding, play essential roles in diverse biological processes [1]. In most cases, they function through numerous interactions with RNA-binding proteins (RBPs) [2]. Identifying in vivo binding sites of these RBPs is critical to understand protein–RNA interaction networks in gene regulation control [2]. UV crosslinking and immunoprecipitation (CLIP) represents one of the most powerful methods to detect direct protein–RNA interactions [3]. Various CLIP methods, such as high-throughput sequencing of RNA isolated by CLIP (HITS-CLIP) [4], photoactivatable ribonucleoside-enhanced CLIP (PAR-CLIP) [5], individual-nucleotide resolution CLIP (iCLIP) [6], and their derivatives [7], [8], [9], have been developed to profile transcriptome-wide protein–RNA interactions. However, one major limitation for current CLIP methods is the material loss during the gel purification process when enriching UV-crosslinked protein–RNA complexes [2]. In addition, another limitation is that most CLIP methods rely on radioisotopes to visualize purified protein–RNA complexes, with the exception of the infrared-CLIP (irCLIP) [8] and UV crosslinking and analysis of cDNAs (CRAC) [10].
Purification of UV-crosslinked protein–RNA complexes is one of the key steps to ensure the specificity of the CLIP experiment. In most CLIP protocols, purification of these complexes is processed via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) separation followed by nitrocellulose membrane transfer [3]. In brief, immunopurified covalent protein–RNA complexes are first separated from non-specific RNAs through a denaturing SDS–PAGE process [3]. Next, the size-resolved complexes are transferred to nitrocellulose membranes. Since the nitrocellulose only binds to proteins, but not to nucleic acids, RNAs that remain bound to the membranes must be attached to proteins through covalent bonds formed by UV crosslinking [3]. In addition to the material loss during various purification steps, there are also experimental variations likely due to the imprecise size selection on nitrocellulose membranes.
To overcome these issues and minimize the purification steps, we set out to explore alternative approaches to achieve equally stringent purification by leaving out the gel steps. Here, we developed a method named gel-omitted ligation-dependent CLIP (GoldCLIP) that omits all gel purification steps. GoldCLIP allows us to perform highly reproducible CLIP experiments that are compatible with diverse crosslinking conditions.
Materials and methods
Construction of HaloTag fusion protein plasmids
The coding sequence of human PTBP1 with stop codon, which encodes polypyrimidine-tract binding protein (PTB), was cloned into the pENTR4 vector using Gibson Assembly (catalog No. E2611; New England Biolabs). Then LR recombination reaction (Invitrogen, Catalog No. 11791020) was applied to transfer the entry sequence into a destination vector, which is engineered in house from a pMSCV-puro (Clontech, PT3303-5) plasmid with inserted N-terminal HaloTag, followed by 3× TEV cleavage sites, 2× StrepII, and a Gateway™ recombination cassette. The sequence of pMSCV-Halo-3× TEV-2× StrepII-PTB-puro was confirmed by sequencing. Halo-YFP was constructed similarly.
Expression and detection of the HaloTag fusion protein
pMSCV-Halo-3× TEV-2× StrepII-PTB-puro and VSVg were co-transfected into the Ecotropic Phoenix HEK293T cells using Lipofectamine® 2000 Transfection Reagent (catalog No.11668019; Invitrogen) according to the manufacturer’s instruction. 48–72 h post transfection, media containing viruses were collected and filtered before infecting a HEK 293T cell line. 72 h after infection, puromycin (1 μg/ml) was added for the selection of stable cell lines. To check HaloTag fusion protein expression, approximately one million cells were collected and lyzed in 100 μl lysis buffer containing 50 mM Tris–HCl (pH 7.4), 100 mM NaCl, 1 mM DTT, 1% Triton X-100, 10% glycerol, and 1× protease inhibitor cocktail (catalog No. G6521; Promega) on ice for 15 min. The insoluble fractions were removed by centrifugation at maximum speed at 4 °C for 10 min using Centrifuge 5424R (Eppendorf) and the supernatant was incubated with HaloTag® Alexa Fluor® 660 Ligand at 1 μM final concentration (catalog No. G8471; Promega) at room temperature for 15 min. Then samples were heated with 4× SDS gel loading buffer at 70 °C for 10 min and loaded onto SDS–PAGE gels. After electrophoresis, the SDS–PAGE gels were directly scanned using the Odyssey® CLx Imaging System at 700 nm.
