Abstract
Colitic lesions are much more severe in C3H/HeJBir (C3H) than C57BL/6J (B6) mice after 10 backcrosses of a disrupted interleukin-10 (Il10) gene. This study identified cytokine deficiency-induced colitis susceptibility (Cdcs) modifiers by using quantitative trait locus (QTL) analysis. A segregating F2 population (n = 408) of IL-10-deficient mice was genotyped and necropsied at 6 weeks of age. A major C3H-derived colitogenic QTL (Cdcs1) on chromosome (Chr.) 3 contributed to lesions in both cecum [logarithm of odds ratio (LOD) = 14.6)] and colon (LOD = 26.5) as well as colitis-related phenotypes such as spleen/body weight ratio, mesenteric lymph node/body weight ratio, and secretory IgA levels. Evidence for other C3H QTL on Chr. 1 (Cdcs2) and Chr. 2 (Cdcs3) was obtained. Cdcs1 interacted epistatically or contributed additively with loci on other chromosomes. The resistant B6 background also contributed colitogenic QTL: Cdcs4 (Chr. 8), Cdcs5 (Chr. 17, MHC), and Cdcs6 (Chr. 18). Epistatic interactions between B6 QTL on Chr. 8 and 18 contributing to cecum hyperplasia were particularly striking. In conclusion, a colitogenic susceptibility QTL on Chr. 3 has been shown to exacerbate colitis in combination with modifiers contributed from both parental genomes. The complex nature of interactions among loci in this mouse model system, coupled with separate deleterious contributions from both parental strains, illustrates why detection of human inflammatory bowel disease linkages has proven to be so difficult. A human ortholog of the Chr. 3 QTL, if one exists, would map to Chr. 4q or 1p.
Crohn's disease and ulcerative colitis are the primary forms of a heterogeneous group of diseases known collectively as chronic inflammatory bowel disease (IBD). The pathogenesis of IBD most likely results from an inability to down-regulate inflammatory responses to normal enteric flora (1). IBD is a disorder of multifactorial etiology comprising immunologic, environmental, and genetic components. A strong genetic influence is supported by increased concordance in monozygotic twins (2) and a high prevalence of disease in first degree relatives (3). Genetic clustering in families (λs) was estimated to be 36.5 for Crohn's disease, 16.6 for ulcerative colitis, and 24.7 for all IBD families (4). Comparatively, ulcerative colitis and insulin-dependent diabetes mellitus (λs = 15), which are alike in terms of their genetic heterogeneity, have similar λs (5). Unlike insulin-dependent diabetes mellitus, for which the MHC provides the major contribution to familial risk, MHC contributions in IBD seem much more modest (6). In both diseases, a large number of non-MHC susceptibility loci have been indicated in family studies. Several genome-wide screens have revealed susceptibility loci for IBD on 10 different chromosomes. Of these, the most frequently replicated is Crohn's disease-associated IBD1 on chromosome (Chr.) 16 (7–9). Evidence exists for additional loci on Chr. 1, 3, 4, 6, 7, 10, 12, 22, and X (8–11). The genetic complexity underlying IBD in humans is illustrated by epistatic interaction reported between IBD1 on Chr. 16 and another locus on Chr. 1p (9).
Mouse models of colitis offer an avenue for identifying IBD genes or pathways that may lead to identification of the human orthologs. Targeted mutations in a variety of mouse genes produce colitis (12). Mice homozygous for a disrupted interleukin-10 (Il10) gene (13) support the hypothesis that a dysregulated immune response to enteric flora can trigger IBD. The severity of the colitis depends on the inbred strain background in which the disrupted gene is placed (14). The C57BL/6J (B6) strain background proved relatively resistant to IL-10 deficiency-induced colitis, whereas the 129/SvEv and BALB/c strains were highly susceptible. The C3H/HeJBir (C3H) strain is a genetic background that is highly susceptible to several experimentally induced forms of IBD, whereas the B6 background is resistant (15–17). The genetic basis for susceptibility to colitis induced by dextran sulfate sodium was elucidated previously by a C3H × B6 outcross (18). In this cross, five loci controlling susceptibility were identified, with colitogenic contributions identified from both C3H and B6 progenitors on Chr. 1, 2, 5, 11, and 18 (18). The IL-10 molecule is a T-helper 2 (Th2) cytokine important in the suppression of T cell responses against intestinal antigens (19). Because of the known predisposition of the C3H background to experimentally induced colitis, the disrupted Il10 gene was transferred simultaneously from the B6,129 chimeric stock onto the C3H and B6 inbred backgrounds for comparison (15). Severe lesions of the cecum and colon were detected in C3H.Il10−/− mice as early as 4 weeks of age, whereas B6.Il10−/− mice developed mild lesions that did not progress in severity (15). Colonic and cecal lesions were intermediate in reciprocal F1 hybrids between the parental strains at 6 weeks of age, indicating heritability of colitis and likely complex multigenic control by background modifier loci (15).
