Abstract
Patient-derived organoids (PDOs) have recently emerged as robust pre-clinical models, however, their potential to predict patient clinical outcomes remain unclear. We report a living biobank of PDOs from metastatic, heavily-pretreated colorectal and gastroesophageal cancer patients recruited in phase I/II clinical trials. Phenotypic and genotypic profiling of PDOs showed a high-degree of similarity to the original patient tumor. Molecular profiling of tumor organoids was matched to drug screening results, suggesting PDOs could complement existing approaches in defining cancer vulnerabilities and improving treatment responses. We compared ex vivo organoid responses to anticancer agents, and PDO-based orthotopic mouse tumor xenograft models to the response of the patient in clinical trials. Our data suggest that PDOs can recapitulate patient responses in the clinic, and have the potential to be implemented in personalized medicine programs.
High-throughput sequencing has been extensively used in precision medicine to identify somatic mutations that can be exploited for cancer treatment and drug development (1). However, the limited role of genomic profiling in predicting response to targeted therapies, and limitations of pre-clinical models currently used for drug validation, represent important obstacles hampering the success of personalized medicine (2). Co-clinical trials are defined as parallel studies where drug response in patients are matched to laboratory pre-clinical models, in order to personalize treatment and understand mechanisms of chemo-sensitivity through functional genomics and reverse translation (3). Most co-clinical trials rely on the use of genetically engineered mouse models or patient-derived xenografts, posing logistic, ethical, and economic issues (4).
LGR5+ stem cells can be isolated from a number of organs and propagated as epithelial organoids in vitro to study physiology and neoplastic transformation (5). Most studies on human colorectal cancer (CRC) organoids have been conducted on cultures derived from primary tumors (6). In contrast, examples of PDOs from metastatic cancer sites remain sparse (7–9). Furthermore, very limited evidence is available on the ability of PDOs to predict response to treatment in the clinic (10). Here we present a living biobank of PDOs from heavily-pretreated metastatic gastrointestinal cancer patients, and show examples of how these cancer organoids can be used to compare drug responses to those of the actual patient.
A total of 110 fresh biopsies from 71 patients enrolled in four prospective phase I/II clinical trials were processed between October 2014 and February 2017. In line with previous data (7), PDOs were grown from 70% of biopsies with a cellularity of 2+ and above, and their establishment rate strongly correlated with tumor cellularity in the parental biopsy (χ2 p<0.0001). No inverse correlation was observed between PDO establishment rate and presence of necrosis (cut-off ≥20%). Tumor percentage is a key limiting factor for genomic and transcriptomic analyses. When the 60% threshold used in large sequencing studies of primary CRC (11) or gastroesophageal cancers (GOC) (12) was applied in our cohort, we found no correlation between PDO take-up rate and tumor percentage, suggesting that PDOs can also be established in cases of a low tumor/stroma ratio thus allowing the ex vivo expansion of the cancer population in samples that would have otherwise failed quality control for next generation sequencing (NGS).
PDOs presented in this study were derived from ultrasound (n=20), computer-tomography (CT)-guided (n=7) or endoscopic (n=2) biopsies of metastatic CRC [mCRC; (n=16)], metastatic GOC [mGOC; (n=4)], and metastatic cholangiocarcinoma (n=1) patients (fig. S1). Liver, pelvic, peritoneal, and nodal metastases of chemo-refractory patients were used to establish PDOs. In several cases PDOs were established from sequential biopsies at baseline (BL), at the time of best response [partial response (PR) or stable disease (SD)], and at the time of disease progression (PD), as well as from multi-region biopsies (table S1).