GoldCLIP-seq library preparation
In a typical GoldCLIP experiment, ∼1 × 107 HEK 293T cells expressing the Halo-PTB fusion protein were crosslinked using UVP crosslinker at either UVC (254 nm, 400 mJ/cm2) or UVA (365 nm, 400 mJ/cm2, pre-incubated for 16 h with media containing 100 μM 4-thiouridine). Crosslinked cells were then scraped off the plates and mixed with ∼5 × 105 of Drosophila S2 cells expressing a Halo-CG7544 fusion protein (serving as an internal normalizing control), dounced with type B pestle in lysis buffer (see above) and digested using micrococcal nuclease (1:1000; catalog No. M0247S; New England Biolabs) for 3 min at 37 °C. Magne® HaloTag® Beads (catalog No. G7281; Promega) were incubated with the lysates with rotation at 4 °C for about 10–16 h. Beads associated with Halo-PTB complexes were first washed with PBST (PBS + 0.1% Triton X-100), dephosphorylated with calf intestinal phosphatase (catalog No. M0290S; New England Biolabs) at 37 °C for 30 min. Then the beads were washed with Trizol LS reagent and equilibrated with 8 M urea. The beads were then washed five times with PNK buffer containing 50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, and 1% Triton X-100. The RNAs crosslinked with the PTB proteins were ligated with an RNA adapter (/5′P/AGGTCGGAAGAGCGGTTCAG/3ddC/) at 3′ end using T4 RNA Ligase I (catalog No. AM2141; Ambion) on beads at 16 °C overnight. Then, further denaturing washes using the buffers containing either 8 M guanidine, 8 M urea or 10% SDS were applied to the beads to completely remove non-covalent contaminants. Finally, PTB–RNA complexes were cleaved off the beads by TEV protease and digested with protease K (catalog No. P8102S; New England Biolabs) at 37 °C for 30 min. The RNA–peptide adducts were cloned following the iCLIP library cloning protocol [11].
The detailed protocol for GoldCLIP-seq is provided in File S1.
Western blotting
PVDF or nitrocellulose membranes were blocked by 5% skim milk at room temperature for 1 h. Then the membranes were incubated at 4 °C overnight with the primary antibodies, including anti-HaloTag (1:2000; catalog No. G9211, Promega), anti-tubulin (1:2000; catalog No.9099; Cell Signaling Technology), anti-StrepII (1:2500; catalog No. B1195; Biodragon Beijing, China), and anti-PTB (1:2000; mAb BB7, ATCC® CRL-2501) [12]. After a brief wash, secondary antibodies (1:2000) were incubated with the membrane at room temperature for 1 h and washed with TBST (TBS + 0.1% Tween) three times. The immunoblots were developed using Pierce ECL reagent (catalog No. 35050, Thermo Scientific) and imaged with Tanon 5200 (Tanon, Shanghai, China).
Immunofluorescence microscopy
Cells were fixed in 4% paraformaldehyde diluted in PBS for 30 min. After three PBS washes, cells were permeabilized with PBS containing 0.5% Triton X-100 for 1 h at 4 °C. The cells were blocked with 1% bovine serum albumin in PBST (PBS + 0.1% Triton) for 1 h. HaloTag® TMR Ligand (1:2000; catalog No. G8251; Promega) and anti-PTB [12] (1:2500) were applied for 1 h at 4 °C in blocking solution, respectively. Then secondary antibody conjugated with Alexa Fluor 488 (catalog No A11034; Thermo Fisher Scientific) was incubated with cells for 1 h at room temperature. After three washes, cells were counterstained with DAPI (catalog No P36931; Thermo Fisher Scientific) at room temperature for 5 min. After three washes, the cells were imaged with confocal laser scanning microscope (Olympus FV1000).