In this study, we use a quantitative trait locus (QTL) mapping approach in identifying modifiers of cytokine deficiency-induced colitis susceptibility (Cdcs) loci responsible for colitis susceptibility in a segregating F2 population from outcross of the C3H.Il10−/− and B6.Il10−/−stocks.
Materials and Methods
Mice.
Development of the C3H and B6 stocks homozygous for a targeted mutation of the Il10 gene (formal designation Il10tm1Cgn) and their reciprocal F1 hybrids have been described previously (15). In the present study, F1 offspring were intercrossed to generate a total of 203 (102 females and 101 males) (C3H.Il10−/− × B6.Il10−/−)F2 and 208 (103 females and 105 males) (B6.Il10−/− × C3H.Il10−/−)F2 mice. Mice were maintained in a humidity-, temperature-, and light cycle (12:12)-controlled vivarium under specific pathogen-free conditions. Opportunistic microbes present in the room were Helicobacter sp. and Pasteurella pneumotropica. Mice were caged in double-pen polycarbonate cages (330-cm2 floor area) and separated by sex at a maximum capacity of five mice per pen. Mice were allowed free access to autoclaved food (NIH diet 31, 6% fat) and acidified water (pH 2.8–3.2). At 6 weeks of age, females and males were necropsied for tissue collection and analyzed for various phenotypes positively correlated with the progression of colitis. This time point was chosen because the parental strains already showed highly significant differences in histopathologic lesions at this time point (15).
The Il10 gene is located on Chr. 1 at 69.9 centimorgans (cM). Because the gene targeting was done in 129/Ola embryonic stem cells (13) and because the 129 inbred strain background is known to harbor colitis susceptibility modifier genes (14), it was essential to define the length of the congenic segment carrying strain 129 alleles in linkage disequilibrium to the disrupted Il10 gene in both C3H and B6 congenic stocks. The strain 129-derived segment in the C3H congenic stock spanned at least a 43.4-cM region from D1Mit10 (56.6 cM) to D1Mit166 (100 cM). In the B6 congenic stock, the strain 129-derived region spanned at least an 18.6-cM region from D1Mit415 (51.4 cM) to D1Mit445 (70 cM).
Histology and Colitis Assessment.
Once the mice reached 6 weeks of age, they were necropsied after CO2 asphyxiation and prepared for histology as described previously (17). The cecum and colon from each mouse were separated, then distended by intraluminal injection using Telly's acid alcohol formalin, and immersed in fixative overnight. Two hematoxylin and eosin-stained sections of the cecum and one of the colon were coded and reviewed by a pathologist (J.P.S.) who had no prior knowledge of the code. Histology of the cecum and colon was scored separately by using criteria described previously (15); these criteria included severity, hyperplasia, ulceration, and the percentage of area involved. Further, the colon was examined and graded as three individual areas: proximal, middle, and distal. Total scores were determined for the cecum and colon by adding the values of lesions associated with the severity, hyperplasia, ulceration, and percentage of tissue involved. Cecum total score ranged from 0 to 12. Colon total score ranged from 0 to 36, because individual scores from the proximal (0–12), middle (0–12), and distal (0–12) regions of the colon were combined. The lesion severity, hyperplasia, ulceration, and percentage of area involved were graded as follows: 0 = normal, 1 = mild, 2 = moderate, and 3 = severe.
Genotyping.
DNA was isolated from 5-mm tail clips and frozen kidneys by using phenol/chloroform extraction. DNAs for all F2 segregants were genotyped by PCR using microsatellite markers purchased from Research Genetics (Huntsville, AL). Cyclers (MJ Research, Cambridge, MA) were used, and products were separated on 4% Metaphor/LE (3:1) agarose gels (BMA Biomedicals). The Jackson Laboratory's allele typing service also provided multiplex genotyping data by using ABI 370 instrumentation. Ninety-four informative microsatellite markers distinguishing C3H and B6 were used, providing coverage at least every 20 cM on each autosome and Chr. X (18). The list of markers is available on request. The linkage maps and marker positions reported are based on The Jackson Laboratory's online Mouse Genome Informatics resource (www.informatics.jax.org).
Flow Cytometric Analysis of Blood Leukocytes.