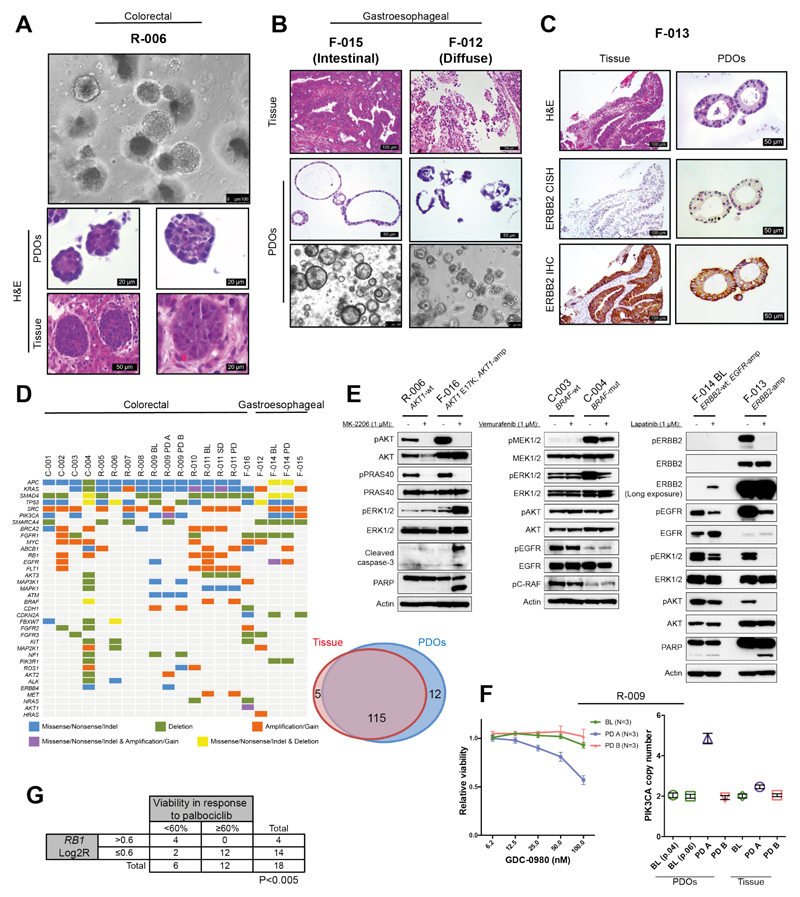
Histological evaluation revealed significant morphological similarities between PDOs and the patient biopsies from which they were originally-derived (Fig. 1A, Fig. 1B, fig. S2A and fig. S2B). Immunohistochemistry markers routinely used in the diagnosis of CRC (CDX-2, CK7) showed that the parental tumor’s expression pattern was maintained in PDOs, even when derived from sequential biopsies during treatment (fig. S2C, fig. S2D and fig. S2E). Similarly, amplification of oncogenic drivers such as ERBB2 (Fig. 1C and fig. S2F) or rearrangements in FGFR2 (fig. S2G) were retained in PDOs from mGOC and metastatic cholangiocarcinoma respectively.
Fig. 1. Histopathological, molecular, and functional characterization of patient-derived organoids (PDOs).
(A) Phase-contrast image of a mCRC PDO culture, and H&E staining comparing organoids to their matching patient biopsy. (B) Diffuse and intestinal growth patterns are retained in mGOC PDOs. (C) ERBB2 amplification and over-expression in mGOC PDOs and parental tissue biopsy; CISH= chromogenic in situ hybridization. (D) Heatmap displaying the most frequently mutated and/or copy number altered genes in PDOs (left). Venn diagram demonstrating 96% mutational overlap between PDOs and parental tissue biopsies (right). (E) Target engagement in genotype-drug phenotype combinations: pathway analysis downstream of ERBB2 in ERBB2-amplified and non-amplified PDOs treated with lapatinib (24h) (right panel); BRAF inhibition (24h) (central panel); AKT inhibition (4h) (left panel). (F) Dose-dependent effect to the dual PI3K/mTOR inhibitor GDC-0980 in three PDOs from patient R-009, all carrying an acquired PIK3CA mutation (H1047R). PDOs established from a liver metastasis biopsied at disease progression (R-009 PD A) that also harbored PIK3CA amplification showed dose-dependent response to GDC-0980. PIK3CA-mutant but non-amplified PDOs established prior to regorafenib treatment (R-009 BL) or from a different liver metastasis biopsied at disease progression (R-009 PD B) did not respond to GDC-0980. Viability data show mean ± SEM of indicated independent experiments. (G) Correlation (Fisher’s exact test) between presence of RB1 amplification in PDOs (panel 1D) and response to the CDK4/CDK6 inhibitor palbociclib in the reported drug screen (fig. S9A). Abbreviations: BL= baseline; SD= stable disease; PD= post-treatment/progressive disease.
NGS was used to profile 151 cancer-related genes in both PDOs (n=23) and their parental biopsies; archival material from primary cancer or pre-treatment diagnostic biopsy was also sequenced for 8 patients, and whole-genome sequencing (WGS) was performed for one PDO (table S2 and table S3). The molecular landscape of our PDOs (Fig. 1D) largely overlapped that reported for mCRC and mGOC in the MSK-IMPACT study (1), with the exception of SRC and EGFR amplifications and ATM and BRCA2 mutations that were more frequent in our mCRC PDO cohort (table S4). Overall, 96% overlap in mutational spectrum was observed between PDOs and their parental biopsies (Fig. 1D), while intra-tumor heterogeneity was observed between archival material (primary cancer) and metastatic deposits (biopsy or PDOs) (fig. S3A and table S2). Interestingly PDOs were able to capture spatio-temporal intra-tumor heterogeneity when established from multiple biopsies at time of disease progression compared to PDOs at the beginning of treatment (Fig. 1D, fig. S3A and table S2). Similar findings were observed for copy number alterations (CNAs) in PDOs and biopsies collected at different time-points during treatment (fig. S3B and fig. S4). WGS confirmed CNAs extrapolated from targeted NGS of PDOs or PDO-derived orthotopic tumors (PDO-xenografts) (fig. S3B and fig. S4); CNAs detected in key oncogenic drivers were further validated by digital-droplet PCR (fig. S5). High concordance was observed in mutational, CNA, and transcriptomic profiling over successive passages when PDOs were tested before and after several months of continuous culture (passage range: 5-13); mutations: R2=0.96 p<0.0001; CNA: R2=0.97 p<0.0001; gene expression (RNA-Seq): R2=0.7 p<0.001 (fig. S6).