Radioisotope labeling
To demonstrate the purity of UV crosslinked PTB–RNA complexes, Halo-PTB/RNA complexes (with or without UV crosslinking) bound to Magne® HaloTag® Beads were dephosphorylated by calf intestinal phosphatase (New England Biolabs). The complexes were washed on beads following GoldCLIP denaturing conditions and labeled with [γ-32P] ATP using T4 polynucleotide kinase (catalog No. M0201; New England Biolabs). The PTB–RNA complexes were cleaved off the beads using TEV protease and separated with 10% SDS–PAGE gels. The 32P-labeled RNA was visualized by Typhoon FLA 7000 (GE Healthcare).
Reads pre-processing
RNA samples were sequenced on either Illumina HiSeq X Ten or HiSeq 2500 platform. Sequencing data were pre-processed using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit). GoldCLIP libraries were de-multiplexed using the sample barcodes located at positions 4–7 and potential PCR duplicates were removed using the random barcodes at the positions 1–3 and 8–9. The 3′ adapter sequence and low-quality bases at 3′ end of reads were trimmed using Cutadapt (v1.10) [13] with command ‘cutadapt --adapter = AGATCGGAAGAGCGGTTCAG --error-rate = 0.2 --quality-cutoff = 20 --minimum-length = 24′ and the reads shorter than 24 bp were discarded. Next, reads with identical sequences were collapsed to remove potential PCR duplicates. Afterward, the random barcode and experimental barcode (positions 1–9) were removed.
Reads mapping
The processed reads (read 1 for paired-end reads) were first mapped to the spike-in fly genome (dm3, download from UCSC) using Bowtie [14] (v1.1.2; bowtie -v 2 -k 1 --best --un -S -p 8 -f). Reads mapped in this step were discarded. The remaining unmapped reads were then mapped against a library of mature tRNAs and rRNAs, as well as the mitochondrial genome (tRNA sequences were retrieved from Genomic tRNA Database (http://gtrnadb.ucsc.edu/genomes/eukaryota/Hsapi19/) and CCA was appended to the 3′ end of the sequences; rRNA sequences were retrieved from RefSeq id, NR_023363.1, NR_003285.2, NR_003287.2, and NR_003286.2). Mapping was performed using Bowtie [14] (v1.1.2; bowtie -v 2 -k 1 --best --un -S -p 8 -f) and the reads mapped were discarded from further analysis. The remaining unmapped reads were mapped against the human genome (hg19, UCSC) using Bowtie (options -v 2 -m 1 --best --strata -S -p 8 -f). The uniquely-mapped reads on human genome were retained for further analysis.
Peak calling and annotation
Peak calling was performed on uniquely-mapped reads using CLIPper [15] with options -s hg19 -o --save-pickle --bonferroni --superlocal --threshold-method binomial. Peaks were assigned to genic regions based on the following priority order: TSS > rRNA > pseudo-gene > ncRNA > 3′ UTR > 5′ UTR > CDS > intron > intergenic region. The software packages pyBEDTools [16] and BEDtools were used to enumerate overlap between peaks and different regions.
Overrepresented PTB-binding motifs
Overrepresented hexamers (6-mers) were identified by comparing the counts in peaks to those in random intervals. The histogram of Z-scores indicates the enrichment of hexamers in GoldCLIP peaks, compared to the randomly-selected similarly-sized regions in the same genes. 100 times random intervals were generated with a custom Python script. Z-scores of the top three 6-mers are indicated. De novo motif finding was performed using HOMER’s findMotifsGenome.pl (http://homer.salk.edu/homer/) script with options ‘-p 8 -rna -S 10 -len 5,6,7′ from the GoldCLIP peak sequences, compared to a set of background ‘peaks’ where three random identically-sized regions were sampled for each real GoldCLIP peak in the same genic regions.
