Abstract
The biogenesis of membrane oligomeric complexes is an intricate process that requires the insertion and assembly of transmembrane (TM) domains into the lipid bilayer. The Oxa1p family plays a key role in this process in organelles and bacteria. Hell et al. (2001, EMBO J., 20, 1281–1288) recently have proposed that Oxa1p could act as part of a general membrane insertion machinery for mitochondrial respiratory complex subunits. We have previously shown that mutations in the TM domain of Cyt1p can partially compensate for the absence of Oxa1p. Here, we demonstrate that a single amino acid substitution in the TM domain of Qcr9p can bypass Oxa1p in yeast. Qcr9p and Cyt1p are two subunits of the respiratory complex bc1 and their relative roles in the assembly of other respiratory complexes have been investigated. The mutations we have isolated in Cyt1p or Qcr9p introduce positively charged amino acids, and we show that the mutant TM domain of Cyt1p mediates the restoration of complex assembly. We propose that the positive charges introduced in Cyt1p and Qcr9p TM domains promote interactions with negatively charged TM domains of other respiratory complex subunits, allowing the coinsertion of both domains into the membrane, in the absence of Oxa1p. This model argues in favor of a role of Oxa1p in the insertion and the lateral exit of less hydrophobic TM domains from the translocation site into the lipid bilayer.
The biogenesis of membrane oligomeric complexes is an intricate process that requires the membrane targeting of subunits, their insertion into the lipid bilayer, folding with cofactors, and the assembly into a functional structure. Numerous studies in bacterial or eukaryotic cells have demonstrated that most membrane proteins insert at the translocon site and that this process is driven by hydrophobic transmembrane (TM) domains (1, 2). However, very little is known about the lateral exit of TM domains from the site of insertion into the lipid bilayer and about the mechanistic details of subunit assembly into functional complexes. The mitochondrial respiratory chain enzymes are complexes composed of numerous nonidentical subunits (Fig. 1). Their biogenesis requires the participation of nucleus-encoded factors that are not intrinsic components of the complexes and are imported into mitochondria by using a sophisticated translocation machinery (3–5). Most of these proteins are conserved through evolution and it has been shown that mutations in the genes encoding subunits of the respiratory complexes or assembly assisting proteins are the direct cause of several human neurodegenerative pathologies (ref. 6, for review).
Figure 1.
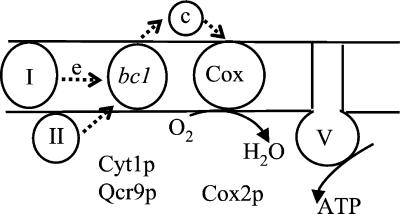
Schematic representation of the mitochondrial respiratory chain. Four respiratory complexes are embedded within the inner membrane of mitochondria: the complex I, III or the bc1 complex, IV or the cytochrome c oxidase complex (Cox) and the insoluble part of complex V or ATP synthase. c represents cytochrome c. bc1 and Cox complexes each contain about 10 subunits. Cyt1p and Qcr9p are nucleus-encoded subunits of bc1 complex and have one TM domain each. Cox2p is a mitochondria encoded subunit of Cox complex with two TM domains. The small arrows indicate the electron (e) flow.
Oxa1p, a membrane protein conserved from bacteria to eukaryotic organelles, is a key component of the insertion machinery of membrane proteins. The Escherichia coli homologue YidC interacts with the Sec translocase and mediates the insertion of a subset of proteins into the bacterial membrane (7, 8). In yeast mitochondria, the absence of Oxa1p leads to a complete loss of respiration associated with severe defects in the insertion and assembly of subunits of the Cox and ATPase complexes and with minor defects in the assembly of the bc1 complex (9–11). The translocation and maturation of the Cox subunit, Cox2p, is almost completely blocked (12, 13) and several other membrane subunits are degraded (14). Oxa1p interacts with nascent mitochondrial polypeptides and it has been proposed that it could act as part of a general membrane insertion machinery (15). Oxa1p is functionally conserved in fission yeast, human and plant mitochondria (16–18) and in chloroplasts, its homologue Alb3 plays a role in the insertion of the light harvesting chlorophyll-binding proteins into the thylakoid (19, 20).
The functional conservation of Oxa1p through evolution suggests a crucial role in the mechanism of protein insertion into the membrane, but its precise function remains poorly understood. Interestingly, we found that its absence in yeast mitochondria can be partially compensated for by different mutations. First, ATP synthase assembly and activity is fully restored in a strain lacking the mitochondrial protease Yme1p (14). Second, we have isolated several suppressor mutations in the TM domain of cytochrome c1 (Cyt1p), an electron carrier of the respiratory complex bc1 (21). These mutations partially restore the Cox and ATP synthase assembly independently of the electron transfer activity of cytochrome c1, showing that the function of Cyt1p revealed by the suppressor mutations is quite distinct from its well-defined role as an electron carrier.