Next we tested the feasibility of using PDOs derived from metastatic cancers as drug screening tools, and validated the robustness of our approach by identifying several genotype-drug phenotype correlations across the PDO panel. We ran 3D screening assays over a period of two weeks, (fig. S7 and fig. S8) using a library of 55 drugs currently in phase I-III clinical trials or in clinical practice (table S5). The heatmap shown in fig. S9A summarizes screening data; hit validation at lower drug concentrations is reported in fig. S9B. For all 19 screens a very high correlation was observed among each screen’s three replicate assays and controls (fig. S10).
F-013 was the only ERBB2-amplified PDO in our cohort (Fig. 1C), and it exhibited the strongest response to lapatinib (dual ERBB2/EGFR inhibitor); indeed, lapatinib potently inhibited the MAPK and PI3K/AKT signaling downstream of EGFR/ERBB2, inducing apoptosis in the F-013 PDO (Fig. 1E and fig. S9A). Interestingly, in a PDO (F-014) that harbors amplified EGFR but no ERBB2 amplification, lapatinib had no effect on viability and only modestly reduced MAPK and PI3K/AKT signaling (Fig. 1E and fig. S9A).
Similarly, across all PDOs, F-016 was the only tumor carrying an AKT1-amplification and E17K mutation (Fig. 1D), and is the only one strongly responding to both AKT inhibitors present in the drug library (MK-2206, GSK690693) (Fig. 1E and fig. S9A). One mCRC PDO (C-004) harbored a BRAF V600E mutation (Fig. 1D) and is the only PDO that showed significantly decreased viability following treatment with the BRAF inhibitor vemurafenib (fig. S9A). Consistent with this, vemurafenib selectively inhibited MEK/ERK signaling in the C-004 PDO (Fig. 1E), but failed to induce apoptosis in keeping with the lack of efficacy of single agent BRAF inhibitors in mCRC (13).
Overall, PIK3CA mutations were not predictive of response to GDC-0980 (a dual PI3K/mTOR inhibitor) in the PDOs panel (Fig. 1D and fig. S9A). In line with this observation, in a patient where pre- and post- treatment PDOs were established from multiple metastases (R-009 BL, PD-A and PD-B), a PIK3CA H1047R mutation common to all the PDOs was not associated with any response to GDC-0980. However, PDOs carrying a synchronous PIK3CA amplification (R-009 PD-A) showed a dose-dependent reduction in cell viability in response to GDC-0980 (Fig. 1F and fig. S3A). Finally, in keeping with published data (14), a significant correlation was observed between RB1 amplification and sensitivity of PDOs to palbociclib [CDK4/CDK6 inhibitor; (Fig. 1G)].