Coverage of PTB-binding motifs at the crosslink clusters
To evaluate the enrichment of CU-rich motifs at the crosslink clusters, the method described in Haberman et al. [17] was adopted to calculate the coverage of PTB-binding motifs at the crosslink clusters.
Data analysis
Raw files of all libraries generated for this study were submitted to GEO (GEO accession number GSE111406). Data analysis was performed in the Python (2.7.13) or R (3.3.1) environment with frequently used R packages (ggplot2 (2.2.1) and dplyr (0.7.0). Pearson’s correlations between biological replicates were calculated using R.
Results
Characterizing HaloTag for purification of covalent protein–RNA complexes
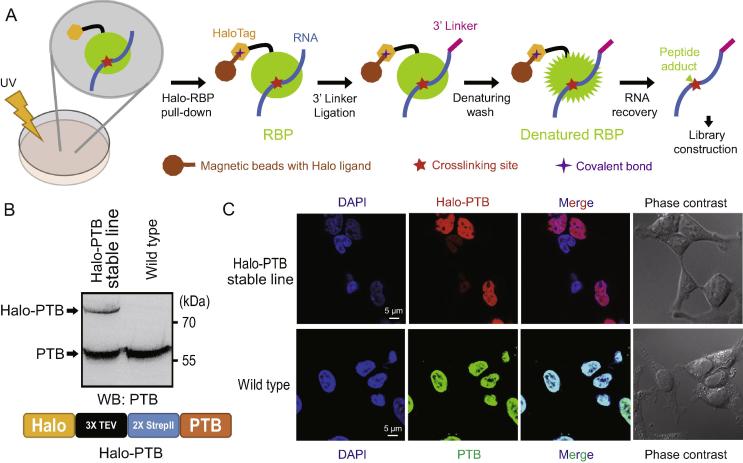
In principle, any protein tag that can form a covalent bond with its binding partner will resist denaturing purification conditions. With the help of such a covalent tag, no gel steps are necessary to isolate crosslinked protein–RNA complexes. To test this idea, we fused a HaloTag to the N-terminus of PTB, a classical RBP [18]. HaloTag is a modified haloalkane dehalogenase designed to covalently bind synthetic chloroalkane ligands irreversibly [19]. The covalent bond formed between HaloTag and its ligand is stable even when heated at 95 °C [19]. Once HaloTag binds covalently to the beads coated with chloroalkane ligands, non-specific contaminants can be readily removed under completely denaturing conditions (Figure 1A and Supplementary File 1). Subsequently, the protein–RNA complexes can be released from the beads via proteases and the resulting purified RNAs can be cloned into libraries for high-throughput sequencing [6] (Figure 1A). First, we established a HEK 293T cell line that stably expresses a Halo-PTB fusion protein with a 3× TEV cleavage site and a 2× StrepII engineered in between (Figure 1B). The epitope-tagged PTB is localized correctly in the nucleus, similar to the endogenous protein (Figure 1C). To specifically purify the Halo-PTB complexes, chloroalkane-ligand coated magnetic beads were used to capture the Halo-tagged fusion proteins. As shown in Figure 2A, the majority of Halo-PTB complexes were depleted from the lysates, suggesting that the Halo-tagged proteins can efficiently bind to the beads.
Figure 1.