In this study, we discovered that a single mutation introducing a positive charge in the TM domain of Qcr9p can also partially restore the insertion and assembly of Cox and ATP synthase in the absence of Oxa1p. Qcr9p is a bc1 complex subunit and the relative roles of Qcr9p and Cyt1p in the assembly of other respiratory complexes have been investigated. Although Qcr9p appears dispensable, we found that cytochrome c1 is essential for Cox assembly and that its TM domain is essential for the suppression activity. We propose that the presence of a positive charge in the TM domain of Qcr9p or Cyt1p promotes interactions with a negatively charged TM domain of other respiratory complex subunit, allowing the coinsertion of these two TM domains into the lipid bilayer even in absence of Oxa1p. This model argues in favor of a role for Oxa1p in the lateral exit of less hydrophobic TM domains, e.g., negatively charged TM domains, from their site of translocation within the membrane.
Materials and Methods
Strains, Media, Genetic Methods, and Plasmids.
All of the strains had the same nuclear background ade2–1 ura3–1 his3–11,-15 trp1–1 leu2–3, -112 can1–100. Yeast media, genetic methods, and the strains CW30, NBT2, and R101 were described in ref. 21. Ethanol/glycerol medium used was 1% yeast extract, 1% casamino acids, 0.05 M sodium potassium phosphate (pH 6.25), 3% ethanol, and 3% glycerol. YEpNB33 is a multicopy vector and pFL61 is a multicopy yeast expression vector; both carry the URA3 gene (21). The double or triple mutant strains were constructed either by transformations or genetic crosses between the various single mutants. Strain PHT28 was constructed by transforming R102 (see below) with a BstEII/KpnI fragment carrying the OXA1 gene and selecting for respiratory competent and uracil prototroph transformants.
Isolation and Cloning of Suppressor Mutations.
Genetic suppressors were isolated after UV mutagenesis of the oxa1∷LEU2 (NBT2) and oxa1∷URA3 (NBT3) strains (21). Molecular genetics enabled us to map seven of the suppressor mutations to the CYT1 gene and four to a distinct locus. To clone this gene, a genomic library was constructed from the suppressor strain R102. CsCl-purified nuclear DNA was partially digested with HinDIII and ligated with the HinDIII-digested YEpNB33. The ligated DNA was used to transform electrocompetent Escherichia coli cells to ampicillin and kanamycin resistance. About 380,000 independent transformants were recovered. The library DNA was CsCl-purified and used to transform the NBT2 strain. Of 400,000 uracil prototroph transformants, 21 were respiratory competent. Plasmids were isolated from these transformants and the inserts were characterized.
Inactivation of the QCR9 Gene: Cloning of the QCR9 cDNA.
For QCR9 inactivations, the NsiI–PstI fragment of QCR9 was replaced by a 1.5-kb NsiI fragment carrying the URA3 gene, or the PCR-amplified G418R cassette (22) was cloned at the Klenow-filled NsiI site. The two constructions were used to transform the wild-type strain CW30 to give YST1 (qcr9∷URA3) and PHT21 (qcr9∷G418R). Plasmid YEpYS3 carries the QCR9 cDNA that was cloned by complementing the PHT21 strain with a yeast cDNA library constructed in pFL61.
Epitope Tagging of Qcr9p.
We have tagged Qcr9p at its C terminus with 13 c-myc epitopes by using the HIS5 marker gene as described (23). DNA was amplified and used to transform to histidine prototrophy two yeast strains CW30 and PHT28 carrying the QCR9 or the QCR9–1 allele, respectively. Strains YS18–5A, YS16–1B expressing the wild type, or mutant tagged Qcr9p are respiratory competent, showing that the tagged proteins are functional.
Construction of a Short Form of Cyt1p.
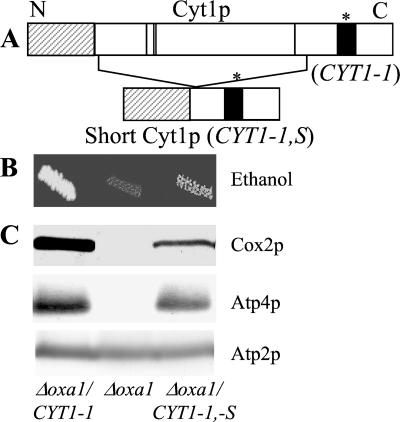
YEpPH225 and YEpYS7 are pFL61-based plasmids expressing either the wild-type CYT1 or the CYT1–1 cDNAs. First, a 450-bp AatII/AvaI internal fragment was deleted from YEpPH225 and YEpYS7. Then, PvuII–Acc65I-digested plasmids were Klenow-treated and self-ligated to give YEpYS12 and YEpYS9. These plasmids enable the expression of a short form of pre-Cyt1p protein in which the central hydrophilic region of Cyt1p (175 aa) is deleted whereas the first 63 aa are fused in-frame to the last 71 aa (see Fig. 6A).
Figure 6.