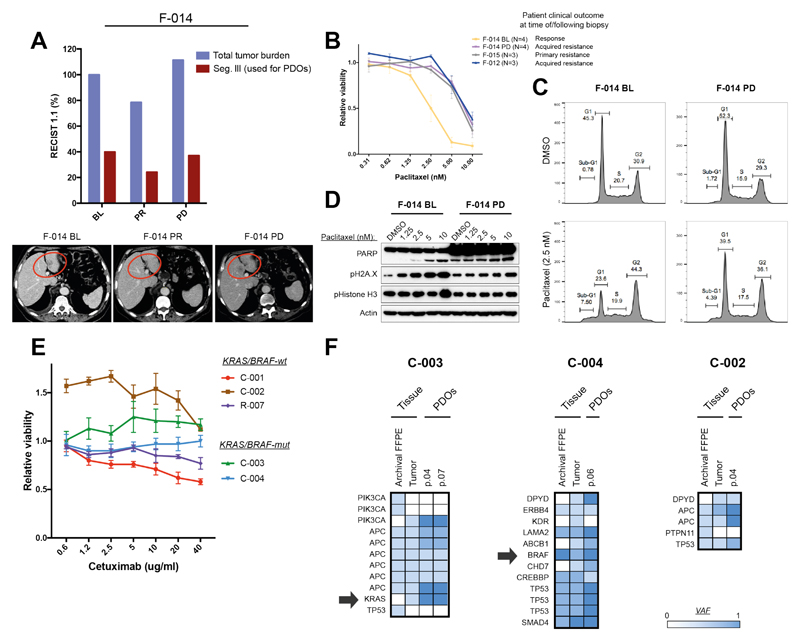
Following extensive molecular and functional characterization of our PDOs, we examined their clinical predictive value by comparing clinical responses observed in patients with ex vivo-response data gathered in organoids in 21 comparisons (table S6). Taxanes are a standard second line treatment option for metastatic gastric cancer, however, efficacy is modest and no predictive biomarkers are available to inform clinical decisions (15). We compared response to paclitaxel in sequential PDOs established before and after treatment in a paclitaxel-sensitive patient (F-014) with PDOs established from liver metastases of two paclitaxel-resistant patients (Fig. 2A and Fig. 2B). PDOs derived from the responsive metastasis showed a ~4-fold lower GI50 for clinically relevant paclitaxel concentrations (16) compared with PDOs from the same patient derived at progression; interestingly, these resistant PDOs demonstrated an identical paclitaxel dose-response profile to the two PDOs established from paclitaxel-refractory patients (Fig. 2B). Cell cycle analysis showed marked apoptosis and G2 arrest upon taxane treatment in the pre-treatment F-014 PDOs, while no significant difference was observed in PDOs established at progression (Fig. 2C and fig. S11A). Similarly, paclitaxel induced dose-dependent DNA damage, mitotic arrest and apoptosis in the pre-treatment F-014 PDOs, but had a much weaker impact on the progression (thus resistant) PDOs (Fig. 2D). Consistent with data observed for second line treatment, a ~10-fold difference in GI50 was observed in response to the combination of 5-fluorouracil and cisplatin in PDOs collected from chemo-sensitive and chemo-refractory mGOC patients receiving first-line treatment (fig. S11B), highlighting the clinical potential of PDOs for treatment selection in cancers of unmet need.
Fig. 2. Patient-derived organoid-based ex vivo co-clinical trials in mGOC and mCRC.
(A) PDOs were generated from sequential biopsies of a liver metastasis (red circle in the bottom panel) of mGOC patient F-014 that showed initial response to paclitaxel (F-014 BL) and subsequently progressed (F-014 PD). Violet bars indicate overall tumor volume (according to RECIST 1.1. criteria) while red bars indicate volume of the target metastasis used to generate PDOs. (B) Cell viability upon paclitaxel treatment was compared in baseline (BL) and progressive disease (PD) PDOs from patient F-014 treated with paclitaxel, and those derived from patients that exhibited primary (F-015) or acquired (F-012) resistance to paclitaxel in the clinic. Viability data show mean ± SEM of indicated independent experiments. (C) Cell cycle analysis upon paclitaxel treatment in the F-014 baseline (BL) PDO compared with the F-014 progressive disease (PD) PDO. (D) Dose-dependent DNA damage was observed in the F-014 baseline (BL) PDO in response to paclitaxel, but not in PDOs from the same patient established at progressive disease (PD). (E) PDOs were established from baseline (BL) (C-003, C-004) and progressive disease (PD) (C-001, C-002) biopsies from patients treated with the anti-EGFR monoclonal antibody cetuximab. PDOs were treated with cetuximab in vitro; data show mean ± SD from independent experiments performed in triplicate. (F) Molecular analysis of baseline (BL) and progressive disease (PD) PDOs, matching biopsy (tumor), and primary bowel cancer (archival); arrows indicate the presence of clonal or sub-clonal mutations in BRAF and KRAS respectively in two patients.
Anti-EGFR monoclonal antibodies, regorafenib, and TAS-102 represent FDA approved options for treatment of chemo-refractory mCRC, however, with the exception of RAS pathway mutations for anti-EGFR therapy, there are no validated clinical biomarkers for patient selection in this setting. We initially tested the predictive value of PDOs in mCRC by comparing response to anti-EGFR treatment with cetuximab in five PDOs and their respective patients (Fig. 2E). Two PDOs established from baseline biopsies prior to anti-EGFR treatment in the PROSPECT-C trial showed no response to cetuximab, in keeping with the primary resistance observed in these two patients in the clinic. Unsurprisingly (17), both PDOs and their respective patient biopsies harbored either KRAS G12D (sub-clonal) or BRAF V600E (clonal) mutations (Fig. 2F). The third cetuximab-resistant PDO (C-002) was established from the progression biopsy of a patient who initially responded to cetuximab, and, interestingly, it harbored an EGFR amplification (Fig. 1D and fig. S5), no RAS pathway mutational aberrations (Fig. 2F), and high amphiregulin mRNA levels. Despite these molecular markers being suggestive of responsiveness to cetuximab, the C-002 PDO showed no response (and in fact paradoxical enhanced proliferation) upon cetuximab treatment in line with the respective patient’s clinical outcome, thus highlighting the potential of PDOs to better predict clinical outcomes compared to molecular pathology alone. Another KRAS-wild type PDO derived from a slow growing progressing metastasis in a patient with otherwise stable disease [C-001; (fig. S11C)] showed a marginal response to cetuximab. Finally, the KRAS-wild type PDO established from a BL biopsy of a patient enrolled in the PROSPECT-R trial [R-007; (fig. S11C)] showed response to cetuximab at doses higher than 5 µg/ml; this, however, could not be compared with clinical response as the patient did not receive anti-EGFR mAbs.