One day before necropsy, ≈150 μl of blood was collected from the retro-orbital venous plexus for analysis of leukocyte populations by flow cytometry. Whole blood was placed in 1 ml of Hanks' buffered salt solution containing 5 mM EDTA and then put on ice. Red blood cells were lysed in 3 ml of Gey's solution, centrifuged at 1,500 rpm for 5 min (Sorvall model RT6000), and repeated a second time. The following monoclonal antibodies were used in combination for leukocyte staining (PharMingen or The Jackson Laboratory flow cytometry service): cy 3-labeled anti-CD4 mAb (GK1.5-Jackson), FITC-labeled anti-CD8a mAb (53–6.72), and phycoerythrin-labeled anti-CD3 mAb (145–2C11-Jackson); FITC-labeled anti-B220 mAb (RA3–6B2), phycoerythrin-labeled anti-IgM mAb (R6–60.2); and FITC-labeled anti-Mac-1 mAb (M1/70), phycoerythrin-labeled anti-Gr-1 mAb (RB6–8C5). Propidium iodide was added to each sample to identify dead cells. The percentages of viable CD4+, CD8+ T cells, B220+IgM+ B cells, Mac-1+Gr-1− monocytes, and Mac-1+Gr-1+ granulocytes were distinguished by the laboratory's FACScan flow cytometer (Becton Dickinson) with analysis performed by using the CELLQUEST software.
Other Colitis-Associated Subphenotypes.
On the day of necropsy, feces were assigned a numerical value for consistency (0 = normal, 1 = soft, and 2 = diarrhea). In addition, feces were collected and prepared for secretory IgA measures via sandwich ELISA as described previously (15), with data expressed as ng of IgA/ml. Serum was also collected, and levels of total IgG reactivity against Escherichia coli membrane (ECM) antigens as well as isotype-specific IgG2a were measured by ELISA as described previously (20). Spleens and mesenteric lymph nodes were weighed and expressed as a ratio to body weight.
Statistical Analysis.
Genome scans were carried out by using both marker regression and interval mapping analysis. Because similar results were obtained by using both methods, only the results of interval mapping are presented. Logarithm of odds ratio (LOD) scores for main effects were computed at 2-cM intervals across the autosomal genome, and LOD scores for pairwise scans were done at 5-cM resolution. The data generated by this project included 22 quantities related to severity of colitis. Some of these quantities are derived (e.g., total scores). There is moderate to high correlation among the various measurements, and thus it is difficult to assess the effective number of independent factors. Some analyses were carried out by using multivariate genome scans and also principle components analysis to reduce the dimensionality. Results obtained by these analyses were largely concordant with the individual trait genome scans. The interpretation of results was simplified by focusing on the individual trait genome scans. Significance of QTL was established by permutation analysis. Pairwise interactions among QTL were assessed by using genome-wide scans as described by Sugiyama et al. (21) and also by Sen and Churchill (22). Briefly, the significance of a QTL pair was assessed first by using an overall F test (with 8 and 399 degrees of freedom) for a full two-way ANOVA model of genotypic effects. When this test exceeded the genome-wide threshold as established by permutation analysis, secondary tests were carried out to test for an interaction effect. If an interaction was indicated, testing was stopped and an interacting QTL pair was declared. If no significant evidence for interaction was uncovered, each of the single QTL models was compared with the two QTL additive model. Both tests must be significant to indicate an additive QTL pair. These secondary tests were carried out at a nominal 0.005 level to account for testing of more than one but fewer than 10 potential QTL pairs. All software used in this analysis is available at www.jax.org/research/churchill or by request from G.A.C.
Results
Distribution of Colitis Lesions.
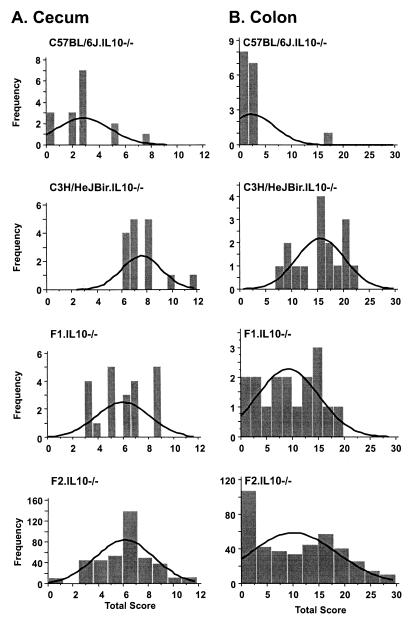
Fig. 1 illustrates the frequency distribution of cecum and colon total scores for the entire F2 segregating population (both sexes) compared with the parental and F1 strains (n = 408). The parental and F1 data shown were reported previously (15) and are reproduced here for comparison to the F2 distribution. As reported previously in the F1 generation (15), a cluster of colitis-related subphenotypes exhibited positive correlation with the histopathologic colon parameters scored in the segregating F2 population. These parameters included spleen/body weight ratio (r = 0.71), mesenteric lymph node/body weight ratio (r = 0.54), and diarrhea grade (r = 0.69). Of the various hematological subphenotypes measured by flow cytometry (percentage of B220+ and/or IgM+ B cells, CD4+ T cells, CD8+ T cells, granulocytes, and monocytes), none showed a strong positive or negative correlation with histopathologic scores (data not shown). Neither secretory IgA nor anti-ECM IgG (total) or anti-ECM IgG2a exhibited positive or negative correlations with histopathology scores.