The mutant membrane domain of Cyt1p mediates suppression. (A) Schematic representation of the mutant pre-Cyt1p protein and the short form (see Materials and Methods for constructions). The black box represents the hydrophobic domain anchoring the protein to the membrane, and * indicates the position of the CYT1–1 mutation. The dashed box represents the 61-aa presequence that is cleaved to give the mature form Cyt1p and the four vertical lines indicate the positions of the heme ligands within the hydrophilic central domain. (B and C) The Δoxa1 mutant (NBT2) was transformed with three plasmids. YEpYS7 expresses the mutant Cyt1p protein (Δoxa1 CYT1–1), pFL61 is the control vector (Δoxa1), and YEpYS9 expresses the short form of the mutant Cyt1p (Δoxa1 CYT1–1,-S). (B) Transformants were grown on ethanol medium and incubated for a week at 28°C. (C) Mitochondria were purified from transformants grown on minimal medium to select for the presence of the plasmid. Cox2p, Atp4p, and Atp2p were immunodetected as described in Fig. 2C.
Isolation of Mitochondria, Carbonate Extraction, and Respiratory Chain Activities.
Mitochondria were purified from cells grown in complete or minimal galactose medium and carbonate extractions were performed as in ref. 14. Cytochrome c oxidase activity was measured according to ref. 21, ATPase activity according to ref. 24, and ubiquinol cytochrome c oxidoreductase activity according to ref. 25.
Electrophoresis and Immunoblotting.
They were performed as described (14). The anti-yeast Cox2p was purchased from Molecular Probes whereas the anti-Atp2p, anti-Atp4p, and anti-c-myc were generous gifts from J. Velours (Institut de Biochimie et Génétique Cellulaire, Bordeaux, France) and J. M. Galan (Institut J. Monod, Paris), respectively.
Results
A Single Amino Acid Substitution Introducing a Positive Charge in the TM Domain of Qcr9p Partially Compensates for the Absence of Oxa1p.
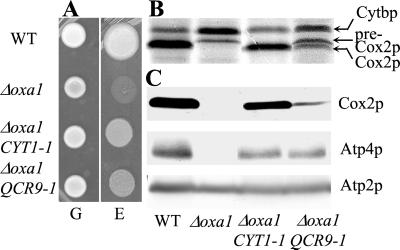
Yeast is a facultative aerobe organism that can derive energy from fermentation. Yeast cells carrying an inactivated Δoxa1 gene are thus viable but respiratory deficient. In consequence, they grow on a fermentable substrate such as glucose but are unable to grow on a nonfermentable medium containing ethanol or glycerol as respiratory substrate. In the search for partners of Oxa1p, we have isolated genetic suppressors that restore the respiratory growth of cells in the absence of Oxa1p (Fig. 2A and Materials and Methods). Among 11 independent dominant nuclear suppressor mutations, seven mapped to the CYT1 gene (21) and four to another gene. To identify this second gene, we constructed a genomic library from the strain R102 and used it to transform the Δoxa1 mutant. All of the respiratory-competent transformants recovered and harbored the QCR9 gene that encodes a noncatalytic subunit of the bc1 complex required for electron transfer activity (26, 27). We have shown that all four independent suppressors carry the same single nucleotide substitution (GGT>CGT:QCR9–1) at codon 23 of the QCR9 ORF, replacing a glycine residue by an arginine. Qcr9p is a small protein composed of 65 aa with a short C-terminal domain located in the intermembrane space and a TM domain where the G23K suppressor is located. As a control, we showed that overexpression of the wild-type QCR9 gene does not compensate for the absence of Oxa1p, demonstrating that the mutation is required for the suppression (data not shown).
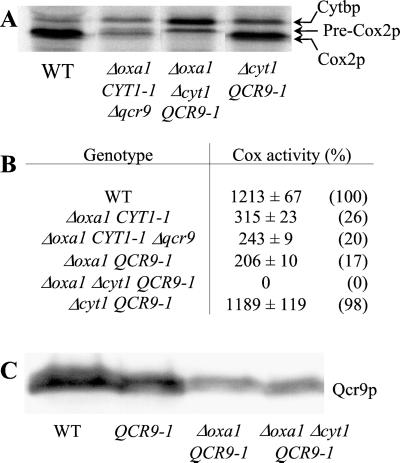
Figure 2.
The QCR9–1 mutation partially compensates for the respiratory defect caused by the absence of Oxa1p. WT (CW30); Δoxa1 (NBT2); Δoxa1 CYT1–1 (R101); Δoxa1 QCR9–1 (R102). See Materials and Methods and Hamel et al. (21). (A) Strains were grown on glucose (G) and ethanol/glycerol (E) media (see Materials and Methods) and incubated for 7 days at 28°C. (B) The mitochondrial translation products were 35S-labeled in vivo in the presence of cycloheximide as described (28). The proteins were separated on 16% SDS-polyacrylamide gels. Arrowheads indicate apocytochrome b (Cytbp) of bc1 complex, pre-Cox2p, and mature Cox2p of cytochrome c oxidase. (C) Mitochondria were purified from cells grown on complete galactose medium as described in Materials and Methods. One hundred micrograms of purified mitochondrial proteins was separated on 12% SDS-polyacrylamide gels, and the accumulation of Cox2p, Atp4p, and Atp2p subunits was analyzed with specific antibodies by Western blotting. Atp2p is used as an internal control as we have previously shown that the accumulation of Atp2p does not depend on Oxa1p (14).