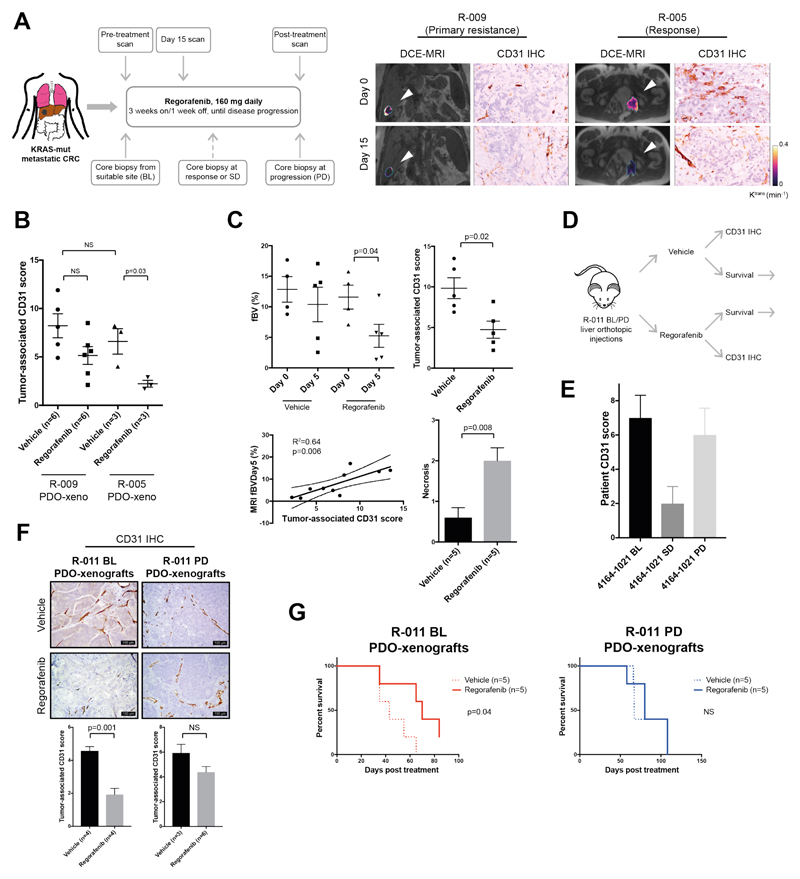
Next we tested the ability of PDOs to recapitulate response to regorafenib, a multiple tyrosine kinase inhibitor blocking oncogenic and angiogenic signaling pathways. No response to regorafenib was observed in our 3D ex vivo screening assays (fig. S9A), an observation in keeping with our recently reported clinical results from the PROSPECT-R trial (18) suggesting that response to regorafenib is mainly driven by its anti-angiogenic effect (Fig. 3A).
Fig. 3. Patient-derived organoid-based co-clinical trials mimic primary and acquired resistance to regorafenib in mice.