Figure 1.
Frequency distribution of lesions in cecum (A) and colon (B) in parental, F1, and F2 generations.
Genetic Linkage Analysis: Main Effects Colitogenic QTL from C3H.
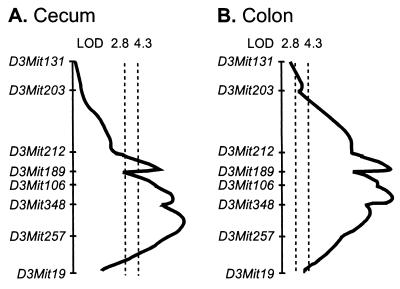
Table 1 lists all loci with significant (genome-wide level of 0.01) or suggestive (genome-wide level of 0.05) linkage to colitis histopathology. A highly significant colitogenic QTL contributed by the C3H genome was identified on the distal part of Chr. 3. This locus, provisionally designated Cdcs1, was linked to all the histopathologic parameters except for cecum ulceration and distal colon ulceration. It contributed 15% of the genetic variance for total cecum score and 26% of the variance for total colon score. This QTL was also significantly linked to the colitis-associated subphenotypes of diarrhea, spleen/body weight ratio, mesenteric lymph node/body weight ratio, and the poorly correlated subphenotypes of secretory IgA and percentage of IgM+ B cells in blood (Table 2). The C3H allele marked by D3Mit348 conferred significantly higher spleen/body weight ratio, mesenteric lymph node/body weight ratio, diarrhea grade, and secretory IgA in feces. Cdcs1 appeared to contribute to most of the colitogenic phenotypes in an additive manner. As shown in Table 1, D3Mit348 (61.8 cM), a marker allele for Cdcs1, provided an LOD score of 14.6 for cecum total score and an LOD score of 26.5 for colon total score. As shown in Fig. 2, interval mapping for cecum and colon total score indicated a broad peak spanning 22.1 cM. Almost all histopathologic subphenotypes in both cecum and colon were linked to this QTL; the few exceptions were ulceration in cecum and in distal colon. Suggestive evidence for linkage of this QTL to the traits of anti-ECM IgG, secretory IgA, and percentage of granulocytes was obtained also. A 99% confidence interval for the location of this QTL extends from 65–75 cM along the genetic map of Chr. 3.
Table 1.
LOD scores denoting significant (≥4.3) or suggestive (≥2.8) linkages detected by genome scans for main effects
| Marker position (susceptibility) | D1Mit156 32.8 cM (C3H) | D2Mit62 65 cM (C3H) | D3Mit348 61.8 cM (C3H) | D8Mit191 21 cM (B6) | D17Nds3 19.06 cM (B6) |
|---|---|---|---|---|---|
| Cecum severity | 8.9 | ||||
| Cecum hyperplasia | 8.2 | ||||
| Cecum ulceration | — | ||||
| Percentage of Cecum DA | 32.9 | ||||
| Cecum total score | 14.6 | 3.33 | |||
| Colon percentage of Prox DA | 30.4 | ||||
| Colon prox severity | 21.0 | ||||
| Colon prox hyperplasia | 19.8 | ||||
| Colon prox ulceration | 4.6 | ||||
| Colon percentage of Mid DA | 25.4 | ||||
| Colon mid severity | 18.5 | ||||
| Colon mid hyperplasia | 19.7 | ||||
| Colon mid ulceration | 3.56 | 3.9 | |||
| Colon percentage of Dist DA | 16.6 | ||||
| Colon dist severity | 4.1 | 13.3 | |||
| Colon dist hyperplasia | 3.5 | 3.8 | 14.4 | ||
| Colon dist ulceration | 4.0 | — | |||
| Prox colon score | 21.8 | ||||
| Mid colon score | 20.9 | ||||
| Dist colon score | 4.2 | 4.3 | 16.2 | ||
| Colon total score | 26.5 |
DA, damaged area; prox, proximal; dist, distal.
Table 2.