HaloTag based GoldCLIP technology
A. Schematic flow chart of GoldCLIP technology. Cells stably expressing Halo-tagged fusion RBPs are crosslinked by UV irradiation. After cell lysis, Halo-RBP complexes are then captured by magnetic beads coated with Halo ligand under native conditions and a specific 3′ linker is ligated to RNAs bound by RBPs. Following denaturing washes, purified RNAs are cloned via an iCLIP protocol for high-throughput sequencing. B. Western blot analysis showing the expression level of Halo-PTB in the HEK 293T Halo-PTB stable cells compared to endogenous PTB using a monoclonal anti-PTB antibody (BB7). Non-transfected HEK 293T cells are used as control. A diagram of Halo-PTB fusion protein is shown below. C. Localization of Halo-PTB fusion proteins in 293T cell line. HaloTag TMR ligand staining of Halo-PTB fusion protein is shown in the top panel, and immunofluorescent staining of endogenous PTB using a monoclonal PTB antibody (BB7) is shown in the bottom panel. RBP, RNA-binding protein; iCLIP, individual-nucleotide resolution CLIP; PTB, polypyrimidine tract-binding protein; TMR, tetramethylrhodamine; TEV, tobacco etch virus.
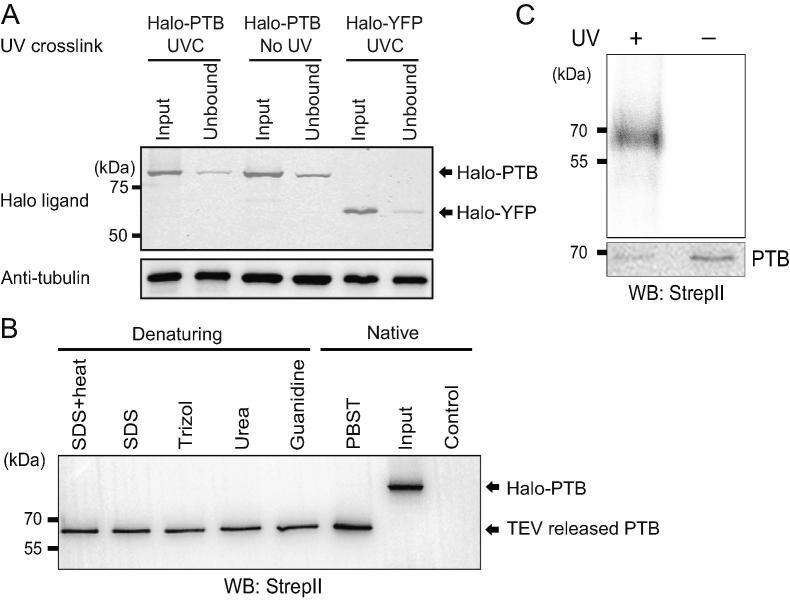
Figure 2.
Isolation of PTB–RNA complexes without gel purifications
A. Comparison of Halo fusion proteins (input vs. unbound fractions) for the indicated samples. The Halo fusion proteins are labeled with HaloTag Alexa Fluor® 660 ligand and resolved on SDS–PAGE. Bottom panel, Western blot analysis of tubulin as a loading control. B. Western blot analysis showing that similar amounts of PTB–RNA complexes are released by TEV protease following the indicated washing conditions after Halo bead isolation from the HEK 293T Halo-PTB stable cells. Non-transfected HEK 293T cells are used as control. C. Autoradiogram (upper panel) of 32P-labeled RNA crosslinked to PTB purified by HaloTag and released by TEV protease. RNA–protein complexes of 60–70 kD are seen only with the UV crosslinking condition. Western blot analysis (bottom panel) of TEV-released PTB proteins from equal amounts of samples prepared from the lysate of the Halo-PTB stable cell line.