(A) mCRC patients on regorafenib treatment underwent biopsies at baseline (BL), partial response/stable disease stage (PR/SD), or post-treatment (PD). An early reduction (15 days) in functional imaging (DCE-MRI) parameters correlated with changes in micro-vasculature assessed by CD31 staining and clinical benefit from regorafenib (right panel). (B) Changes in micro-vasculature in response to regorafenib were assessed in PDO mouse xenografts by quantification of tumor-associated CD31-positive vessels. Data show PDO xenografts from a primary resistant (R-009) and a long-term responder (R-005) to regorafenib. Mean ± SD from indicated number of mice (n) in a representative experiment is shown; significance was determined using Student’s unpaired t-test. (C) Reduction in fractional blood volume (fBV) in regorafenib-treated mice carrying long-term regorafenib responder (R-005) PDO-xenografts. A total of ten animals were analyzed (five in each arm); data represent the mean ± SD of an individual experiment. Day 0 fBV values could not be obtained for two animals due to respiratory movement. Significance was determined using Student’s paired t-test for fBV and unpaired t-test for CD31 and necrosis. (D) Schematic representation of animal experiment using PDOs from patient R-011, established pre- and post-treatment with regorafenib. Mice carrying liver orthotopic R-011 pre-treatment (BL) and post-treatment (PD) PDO-xenografts were randomized to control and treatment arms, and treated with vehicle or regorafenib for 10 days. Following-treatment, each arm was further randomized to a cohort culled for histopathological analysis and a survival cohort which was monitored over time. (E) CD31 immunostaining in the parental patient baseline (BL), stable disease (SD), and post-treatment (PD) biopsies, demonstrating an initial reduction in tumor microvasculature in response to regorafenib. Data represent mean ± SD calculated by scoring ten high-power field tumor areas. (F) Representative images of CD31 immunostaining in the baseline (BL) and post-treatmen (PD) R-011 PDO-xenografts. Data represent mean ± SD calculated by scoring at least ten high-power field tumor areas per animal in an individual experiment; n= number of animals analyzed in each group. Significance was determined using Student’s unpaired t-test. (G) Kaplan-Mayer curves of regorafenib- or vehicle-treated mice bearing baseline (BL) and post-treatment (PD) PDO-xenografts from patient R-011 from an individual experiment. (n= number of mice analyzed). Significance was determined using the Mantel-Cox log-rank test.
In order to match response to regorafenib in the clinic and in aligned PDOs we established an orthotopic human tumor xenograft model by implanting luciferase-expressing (Luc+) PDOs in the liver of NSG mice [PDO-xenografts; (fig. S12A)]. We initially compared response to regorafenib in PDO-xenografts from a patient with primary resistance [R-009 (n=11)] and from a patient who achieved a durable (10 months) response [R-005 (n=6)] to regorafenib (fig. S12B and fig. S12C). In keeping with clinical response (Fig. 3A), PDO-xenografts from the regorafenib-sensitive patient displayed a significant (p=0.03) reduction in their micro-vasculature in response to regorafenib as revealed by CD31 immunostaining; in contrast, no significant changes were observed in PDO-xenografts from the regorafenib-resistant patient (Fig. 3B). In order to mimic our clinical observations, we performed functional susceptibility-contrast magnetic resonance imaging (MRI) in PDO-xenografts of the responder patient [R-005 (n=10)] before and after treatment (fig. S12D). In line with dynamic contrast-enhanced MRI (DCE-MRI) results in patients (Fig. 3A), susceptibility-contrast MRI revealed a significant reduction in tumor fractional blood volume (fBV) in regorafenib-treated mice (Fig. 3C). These changes were associated with a reduction in CD31 staining and increased necrosis (Fig. 3C). Remarkably, across all animals, a robust correlation was observed between the fBV values obtained from susceptibility contrast MRI and the micro-vasculature assessment [CD31 (R2=0.64 p=0.006)] of the same samples (Fig. 3C). Interestingly, in line with our clinical data, changes in micro-vasculature indicative of response appeared to be independent of changes in tumor volume (fig. S12E) (18). Three different histopathological growth patterns [(HGPs); desmoplastic HGP; pushing HGP; replacement HGP] have been associated with different degrees of response to anti-angiogenic drugs, with the replacement HGP being frequently associated with vessel co-option and primary resistance (19). In our experiments a predominance of replacement HGP, and thus vessel co-option, was observed in PDO-xenografts from the resistant patient, whilst tumors established from the PDOs of the sensitive patient showed prevalence of desmoplastic and pushing HGPs (fig. S12F), suggesting that vessel co-option might be the mechanism underpinning primary resistance to regorafenib. When the responder to regorafenib (R-005) progressed and received subsequent treatment, he was enrolled in a phase I trial of the ATR inhibitor VX-970. No response was observed in this patient with VX-970 monotherapy, and this was in keeping with lack of response to ATM/ATR inhibitors observed in his PDOs in the drug screening reported in fig. S9A.
In order to test the PDOs’ ability to capture tumor evolution and acquired resistance to treatment, we generated xenografts using PDOs from the same liver metastasis before (BL) and after treatment (PD) in mCRC patient R-011 that exhibited initial response and subsequent progression to regorafenib (fig. S13A). Mice were randomized to treatment and control arms, and, following treatment, each arm was further randomized for survival or functional analysis (Fig. 3D). In line with clinical findings (Fig. 3E) (18), CD31 immunostaining revealed a ~60% reduction in micro-vasculature in response to regorafenib in BL PDO-xenografts, while no significant change was observed in PD PDO-xenografts [p:0.001; (Fig. 3F)]. More importantly, regorafenib treatment offered a selective survival benefit in mice carrying BL PDO-xenografts (Fig. 3G and fig. S13B), confirming the predictive value of PDOs and their ability to reflect cancer evolution upon treatment.