Significant (LOD ≥4.3) linkage for colitis-related phenotypes detected by genome scans for main effects
| Marker location | D3Mit348 61.8 cM | D12Nds2 59 cM | D17Nds3*/ D17Mit34† 19.06/18.8 cM |
|---|---|---|---|
| Spleen/body weight ratio | 15.9 | 5.9* | |
| Mesenteric lymph node/body weight ratio | 7.4 | ||
| Diarrhea | 18.1 | ||
| sIgA (ng/ml) | 6.6 | 4.3 | |
| Percentage of IgM+ B cells | 7.1 | ||
| Percentage of CD8+ T cells | 6.1† |
Peak LOD score at D17Nds3.
Peak LOD score at D17Mit34.
Figure 2.
Complexity of the broad QTL on Chr. 3 for histopathologic and colitis-related subphenotypes.
A second C3H-derived colitogenic linkage with apparent additive effects was detected on Chr. 1, located proximal to and clearly separable from the congenic region derived from the colitis-susceptible 129 strain contributing the targeted Il10 mutation. The strongest effects of this locus, provisionally designated Cdcs2 and marked by D1Mit156 (32.8 cM), were observed in the distal colon (distal colon score, distal colon severity, and distal colon hyperplasia). Attribution of the colitogenic allele to C3H was enabled by the ability to distinguish among C3H, 129, and B6 alleles at this locus and also by the observation that this QTL interacted additively with Cdcs1 (see below). In the C3H.Il10−/− congenic stock, we confirmed the presence of C3H alleles between the centromere and D1Mit46 at 43.1 cM, a marker at least 10.3 cM distal to the colitogenic D1Mit156 marker for Cdcs2. Similarly, typing of informative microsatellite markers distinguishing strain 129 from B6 alleles in the B6.l10−/− stock unambiguously confirmed the B6 origin of D1Mit156 in that stock; the most proximal 129-contributed allele detected (D1Mit49) was ≈9 cM distal to D1Mit156. Thus, in both stocks, the relevant parental strains and not strain 129 contributed genetic material that included the D1Mit156 marker for Cdcs2.
Cdcs3 provisionally denotes a C3H-derived QTL on Chr. 2 (peak marker D2Mit62, 65 cM) that contributed in an apparent recessive fashion. The observed effects of this QTL were linked to traits (hyperplasia and ulceration) in the distal colon (Table 1). Significant evidence for linkage was observed for distal colon score; suggestive evidence was obtained for linkage to distal colon severity.
Genetic Linkage Analysis: Main Effects Colitogenic QTL from B6.
Significant evidence for linkage for certain of the subphenotypes was detected on Chr. 8 (provisionally Cdcs4) and Chr. 17 (provisionally Cdcs5; Table 1). Both of these colitogenic contributions came from the relatively IBD-resistant B6 parental genome and appeared dominant. The significant main effect of the B6-derived Cdcs4 locus, marked by D8Mit191 (21 cM), was limited to cecum total score. However, as noted below, Cdcs4 affected other colitogenic subphenotypes through epistatic interactions with multiple other loci. Two of the Cdcs5 markers on Chr. 17 were intra-MHC markers [D17Mit34 (18.8 cM, MHC class I H2-K region) and D17Nds3 (within the Tnfa locus, 19.06 cM)], whereas another was more distal [D17Mit49 (23.2 cM)]; all showed strong dominant effects for spleen/body weight ratio. Homozygosity for C3H markers at either D17Mit34 (18.8 cM) or D17Mit49 (23 cM) was associated with increased mean CD8+ T cell percentage in peripheral blood (8% in C3H homozygotes versus 5.3% in heterozygotes and 4.8% in B6 homozygotes, respectively); however, this phenotype did not correlate well with the histopathology.
Additive and Epistatic Interactions Among Cdcs QTL.
Separately, the QTL on Chr. 1, 2, 3, 8, and 17 contributed significant main effects to the histopathologic subphenotypes and other of the colitis-related blood or fecal parameters. When marker alleles for Cdcs1 conferring the major component of colitogenic susceptibility were paired with markers for the other Cdcs QTL with the main effects noted above, additive contributions were noted for most. In addition, pairwise testing across all markers for significant interactions enabled identification of multiple gene × gene epistatic interactions for histopathologic lesions as well as for the other aforementioned phenotypes. The pairs of loci showing strongest evidence for interactions are detailed in Table 3. Two markers on Chr. 18 exhibit their effects as part of interacting pairs with D3Mit348 and D8Mit94. Cdcs1, the major C3H susceptibility QTL on Chr. 3 marked by D3Mit348, contributed epistatically with the B6-derived QTL on Chr. 18 (marked by D18Mit7) for the subphenotype cecum hyperplasia. Two B6-contributed colitogenic QTL associated with both cecum hyperplasia and cecum total score were detected by pairwise analysis for epistatic interaction. The Chr. 8 marker was D8Mit94 (13 cM), positioned 8 cM proximal to the Cdcs4 marker (D8Mit191) associated with the main effects. The Chr. 18 marker (D18Mit124 at 32 cM) shown to interact epistatically with the Chr. 8 QTL was positioned 18 cM proximal to the marker shown to interact epistatically with Cdcs1 on Chr. 3. Because significant Chr. 18 contributions were detected through epistatic interactions with two different chromosomes (Chr. 3 from C3H and Chr. 8 from B6), a colitogenic QTL on this chromosome was provisionally designated Cdcs6.