Next, we tested the stability of the covalent bond formed between the chloroalkane-ligands and the Halo-PTB fusions. The beads pre-bound with Halo-PTB proteins were rigorously washed using various denaturing conditions such as SDS with heat, guanidine, urea, or Trizol. Compared to a native washing condition (PBST), significant amount of PTB proteins were retained and cleaved off the beads using TEV proteases after various denaturing washes (Figure 2B). Therefore, the Halo-PTB proteins attached to the beads are resistant to the “hash” washing conditions that we tested. Remarkably, following these washes (see the Methods and File S1), T4 polynucleotide kinase was only able to add radiolabeled phosphate to a single UV-dependent complex that matches the size of PTB (Figure 2C). Accordingly, the TEV-released PTB–RNA complexes are essentially free of any detectable nucleic acid contaminants. Taking advantage of the reverse transcriptase (RT) stops generated during reverse transcription of the RNA–peptide adducts, we produced cDNA libraries for sequencing following a standard iCLIP protocol [6].
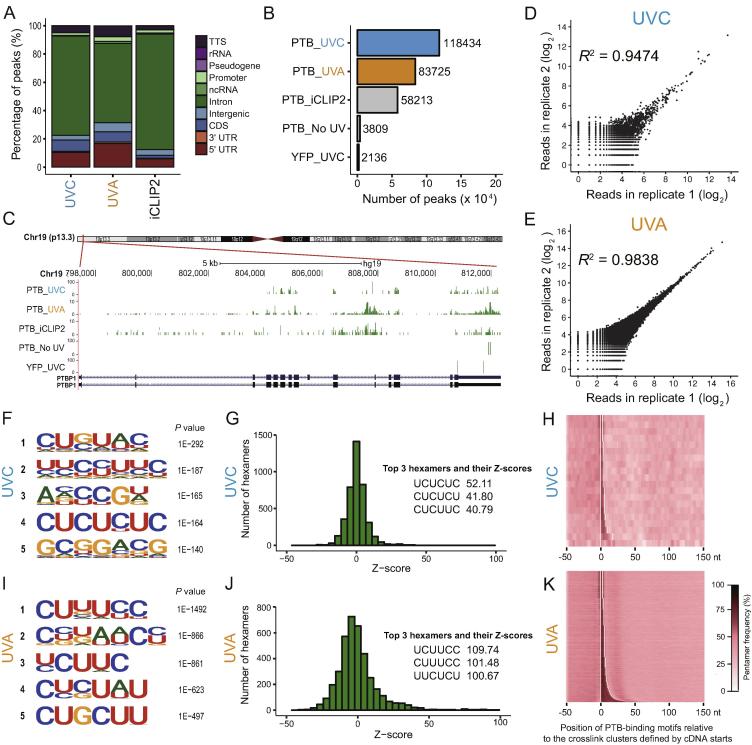
GoldCLIP identifies endogenous PTB targets
We next analyzed the GoldCLIP data adopting the published bioinformatics tools for iCLIP [6]. The majority of peaks are located in intronic regions across the human genome (Figure 3A), similar to the published datasets [20]. As shown in Figure 3B, comparable number of peaks were obtained using cells crosslinked with either UVC (254 nm) or UVA (365 nm, supplemented with photoactivatable 4-thiouridine). Importantly, the peaks identified by GoldCLIP are dependent on UV crosslinking (Figure 3B); only trivial number of peaks can be identified with Halo-YFP, a negative control that does not bind RNAs (Figure 3B). Consistent with previous data [20], GoldCLIP successfully identified PTBP1 pre-mRNAs as one of the PTB targets, suggesting that an autoregulation loop may be in place to control the posttranscriptional processing and/or expression of PTBP1 (Figure 3C).
Figure 3.