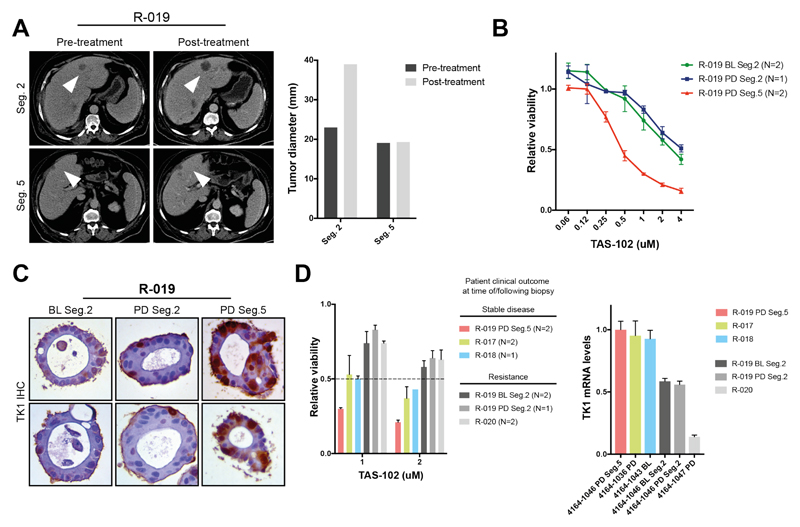
TAS-102, a combination of the nucleoside analog trifluridine and the thymidine phosphorylase inhibitor tipiracil, is approved for the treatment of chemo-refractory mCRC but no validated bio-markers are currently available (20). We compared clinical and pre-clinical response to TAS-102 in 6 organoids from 4 different patients treated with TAS-102. Initially we tested response to TAS-102 in PDOs from a patient (R-019) who had a mixed response, with stability of disease in one of the liver metastases (segment 5) and rapid progression in another one (segment 2) (Fig. 4A). Ex vivo dose-response data showed a ~8-fold difference in GI50 between PDOs derived from the TAS-102 sensitive metastasis and those derived from pre- and post-treatment biopsy of the rapidly progressing metastasis (Fig. 4B), highlighting the ability of PDOs to recapitulate intra-patient heterogeneity. TK1 has been proposed as a potential biomarker of response to TAS-102 (21); interestingly, TK1 protein expression was indeed higher in PDOs from the responding metastasis compared with those from the non-responding site (Fig. 4C). When we extended the TAS-102 sensitivity analysis to 3 other PDOs/patients we confirmed that PDOs from patients who achieved disease control were sensitive to low uM concentrations of TAS-102, while no significant effect on cell viability was observed in PDOs from resistant (primary or acquired) patients (Fig. 4D, left); in line with previous data, TK1 mRNA expression was higher in PDOs from patients that achieved stable disease in response to TAS-102 (Fig. 4D, right).
Fig. 4. Patient-derived organoids recapitulate intra- and inter-patient heterogeneity in response to TAS-102.
(A) PDOs were established from a patient (R-019) with mixed response to TAS-102. While the segment 2 metastasis rapidly progressed, the segment 5 one remained stable upon TAS-102 treatment (white arrows in the CT-scan indicate metastases; bars indicate pre- and post-treatment measurement of the indicated metastases). (B) Ex vivo dose-response curves in baseline (BL) and post-treatment (PD) multi-region PDOs from patient R-019 (with mixed response to TAS-102). N= independent experiments; viability values are expressed as mean ± SEM. (C) TK1 immunohistochemistry (IHC) expression in TAS-102 refractory (segment 2) and sensitive (segment 5) PDOs. BL = baseline; PD = post-treatment/progressive disease. (D) Cell viability (left) and TK1 mRNA expression (right) in PDOs from TAS-102 responsive and refractory patients. BL= baseline; PD= post-treatment/progressive disease. N indicates independent experiments; viability values are expressed as mean ± SEM.
Overall, for the PDOs we analyzed, we found 100% sensitivity, 93% specificity, 88% positive predictive value, and 100% negative predictive value in forecasting response to targeted agents or chemotherapy in patients [Fisher’s exact test p<0.0001; (table S7)]. Our data suggest that PDOs can be exploited for functional genomics to simulate cancer behavior ex vivo and integrate molecular pathology in the decision-making process of early phase clinical trials.
Supplementary Material
One Sentence Summary.
Patient-derived organoids predict response of metastatic gastrointestinal cancers to therapy.