Table 3.
Significant pairwise epistatic interactions between marker pairs for various phenotypes
| Interacting marker pairs | Phenotypes controlled | F overall (>4.7)* | P | F interaction (>4.7)† | P‡ |
|---|---|---|---|---|---|
| D3Mit348 × D18Mit7 | Cecum hyperplasia | 7.66 | 1.5 × 10−9 | 4.77 | 9.1 × 10−4 |
| D8Mit94 × D18Mit124 | Cecum hyperplasia | 5.04 | 5.6 × 10−6 | 5.24 | 4.0 × 10−4 |
| Cecum total score | 5.71 | 6.8 × 10−7 | 6.60 | 3.8 × 10−5 | |
| D3Mit189 × D17Mit34 | anti-ECM IgG | 5.49 | 1.3 × 10−6 | 6.11 | 8.8 × 10−5 |
| Percentage of IgM+ B cells | 5.36 | 2.1 × 10−6 | 4.97 | 6.4 × 10−4 |
F statistic thresholds for significance at genome-wide level of P < 0.01. The genome-wide 0.01 threshold for the simultaneous pairwise genome scan was estimated to be 4.7 by permutation analysis. The test for interaction is based on the nominal 0.001 significance level, which coincidentally is also 4.7.
Based on tabulated F distribution with 8,399 degrees of freedom.
Based on tabulated F distribution with 4,399 degrees of freedom.
The detection of additive interaction for mid-colon lesions between D1Mit156, the marker for Cdcs2, and D3Mit19 at the distal end of the Cdcs1 support interval supported the existence of a C3H-derived colitogenic QTL on Chr. 1. Although no evidence for a significant main effect for loci on Chr. 19 was found, a putative QTL affecting total colon score and middle colon score was indicated through the detection of a significant epistatic interaction between the colitogenic C3H-derived allele at D3Mit257 and a codominant contribution at D19Mit53. Similarly, epistasis was detected with regard to two Ig phenotypes elevated in the C3H.Il10−/− parental compared with B6.Il10−/− parental mice. For anti-ECM IgG and IgM+ B cell percentage in peripheral blood, the C3H-derived allele at D3Mit189 marking the proximal end of Cdcs1 interacted epistatically with the B6-derived D17Mit34 marker for Cdcs5. For secretory IgA levels, an epistatic interaction was observed between the B6-derived Cdcs4 QTL marked by D8Mit178 and the C3H allele at D12Nds2 (59 cM), a marker within the Ig heavy chain variable chain locus.
Discussion
In this study, we located six major QTL contributing to colitis susceptibility as either main effectors and/or interacting QTL. The most significant linkage, provisionally designated Cdcs1 on Chr. 3, exhibited C3H susceptibility and was associated with nearly all histopathologic parameters and all colitis-related phenotypes. This locus is the major determinant of colitis susceptibility in this intercross population (accounting for 15–30% of total variance in subphenotypes). However, the secondary QTL represent statistically and biologically significant contributions to the overall phenotype. In the absence of segregation at the Chr. 3 locus, these loci and their interactions would represent major QTL. IBD and colitis susceptibility are clearly complex traits. Comparable epistatic interactions to those underlying colitis susceptibility in our mouse model have been reported recently for IBD susceptibility in humans (9). Table 4 provides a summary of the mouse linkages along with potential human homologs. Several attractive Cdcs1 candidate genes are located on Chr. 3 within the 99% confidence interval (65–75 cM). These candidates include the Egf gene (65.2 cM) encoding the epidermal growth factor. The parental strains are polymorphic at this locus; hence, allelic variants could affect mucosal susceptibility to injury and facilitate repair of inflammatory damage. Indeed, therapeutic effects were reported after systemic epidermal growth factor administration to rats with trinitrobenzene sulfonic acid-elicited colitis (23) as well as to humans with gastrointestinal diseases (24). Possibly, epidermal growth factor generates a barrier that limits mucosal injury or functions as a scavenger of reactive oxygen species. In that regard, up-regulation of the Nfkb1 gene (68.9 cM), encoding the NF-κB p105 transcription factor protein, represents an early response to inflammatory stress (25, 26) with induction observed in areas of inflammation where large quantities of superoxide anion are present such as in colitis. Given the attractiveness of Nfkb1 as a Cdcs1 candidate gene, comparative analysis of C3H versus B6 NF-κB activation in response to lipopolysaccharide (LPS) or IL-12 may be informative. The human ortholog for the Cdcs1 locus, if one existed, would be expected on either Chr. 4q or 1p rather than the Crohn's disease-associated IBD1 locus on Chr. 16. However, it is intriguing that the IBD1 candidate gene recently identified, NOD2, is a gene, the product of which activates NF-κB, making the latter responsive to LPS stimulation (27). The reduced LPS sensitivity of C3H mice is linked to a defect of the Toll-like receptor 4 (Tlr4) on Chr. 4. Although the Tlr4 genetic difference did not manifest as a Cdcs QTL, it is known that additional defects in other receptors such as the IL-12Rβ subunit can contribute to LPS insensitivity (28, 29). Because C3H CD4+ T cells activated by cecal bacterial antigens produce high IFN-γ levels and these C3H T cells transfer colitis when adoptively transferred into C3H-SCID recipients (16), this strain may have evolved mechanisms for maintaining higher NF-κB levels despite the LPS insensitivity. Indeed, we reported previously that bacterial antigen-stimulated CD4+ T cells from C3H.IL-10-deficient mice secreted much higher levels of IFN-γ and IL-3 than did CD4+ T cells from B6.IL-10-deficient mice (15).