In the absence of Oxa1p, the translocation and maturation of Cox2p is blocked, the assembly of Cox and ATP synthase is impaired, and several membrane subunits undergo degradation. As shown in Fig. 2 B and C, the QCR9–1 mutation partially restores Cox2p maturation and the accumulation of membrane subunits such as Cox2p and Atp4p. Twenty percent of the wild-type Cox activity is recovered, and the ATPase oligomycin-sensitive activity is fully restored (data not shown). These results indicate that the single glycine to arginine substitution in the TM domain of the bc1 complex subunit Qcr9p partially alleviates the respiratory complex assembly defects caused by the absence of Oxa1p. Interestingly, Qcr9p and Cyt1p are two subunits of the bc1 complex and the QCR9–1 (G23>K) mutation, like the CYT1–1 (L216>K) and CYT1–2 (L219>K) mutations, also introduce a positively charged amino acid within the TM domain of the protein.
Interactions Between the TM Domains of Cyt1p and Qcr9p in the Presence of Oxa1p.
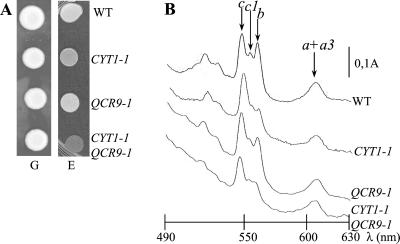
According to the crystal structure of beef and Saccharomyces cerevisiae bc1 complexes (29–30), the two TM domains of Cyt1p and Qcr9p are in close proximity. Thus, we have investigated whether functional interactions between the two TM domains could exist and their possible implication in respiratory complex assembly. We have compared the effects of each single mutation CYT1–1 or QCR9–1 and the combination of both mutations on the biogenesis of the bc1, Cox, and ATPase complexes. Although both single mutants show substantial respiratory growth, the respiratory growth of the double mutant is strongly impaired (Fig. 3A): its duplication time is about 12 h as compared with 2.5 h for the wild type (data not shown). As shown by cytochrome spectra, the amount of assembled bc1 complex is decreased in the three strains (Fig. 3B). The bc1 activity is further decreased in the CYT1–1 QCR9–1 double mutant (18% of the wild-type level) compared with each single mutant (Table 1). A slight diminution of Cox activity also is observed in the double mutant whereas the QCR9–1 and CYT1–1 mutations alone have no detectable effect on this complex. The ATP synthase activity is not affected in any of the strains (data not shown). Finally, the activities of the three respiratory complexes are nearly nil in the triple mutant Δoxa1 CYT1–1 QCR9–1, several subunits being degraded (data not shown).
Figure 3.
Respiratory growth is severely impaired in the double mutant CYT1–1 QCR9–1. WT (CW30); CYT1–1 (PHT2); QCR9–1 (PHT28); QCR9–1 CYT1–1 (CQ2–2B). (A) Strains were grown on glucose (G) and ethanol (E) media as a respiratory substrate and incubated for 3 days at 28°C. (B) Low-temperature cytochrome absorption spectra of whole cells were recorded according to ref. 9. The arrows indicate the absorption maxima of the α bands of cytochromes c (546 nm), c1 (552 nm), b (558 nm), and a + a3 (602 nm).
Table 1.
The enzymatic activities of the bc1 and Cox complexes are affected in the double mutant CYT1-1 QCR9-1
| Genotype | bc1 activity (%) | Cox activity (%) |
|---|---|---|
| WT | 608 ± 61 (100) | 1213 ± 67 (100) |
| CYT1-1 | 456 ± 51 (75) | 1310 ± 62 (108) |
| QCR9-1 | 365 ± 55 (60) | 1322 ± 50 (109) |
| CYT1-1 QCR9-1 | 109 ± 18 (18) | 801 ± 23 (66) |
WT (CW30); CYT1-1 (PHT2); QCR9-1 (PHT28); QCR9-1 CYT1-1 (CQ2-2B). Enzymatic activities were measured on mitochondria purified from galactose grown cells as described in Materials and Methods and in Fig. 4B. The ubiquinol cytochrome c oxido-reductase activity (bc1 activity) is expressed in nmol of reduced cytochrome c/min per mg of protein.
In conclusion, both the assembly and the enzymatic activities of the bc1 complex are affected in the CYT1–1 QCR9–1 double mutant. The two mutations together have a much more deleterious effect on the bc1 complex activity than each single mutation. Also a slight decrease in the Cox activity is only visible in the double mutant. Thus, the two TM domains of Cyt1p and Qcr9p seem to genetically interact and this interaction appears critical for the assembly and/or activity of bc1 and Cox complexes.
Cyt1p Is Required for the Restoration of Cox Assembly in Absence of Oxa1p.
Mutations in the TM domains of Cyt1p and Qcr9p partially restore the insertion and assembly of Cox and ATP synthase in the absence of Oxa1p. However, this restoration is always stronger with the CYT1–1 than with the QCR9–1 mutation (Fig. 2). To establish the relative role of these two subunits in the restoration of respiratory complex assembly, we asked whether either of the suppressor mutations could function in the absence of the other subunit.