Acknowledgments
The data presented in this paper are tabulated in the main text and supplemental materials. Sharing of materials is subject to a material transfer agreement (MTA) with the The Institute of Cancer Research, London (please direct requests to N.V). This work was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre (BRC) at The Royal Marsden NHS Foundation Trust, and The Institute of Cancer Research (grant numbers A62, A100, A101, A159), Cancer Research UK (grant number CEA A18052) and the European Union FP7 (grant number CIG 334261) to N.V; by Cancer Research UK (C52506/A22909) and Wellcome Trust (105104/Z/14/Z) grants to A.S.; by Cancer Research UK Cancer Imaging Centre funding (C1060/A16464) to The Institute of Cancer Research, and by a Bayer Oncology Group Research Grant to D.C. A.S. is supported by The Chris Rokos Fellowship in Evolution and Cancer. I.C. has had advisory roles with Merck Serono, Roche, Sanofi Oncology, Bristol Myers Squibb, Eli-Lilly, Novartis, Gilead Science, and has received research funding from Merck-Serono, Novartis, Roche and Sanofi Oncology, and honoraria from Roche, Sanofi-Oncology, Eli-Lilly, Taiho. D.C. received research funding from: Roche, Amgen, Celgene, Sanofi, Merck Serono, Novartis, AstraZeneca, Bayer, Merrimack and MedImmune.
Footnotes
All other authors declare no conflict of interest.
References
- 1.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voest EE, Bernards R. DNA-guided precision medicine for cancer: a case of irrational exuberance? Cancer Discovery. 2016;6(2):130–132. doi: 10.1158/2159-8290.CD-15-1321. [DOI] [PubMed] [Google Scholar]
- 3.Byrne AT, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17(4):254–268. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
- 4.Clohessy JG, Pandolfi PP. Mouse hospital and co-clinical trial project--from bench to bedside. Nat Rev Clin Oncol. 2015;12(8):491–498. doi: 10.1038/nrclinonc.2015.62. [DOI] [PubMed] [Google Scholar]
- 5.Bredenoord AL, Clevers H, Knoblich JA. Human tissues in a dish: The research and ethical implications of organoid technology. Science. 2017;355(6322):eaaf9414. doi: 10.1126/science.aaf9414. [DOI] [PubMed] [Google Scholar]
- 6.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weeber F, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proceedings of the National Academy of Sciences. 2015;112(43):13308–13311. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii M, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18(6):827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Pauli C, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discovery. 2017;7(5):462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dekkers JF, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Science Translational Medicine. 2016;8(344):344ra84. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman DM, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. New England Journal of Medicine. 2015;373(8):726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discovery. 2016;6(4):353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth EC, et al. Gastric cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2016;27(suppl_5):38–49. doi: 10.1093/annonc/mdw350. [DOI] [PubMed] [Google Scholar]
- 16.Zasadil LM, et al. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Science Translational Medicine. 2014;6:229ra243. doi: 10.1126/scitranslmed.3007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verissimo CS, et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife. 2016;5:e18489. doi: 10.7554/eLife.18489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan K, et al. Functional imaging and circulating biomarkers of response to regorafenib in treatment-refractory metastatic colorectal cancer patients in a prospective phase II study. Gut. 2017 doi: 10.1136/gutjnl-2017-314178. Published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frentzas S, et al. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med. 2016;22(11):1294–1302. doi: 10.1038/nm.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer RJ, et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. New England Journal of Medicine. 2015;372(20):1909–1919. doi: 10.1056/NEJMoa1414325. [DOI] [PubMed] [Google Scholar]
- 21.Kuboki Y, et al. TAS-102 plus bevacizumab for patients with metastatic colorectal cancer refractory to standard therapies (C-TASK FORCE): an investigator-initiated, open-label, single-arm, multicentre, phase 1/2 study. The Lancet Oncology. 2017;18:1172–1181. doi: 10.1016/S1470-2045(17)30425-4. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15(1):182–193. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46(8):912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Münz M, et al. CSN and CAVA: Variant annotation tools for rapid, robust next-generation sequencing analysis in the clinical setting. Genome Medicine. 2015;7(1):76–83. doi: 10.1186/s13073-015-0195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Meth. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotech. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortazavi A, Williams BA, McCue K, Scaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 30.Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotech. 2015;33(3):243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Workman P, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102(11):1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: The ARRIVE guidlines for reporting animal research. PLoS Biol. 2010;8(6):e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson SP, Boult JKR, Vasudev NS, Reynolds AR. Monitoring the vascular response and resistance to sunitinib in renal cell carcinoma in vivo with susceptibility contrast MRI. Cancer Research. 2017;77:4127–4134. doi: 10.1158/0008-5472.CAN-17-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Troprès I, et al. In vivo assessment of tumoral angiogenesis. Magnetic Resonance in Medicine. 2004;51(3):533–541. doi: 10.1002/mrm.20017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.