GoldCLIP identified endogenous RNA targets of PTB
A. Comparison of genomic distribution of the uniquely-mapped reads identified by Halo-PTB GoldCLIP or the published iCLIP datasets. Color code is indicated in the legend box on the right. TTS, transcription termination site. B. Total number of peaks identified by GoldCLIP. Two different crosslinking conditions (UVC in blue, 254 nm and UVA in orange, 365 nm) are shown with two negative controls: Halo-PTB without UV crosslinking (PTB_No UV) and Halo-YFP crosslinked with UVC (YFP_UVC). Peaks identified using the published datasets (PTB_iCLIP2) are shown in gray. C. Comparison of Halo-PTB clusters identified by GoldCLIP at the PTBP1 locus. Genomic tracks of reverse transcriptase stops are shown for the different samples, i.e., Halo-PTB crosslinked with either UVC (PTB_UVC) or UVA (PTB_UVA), iCLIP from endogenous PTB (PTB_iCLIP2), Halo-PTB without UV crosslinking (PTB_No UV) and Halo-YFP crosslinked with UVC (YFP_UVC). D. and E. show the highly correlated Pearson’s coefficient between the number of reads obtained from two biological replicates in a 500 bp window across the whole genome for UVC crosslinking (D) and UVA (E), respectively. F. Top HOMER motifs calculated from the peak reads after UVC crosslinking are shown. G. Over-represented Halo-PTB binding motifs identified by GoldCLIP after UVC crosslinking. Histogram of Z-scores indicates the enrichment of hexamers in GoldCLIP clusters compared to randomly chosen regions of similar sizes in the same genes. Z-scores of the top three hexamers are indicated. H. Heatmap showing the coverage of Halo-PTB binding motifs at crosslink clusters that are defined with a 3-nt clustering window. The clusters are sorted from the shortest to the longest. The nucleotide preceding the start and the nucleotide following the median end of all clusters are marked by white lines in the plot. A color key for the coverage per nucleotide of the PTB-binding motifs is shown on the right. I. Similar to F except UVA crosslinking condition was used. J. Similar to G except UVA crosslinking condition was used. K. Similar to H except UVA crosslinking condition was used.
Since no gel steps were involved, minimum material loss is expected during the HaloTag purification processes (Figure 1A). Consequently, there is no need to cut membranes to isolate covalent protein–RNA complexes. Such membrane-cutting steps might greatly contribute to experimental variabilities, owing to the ambiguity of size-range selection. As a result, data generated by GoldCLIP are highly reproducible between biological replicates (Figure 3D and E).
To determine the accuracy of the GoldCLIP experiments, we performed a de novo motif search using HOMER [21]. The motifs we identified nicely match the known binding consensus of PTB [20], [22] (Figure 3F and I). We also searched for the significantly-enriched 6-mers in the datasets and obtained similar results (Figure 3G and J). The sequence features of the GoldCLIP reads were then compared to the crosslink sites identified as 3′ ends of cDNAs (e.g., RT stops). The most enriched pentamers overlap nicely with the RT stops (Figure 3H and K), further demonstrating that GoldCLIP can accurately pinpoint RBP binding sites at single-nucleotide resolutions [20]. Moreover, comparable results were obtained using either UVC or UVA crosslinking (Figure 3F−H and I–K), suggesting that GoldCLIP is readily compatible with various RBP–RNA fixation conditions.
Discussion
The GoldCLIP method described here skips all gel purification steps and produces highly-reproducible results, which greatly simplifies the previously challenging CLIP-based technologies. In principle, our GoldCLIP method ensures successful construction of sequencing libraries, providing the protein of interest can be crosslinked with its RNA targets in vivo. Moreover, far less amount of materials will certainly be possible with an optimized library construction strategy (i.e., irCLIP [8]). With the help of CRISPR/Cas9, HaloTag can be engineered into the endogenous gene loci [23]. Therefore, GoldCLIP is readily applicable to diverse factors to reveal their endogenous RNA targets at physiological levels. When coupled with barcoded 3′ ligation linkers [24], our bead-based methods can be adopted to perform high-throughput profiling of multiple RBPs simultaneously, facilitating a more comprehensive understanding of “RBP code”.