Table 4.
Provisional nomenclature for colitogenic linkages
| Locus (Chr.) | Susceptibility donor | Mode of action | Phenotype(s) controlled | Candidate genes | Potential human IBD linkage |
|---|---|---|---|---|---|
| Cdcs1 (3) | C3H | Additive:epistatic with multiple loci | Cecum/colon lesions and multiple subphenotypes | Fabpi, Egf, Nfkb1 | 4q or 1p |
| Cdcs2 (1) | C3H | Additive | Distal colon severity and hyperplasia | Casp8, Cd28, Ctla4, Icos | 2q |
| Cdcs3 (2) | C3H | Recessive | Distal colon hyperplasia, severity, ulceration, distal colon score, B cell number | Thbs1, B2m Il1, Pcna | 15q 2q |
| Cdcs4 (8) | B6 | Epistatic | Cecum hyperplasia | Defb, Defcr, Scc8 | 8p |
| Cdcs5 (17) | B6 | Recessive | Proximal colon severity, mid-colon ulceration | H2-Ea, C4/Slp, Tnf | 6p |
| Cdcs6 (18) | B6 | Epistatic | Cecum hyperplasia | Dcc, Apc, Mad2, Mad4 | 18q or 5q |
Our finding that, in hybrid combinations, the nominally resistant B6 parental genome contributed at least three Cdcs linkages to the colitis subphenotypes is particularly relevant in understanding why colitis heritability in outbred human populations is so complex. Multiple intestinal neoplasias (Mins) can develop in response to mutation in the adenomatosis polyposis coli (Apc) gene on Chr. 18. The B6 background is highly permissive to ApcMin-induced neoplasia compared with other inbred strains surveyed (30). This B6-specific susceptibility was linked in part to a null mutation in a secretory phospholipase (Pla2g2a) gene on Chr. 6 that epistatically suppressed ApcMin action in other strains expressing this enzyme (31). In the current study of IL-10 deficiency-induced colitis, a B6 QTL on Chr. 18 marked by D18Mit7 (50 cM) increased the severity of cecum hyperplasia through epistatic interaction with Cdcs1 on Chr. 3. Another B6-derived Chr. 18 marker, D18Mit124 (32 cM), interacted epistatically with the Cdcs4 peak marker, D8Mit94, to enhance cecum total score. This result suggests that Cdcs6 represents a modifier or modifiers of hyperplasia in the cecum. Chr. 18 contains logical candidates for such modifiers including the deleted in colorectal carcinoma (Dcc) locus (45 cM), a logical candidate for an epistatic modulator of hyperplasia. Another logical candidate for a B6 epistatic modifier is found even closer (48 cM) to D18Mit7: the MAD homolog 4 (Madh4) encoding SMAD4, a tumor suppressor factor functioning through the transforming growth factor-β-signaling pathway. Transforming growth factor-β has been shown to have suppressive effects in the development of colon cancer as well as an ability to restore colonic mucosal integrity after inflammatory assault (32). If an ortholog for Cdcs6 exists, it would map to either human Chr. 5q (APC) or 18q (DCC and SMAD). Evidence for a human 5q colitis linkage has been reported (33). A deleted portion of 18q containing DCC is detected in ≈70% of human colorectal cancers (34). Another B6 colitogenic QTL showing a significant additive effect in combination with the C3H-derived segment at Cdcs1 was on Chr. 8 and marked by D8Mit191 (21 cM). This locus affected the traits of cecum hyperplasia, cecum-damaged area, and cecum total score. If Nfkb1 were indeed the correct candidate gene for Cdcs1, an elegant candidate for the B6 contribution on Chr. 8 would be Ikbkb (inhibitor of κB kinase β or IKK), which helps to maintain activated NF-κB. Other interesting candidates in the area are two defensin genes, defensin B (Defb) and the defensin-related cryptdin peptide (Defcr), the fibroblast growth factor receptor 1 (Fgfr1), and a QTL controlling susceptibility to colorectal cancer 8 (Scc8). With regard to Scc8, another QTL controlling responsiveness to carcinogen-induced colorectal carcinoma (Scc7) has been localized to ≈71cM on Chr. 3. Hence, the Chr. 3/Chr. 8 joint effect could entail additive effects of susceptibilities contributed by each parental genome.