We constructed two Δoxa1 strains carrying either the CYT1–1 mutation and the Δqcr9 inactivation, or the QCR9–1 mutation and the Δcyt1 inactivation. As the two subunits Cyt1p and Qcr9p are required for the bc1 electron transfer activity, this activity is nil in the two strains and we found that the ATPase activity is still fully restored (data not shown). As shown in Fig. 4 A and B, Cox2p maturation and Cox activity are still partially restored in the strain carrying the CYT1–1 mutation and the Δqcr9 inactivation, whereas no restoration is detected in the strain carrying the QCR9–1 mutation and the Δcyt1 inactivation. We have ruled out the possibility that the Cox deficiency in the latter strain is caused by the destabilization of Qcr9p in the absence of Cyt1p (Fig. 4C). Moreover, carbonate extractions show that Qcr9p is found predominantly in the membrane fraction in both strains (data not shown). Thus, the lack of restoration of Cox assembly observed in the Δcyt1 QCR9–1 strain is not caused by the destabilization or mislocalization of Qcr9p but is more likely to be caused by the absence of Cyt1p. Hence, Cyt1p is required for the restoration of Cox assembly by the QCR9–1 mutation whereas Qcr9p is not necessary for this restoration by the CYT1–1 mutation. Thus, Cyt1p has a major role in the restoration of Cox assembly in the absence of Oxa1p.
Figure 4.
Cyt1p is essential for the restoration of Cox assembly in the absence of Oxa1p. (A) Mitochondrial translation products were labeled as described in Fig. 2B. WT (CW30); Δoxa1 CYT1–1 Δqcr9 (YST3); Δoxa1 Δcyt1 QCR9–1 (YST4); Δcyt1 QCR9–1 (YSG9–3B). (B) Enzymatic activities were measured in mitochondria purified from galactose grown cells as described in Materials and Methods. The cytochrome c oxidase activity (Cox activity) is expressed in nmol of oxidized cytochrome c/min per mg of protein and percentages of wild-type activities are given in parenthesis. (C) Qcr9p was tagged with the c-myc epitope as described in Materials and Methods. Five hundred micrograms of total yeast proteins, extracted as described (31), was separated on 12% SDS-polyacrylamide gels and immunoblotted with anti-c-myc. WT (YS18–5A), QCR9–1 (YS16–1B), Δoxa1 QCR9–1 (YS16–18D), Δoxa1 Δcyt1 QCR9–1 (YS16–1C).
The Mutant TM Domain of Cyt1p Mediates the Restoration of Complex Assembly.
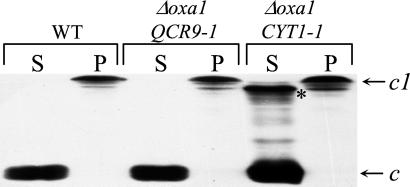
Cyt1p has a bipartite structure. A large N-terminal hydrophilic domain, located in the intermembrane space and binding the heme moiety, is responsible for the electron transfer activity. A short C-terminal hydrophobic domain anchors the molecule to the mitochondrial inner membrane (refs. 32 and 33 for reviews). As shown in Fig. 5, Cyt1p is tightly bound to the membrane and resistant to carbonate extraction in wild-type cells. However, fast-migrating carbonate-extractable forms of cytochrome c1 are systematically observed in the Δoxa1 CYT1–1 strain, and we have previously proposed that they could be responsible for the restoration of complex assembly (21). In the Δoxa1 QCR9–1 strain, no carbonate extractable forms of cytochrome c1 are detected. This finding indicates that these forms do not play a major role in suppression but are likely to be a simple consequence of the CYT1–1 mutation. Indeed, this mutation introduces a positive charge in the TM and thus probably modifies the binding properties of Cyt1p to the membrane. Furthermore, the nature and position of all of the suppressor mutations in the TM domains of Cyt1p and Qcr9p suggests that the TM domain of Cyt1p is responsible for the restoration of complex assembly.
Figure 5.
Carbonate-extractable forms of cytochrome c1 do not accumulate in the Δoxa1 QCR9–1 strain. Mitochondria (equivalent of 300 μg of proteins) were purified from cells grown on galactose complete medium and treated with sodium carbonate as described (14). The supernatants (S; soluble fractions) and pellets (P, membrane fractions) were separated on 12.5% SDS-polyacrylamide gels and transferred to nitrocellulose filters. Detection of cytochromes c and c1 was performed on the membrane as described (34). * indicates the fast-migrating extractable forms of cytochrome c1. WT (CW30); Δoxa1 QCR9–1 (R102); Δoxa1 CYT1–1 (R101).