Authors’ contributions
Yu Y and Hannon GJ conceived the idea. Yu Y and Gu J designed the experiments. Gu J, Qiu D, and Yu Y performed the experiments. Wang M, Yang Y, and Zhang Y performed bioinformatics analysis under the supervision of Zhou Y and Yu Y. Yu Y, Gu J, and Wang M wrote the manuscript. Yu Y, Hannon GJ, Ma J, and Zhou Y provided the advice and reviewed the manuscript. All authors read and approved the final manuscript.
Competing interests
YY and JG are co-inventors on a patent related to the GoldCLIP method. There are no other conflicts of interests to be declared.
Acknowledgments
We thank members of Yu lab for helpful discussions, and Chunyan Fan and Jianli Hu for technical support. We are grateful for Yuanchao Xue for sharing reagents and Hongjie Zhang for support of radioactive isotopes. This work was supported in part by grants from the Ministry of Science and Technology of China (Grant No. 2017YFA0504200 to YY, Grant Nos. 2012CB910502 and 2011CB966304 to JM), and the National Natural Science Foundation of China (Grant Nos. 91640105 and 31770875 to YY, Grant No. 31230041 to JM, and Grant Nos. 91640115 and 31670827 to YZ).
Handled by Yun-Gui Yang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.1016/j.gpb.2018.04.003.
Contributor Information
Jinbiao Ma, Email: majb@fudan.edu.cn.
Yu Zhou, Email: yu.zhou@whu.edu.cn.
Gregory J. Hannon, Email: greg.hannon@cruk.cam.ac.uk.
Yang Yu, Email: yuyang@ibp.ac.cn.
Supplementary material
GoldCLIP-seq protocol
References
- 1.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler E.C., Van Nostrand E.L., Yeo G.W. Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip Rev RNA. 2018;9:e1436. doi: 10.1002/wrna.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 4.Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zarnegar B.J., Flynn R.A., Shen Y., Do B.T., Chang H.Y., Khavari P.A. irCLIP platform for efficient characterization of protein-RNA interactions. Nat Methods. 2016;13:489–492. doi: 10.1038/nmeth.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim B., Jeong K., Kim V.N. Genome-wide mapping of DROSHA cleavage sites on primary microRNAs and noncanonical substrates. Mol Cell. 2017;66:258–269. doi: 10.1016/j.molcel.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Granneman S., Kudla G., Petfalski E., Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci U S A. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B. iCLIP–transcriptome-wide mapping of protein−RNA interactions with individual nucleotide resolution. J Vis Exp. 2011;50:e2638. doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou M.Y., Underwood J.G., Nikolic J., Luu M.H., Black D.L. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- 13.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 14.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovci M.T., Ghanem D., Marr H., Arnold J., Gee S., Parra M. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dale R.K., Pedersen B.S., Quinlan A.R. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haberman N., Huppertz I., Attig J., König J., Wang Z., Hauer C. Insights into the design and interpretation of iCLIP experiments. Genome Biol. 2017;18:7. doi: 10.1186/s13059-016-1130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spellman R., Smith C.W. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Los G.V., Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 20.Haberman N., Huppertz I., Attig J., Konig J., Wang Z., Hauer C. Insights into the design and interpretation of iCLIP experiments. Genome Biol. 2017;18:7. doi: 10.1186/s13059-016-1130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Y., Zhou Y., Wu T., Zhu T., Ji X., Kwon Y.S. Genome-wide analysis of PTB−RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Nostrand E.L., Gelboin-Burkhart C., Wang R., Pratt G.A., Blue S.M., Yeo G.W. CRISPR/Cas9-mediated integration enables TAG-eCLIP of endogenously tagged RNA binding proteins. Methods. 2017;118–119:50–59. doi: 10.1016/j.ymeth.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hafner M., Lianoglou S., Tuschl T., Betel D. Genome-wide identification of miRNA targets by PAR-CLIP. Methods. 2012;58:94–105. doi: 10.1016/j.ymeth.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GoldCLIP-seq protocol