To test this hypothesis, we constructed a shortened Cyt1p (see Materials and Methods) that no longer carries the hydrophilic domain of Cyt1p but only encodes the presequence essential for the correct mitochondrial import and the C-terminal part of the protein with the mutant TM domain (Fig. 6A). Remarkably, the respiratory growth is restored by the short Cyt1p in cells carrying an inactivated Δoxa1 gene together with a wild-type CYT1 gene ensuring full electron transfer activity through the bc1 complex. We found that Cox2p accumulation and Cox activity are still partially recovered whereas the accumulation of Atp4p and ATPase activity are restored to wild-type level (Fig. 6 B and C, and data not shown). However, the restoration mediated by the short Cyt1p does not take place in the absence of wild-type Cyt1p or Qcr9p (data not shown). This finding suggests that this short mutant Cyt1p requires the presence of some assembled bc1 complex, which may be necessary for the correct integration or orientation of the proteins in the membrane. In addition, a short form of Cyt1p with a wild-type TM domain is able to restore neither the respiratory growth nor the complex assembly, stressing the importance of the CYT1–1 mutation in the mechanism of suppression (data not shown). In conclusion, the mutant TM domain of cytochrome c1 restores the assembly of both Cox and ATPase complexes in the absence of Oxa1p.
Discussion
The Oxa1p family plays a pivotal role in the insertion and assembly of membrane proteins into organelles and bacteria. In yeast mitochondria, the absence of Oxa1p leads to severe defects in the biogenesis of the Cox and ATPase complexes and to minor defects in the assembly of the bc1 complex. Unexpectedly, we have shown that single mutations in the TM domains of two subunits of the bc1 complex, Cyt1p and Qcr9p, can bypass the absence of Oxa1p and partially restore the biogenesis of Cox and ATPase assembly (ref. 21 and this study). This restoration is independent of the electron transfer activity of the bc1 complex because we have shown that it also occurs in strains deficient for the bc1 complex activity (see Fig. 4).
Introduction of a Positive Charge in TM Domains of Either of the Two bc1 Subunits Restores the Insertion and Assembly of Cox and ATPase Complexes.
In most cases, the mutations in Cyt1p or Qcr9p that compensate for the absence of Oxa1p correspond to single amino acid substitutions. For Cyt1p, these mutations affect a number of amino acids all located in the TM domain, e.g., the CYT1–1 or CYT1–2 mutations that change the leucine 216 or 219 to a lysine residue (21). In this study, we show that the mutant TM domain of Cyt1p is sufficient on its own for the restoration of complex assembly. For Qcr9p, we have obtained in all four cases, the same substitution changing the glycine 23 to an arginine residue (mutation QCR9–1). This glycine, located in the middle of the TM domain, is conserved in the plant, chicken, and mammal homologues and may play an important role in complex assembly.
According to the crystal structure of the yeast bc1 complex (30, 33), the TM domains of Cyt1p and Qcr9p are in close proximity and our studies on the double mutant CYT1–1 QCR9–1 show that these two domains interact genetically. Moreover, Cyt1p seems to be absolutely required for the restoration of Cox assembly by the mutant Qcr9p, this restoration being in any case weaker than with the mutant Cyt1p itself. Thus, the QCR9–1 mutation seems to act with Cyt1p, at least for restoration of Cox assembly, and glycine 23 appears critical for the interactions between Qcr9p and Cyt1p. The full restoration of ATPase assembly by the QCR9–1 mutation in the absence of Cyt1p is reminiscent of restoration observed in the protease-deficient mitochondria (14) and suggests that the need of Oxa1p for ATPase assembly is slight enough to be bypassed by a number of extragenic mutations.
Restoration of Complex Assembly Could Occur Through Lateral Interactions Between TM Domains of Various Subunits.
It recently was shown that the yeast and mammalian respiratory complexes assemble into supramolecular structures (35, 36). In particular, the bc1 and Cox complexes associate to form a supercomplex and the subunits Qcr9p and Cyt1p both appear important for its formation. We have shown that the double mutation CYT1–1 QCR9–1 not only drastically reduces the bc1 complex activity but also noticeably affects Cox activity. This result together with the fact that the Cyt1p and Qcr9p TM domains are located at the periphery of the bc1 complex (30) suggest that these domains could interact with Cox subunits during the biogenesis of the complexes.
QCR9–1 and the mutations in the Cyt1p TM domain that compensate for the absence of Oxa1p introduce positively charged amino acids in the membrane region. Thus it is tempting to propose that this charge is responsible for the restoration of respiratory complex assembly. This positively charged amino acid could interact with a negatively charged amino acid of another TM domain. Such negatively charged amino acids occur in several TM domains of respiratory complex subunits. In particular, an analysis of the crystal structure of beef Cox complex (37) and sequence alignments between beef and yeast subunits indicates that the beginning of the first TM helix of Cox2p presents several acidic amino acids. Indeed, one of the main consequence of the absence of Oxa1p is a defect of Cox2p insertion and translocation (12, 13). Thus, it is tempting to propose that the negatively charged residues of Cox2p could interact with the positive charge present in the mutant TM of Cyt1p or Qcr9p, this interaction leading to the restoration of Cox2p insertion.
Oxa1p Could Play a Role in the Insertion and Lateral Exit of Less Hydrophobic TM Domains.
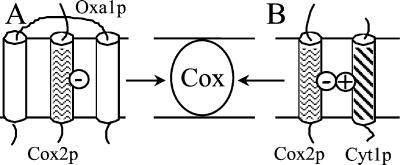
How could such simple membrane interactions bypass the need for Oxa1p, which has been proposed to be essential for the insertion within the inner membrane of respiratory complex subunits? It has been recently shown that the insertion of some membrane subunits, such as Atp9p and Atp8p subunits, is completely Oxa1p-independent (14, 38). Similarly, substantial assembly of the bc1 complex subunits can occur in the absence of Oxa1p (9). Samuelson et al. (7) and Scotti et al. (8) have proposed that the bacterial Oxa1p homologue, YidC, could act like the TRAM protein of the mammalian endoplasmic reticulum membrane. TRAM appears to be required for the insertion and lateral exit of TM segments that are not sufficiently hydrophobic to equilibrate by themselves into the lipid bilayer, these less hydrophobic sequences being sandwiched between the multispanning TRAM protein (39, 40). Oxa1p has five predicted TM segments and may play a similar role in the inner membrane of mitochondria for the less hydrophobic TM segments, such as the first TM helix of Cox2p (see Fig. 7). If this model is true, one can predict that the introduction of charged residue in a TM domain would render it more dependent on Oxa1p. This is the case for the TM domain of Cyt1p for which the presence of a positive charge (e.g., CYT1–1) leads to the accumulation of noninserted (carbonate extractable) forms of Cyt1p in the Δoxa1 mutant, the insertion of wild-type Cyt1p being Oxa1p-independent. In the absence of Oxa1p and in the presence of mutant TM domain of either Qcr9p or Cyt1p, we propose that the positively charged TM domains of the bc1 complex subunit can interact with the negatively charged TM domain of Cox2p or other Oxa1p-dependent subunits. This interaction could allow some lateral exit of the TM domain within the lipid bilayer from their site of insertion, leading to partial restoration of respiratory complex assembly even when Oxa1p is not present. The insertion of less hydrophobic TM domain might play an essential role for the formation of oligomeric membrane complexes because the charged residues located in the TM domain are likely to be important for the folding and stabilization of the complex structure.
Figure 7.
Mode of action of Oxa1p and positively charged TM domains. (A) Oxa1p could allow the membrane insertion of charged or less hydrophobic TM domains (e.g., first TM helix of Cox2p) by sandwiching them until they reach stable integration within the oligomeric complex (Cox complex). (B) Positively charged TM domain (e.g., Cyt1p) could be coinserted with a negatively charged one (e.g., first TM helix of Cox2p).
Several additional proteins might be also involved in the translocation/insertion machinery of mitochondrial membrane proteins (41–44). In particular, Preuss et al. (44) have shown that the double mutant Δmba1 Δoxa1 exhibits a strong growth defect even on glucose, suggesting that Oxa1p and Mba1p have an important overlapping function. However, the fact that the CYT1–1 mutation does not suppress the double mutant Δmba1 Δoxa1 (data not shown) suggests that Mba1p acts at a different step of the translocation machinery. This work provides a genetic demonstration that the TM domain of subunits of different complexes interact during the biogenesis and reinforces the recent view that respiratory enzymes are assembled into supramolecular structures (35, 36).
Acknowledgments
We thank N. Lachacinsky for technical assistance. We thank Dr. J. Herrmann (Institute for Physiological Chemistry, Munich) for the mba1 mutant and Drs. J. Velours and J. M. Galan for the gift of antisera. We are grateful to Drs. N. Bonnefoy, O. Groudinsky, and C. J. Herbert for discussions and critical reading of the manuscript and C. J. Herbert for checking the English. This work was supported in part by a grant from the Association Française contre les Myopathies and by a fellowship from Ligue contre le Cancer de l'Essonne (to P.H.).
Abbreviation
- TM
transmembrane
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.de Gier J-W, Luirink J. Mol Microbiol. 2001;40:314–322. doi: 10.1046/j.1365-2958.2001.02392.x. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport T A, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 3.Grivell L A. Crit Rev Biochem Mol Biol. 1995;30:121–164. doi: 10.3109/10409239509085141. [DOI] [PubMed] [Google Scholar]
- 4.Herrmann J, Neupert W. Curr Opin Microbiol. 2000;3:210–214. doi: 10.1016/s1369-5274(00)00077-1. [DOI] [PubMed] [Google Scholar]
- 5.Tokatlidis K, Schatz G. J Biol Chem. 1999;274:35285–35288. doi: 10.1074/jbc.274.50.35285. [DOI] [PubMed] [Google Scholar]
- 6.Sue C M, Schon E A. Brain Pathol. 2000;10:442–450. doi: 10.1111/j.1750-3639.2000.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuelson J C, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips G J, Dalbey R E. Nature (London) 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 8.Scotti P A, Urbanus M L, Brunner J, de Gier J-W L, von Heijne G, van der Does C, Driessen A J M, Oudega B, Luirink J. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnefoy N, Chalvet F, Hamel P, Slonimski P P, Dujardin G. J Mol Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- 10.Bauer M, Behrens M, Esser K, Michaelis G, Pratje E. Mol Gen Genet. 1994;245:272–278. doi: 10.1007/BF00290106. [DOI] [PubMed] [Google Scholar]
- 11.Altamura N, Capitanio N, Bonnefoy N, Papa S, Dujardin G. FEBS Lett. 1996;382:111–115. doi: 10.1016/0014-5793(96)00165-2. [DOI] [PubMed] [Google Scholar]
- 12.Hell K, Herrman J, Pratje E, Neupert W, Stuart R A. Proc Natl Acad Sci USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He S, Fox T D. Mol Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemaire C, Hamel P, Velours J, Dujardin G. J Biol Chem. 2000;275:23471–23475. doi: 10.1074/jbc.M002045200. [DOI] [PubMed] [Google Scholar]
- 15.Hell K, Neupert W, Stuart R A. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonnefoy N, Kermorgant M, Groudinsky O, Dujardin G. Mol Microbiol. 2000;35:1135–1145. doi: 10.1046/j.1365-2958.2000.01781.x. [DOI] [PubMed] [Google Scholar]
- 17.Bonnefoy N, Kermorgant M, Groudinsky O, Minet M, Slonimski P P, Dujardin G. Proc Natl Acad Sci USA. 1994;91:11978–11982. doi: 10.1073/pnas.91.25.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamel P, Sakamoto W, Wintz H, Dujardin G. Plant J. 1997;12:101–109. doi: 10.1046/j.1365-313x.1997.12061319.x. [DOI] [PubMed] [Google Scholar]
- 19.Sundberg E, Slagter J G, Friborg I, Cleary S P, Robinson C, Coupland G. Plant Cell. 1997;9:717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore M, Harrison M S, Peterson E C, Henry R. J Biol Chem. 2000;275:1529–1532. doi: 10.1074/jbc.275.3.1529. [DOI] [PubMed] [Google Scholar]
- 21.Hamel P, Lemaire C, Bonnefoy N, Brivet-Chevillotte P, Dujardin G. Genetics. 1998;150:601–611. doi: 10.1093/genetics/150.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wach A, Brachat A, Pohlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 23.Longtine M S, McKenzie A, III, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 24.Spannagel C, Vaillier J, Chaignepain S, Velours J. Biochemistry. 1998;37:615–621. doi: 10.1021/bi9714971. [DOI] [PubMed] [Google Scholar]
- 25.Brasseur G, Coppée J, Colson A-M, Brivet-Chevillotte P. J Biol Chem. 1995;270:29356–29364. doi: 10.1074/jbc.270.49.29356. [DOI] [PubMed] [Google Scholar]
- 26.Phillips J D, Schmitt M E, Brown T A, Beckmann J D, Trumpower B L. J Biol Chem. 1990;265:20813–20821. [PubMed] [Google Scholar]
- 27.Graham L A, Phillips J D, Trumpower B L. FEBS Lett. 1992;313:251–254. doi: 10.1016/0014-5793(92)81203-x. [DOI] [PubMed] [Google Scholar]
- 28.van Dyck L, Neupert W, Langer T. Genes Dev. 1998;12:1515–1524. doi: 10.1101/gad.12.10.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia D, Yu C-A, Kim H, Xia J-Z, Kachurin A M, Zhang L, Yu L, Deinsenhofer J. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure (London) 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 31.Yaffe M P. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- 32.Trumpower B. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berry E, Guergova-Kuras M, Huand L, Crofts A. Annu Rev Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- 34.Vargas C, McEwan A G, Downie J A. Anal Biochem. 1993;209:323–326. doi: 10.1006/abio.1993.1127. [DOI] [PubMed] [Google Scholar]
- 35.Schagger H, Pfeiffer K. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruciat C-M, Brunner S, Baumann F, Neupert W, Stuart R A. J Biol Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- 37.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 38.Li M, Mihara K. J Biol Chem. 2001;276:24704–24712. doi: 10.1074/jbc.M102584200. [DOI] [PubMed] [Google Scholar]
- 39.Matlack K E S, Mothes W, Rapoport T A. Cell. 1998;92:381–390. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- 40.Heinrich S U, Mothes W, Brunner J, Rapoport T A. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 41.Rep M, Nooy J, Guelin E, Grivell L A. Curr Genet. 1996;30:206–211. doi: 10.1007/s002940050122. [DOI] [PubMed] [Google Scholar]
- 42.He S, Fox T D. Mol Cell Biol. 1999;19:6598–6607. doi: 10.1128/mcb.19.10.6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Souza R L, Green-Willms N S, Fox T D, Tzagoloff A, Nobrega F G. J Biol Chem. 2000;275:14898–14902. doi: 10.1074/jbc.275.20.14898. [DOI] [PubMed] [Google Scholar]
- 44.Preuss M, Leonhard K, Hell K, Stuart R A, Neupert W, Herrmann J M. J Cell Biol. 2001;153:1085–1095. doi: 10.1083/jcb.153.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]