It was unexpected that with a main effect of mid-colon ulceration on Chr. 17 (mid-colon ulceration) was derived from the B6 rather than C3H genome. Activated peritoneal macrophages from C3H/HeJ were reported to secrete more tumor necrosis factor-α than those from B6; this phenotype was associated with polymorphisms in the Tnfa promoter region (35). Significantly higher levels of CD8+ T cells circulating in the peripheral blood of our F2 population was associated also with homozygosity for the C3H alleles at D17Mit34 and D17Nds3. However, the MHC haplotype (H2k) of C3H is associated with deletions of genes in the class I Qa2 cluster required for selection of CD8αα cells in the intestinal epithelial lymphocyte compartment (36). Possibly, the presence of functional B6-derived alleles in the Qa2 region represented the increased colitogenic contribution of this haplotype (H2b). It should be noted that IL-10 deficiency in the strain 129 background, which also expresses the H2b MHC haplotype, also develops severe colitis (14).
In summary, the complexity shown in this analysis of cytokine deficiency-induced colitis in mice underscores the difficulty in elucidating the complex genetics underlying susceptibility to IBD development in humans. We are developing interval-specific congenic stocks (37) to guide analysis of relevant candidate genes in this mouse model system.
Acknowledgments
This work was supported by National Institutes of Health Grant PPG-44240. Institutional shared services (The Jackson Laboratory) were supported by National Cancer Institute Cancer Center Support Grant CA-34196.
Abbreviations
- IBD
inflammatory bowel disease
- Chr.
chromosome
- B6
C57BL/6J
- C3H
C3H/HeJBir
- QTL
quantitative trait locus/loci
- Cdcs
cytokine deficiency-induced colitis susceptibility
- cM
centimorgan
- ECM
Escherichia coli membrane
- LOD
logarithm of odds ratio
- LPS
lipopolysaccharide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Elson C O, Cong Y, Brandwein S, Weaver C T, McCabe R P, Mahler M, Sundberg J P, Leiter E H. Ann NY Acad Sci. 1998;859:85–95. doi: 10.1111/j.1749-6632.1998.tb11113.x. [DOI] [PubMed] [Google Scholar]
- 2.Tysk C, Lindberg E, Jarnerot G, Floderus M B. Gut. 1988;29:990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayless T M, Tokayer A Z, Polito J M, Quaskey S A, Mellits E D, Harris M L. Gastroenterology. 1996;111:573–579. doi: 10.1053/gast.1996.v111.pm8780559. [DOI] [PubMed] [Google Scholar]
- 4.Satsangi J, Jewell D P, Bell J I. Gut. 1997;40:572–574. doi: 10.1136/gut.40.5.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todd J. Proc Natl Acad Sci USA. 1995;92:8560–8565. doi: 10.1073/pnas.92.19.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouma G, Oudkerk Pool M, Crusius J B A, Schreuder G M T, Hellemans H P R, Meijer B U G A, Kostense P J, Giphart M J, Meuwissen S G M, Pena A S. Clin Exp Immunol. 1997;109:175–179. doi: 10.1046/j.1365-2249.1997.4121510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohmen J D, Yang H Y, Yamamoto K K, Zhao H Y, Ma Y, Bentley L G, Huang Z, Gerwehr S, Pressman S, McElree C, et al. Hum Mol Genet. 1996;5:1679–1684. doi: 10.1093/hmg/5.10.1679. [DOI] [PubMed] [Google Scholar]
- 8.Hugot J P, Laurent-Puig P, Gower-Rousseau C, Olson J M, Lee J C, Beaugerie L, Naom I, Dupas J L, Van Gossum A, Orholm M, et al. Nature (London) 1996;379:821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 9.Cho J H, Nicolae D L, Gold L H, Fields C T, LaBuda M C, Rohal P M, Pickles M R, Qin L, Fu Y, Mann J S, et al. Proc Natl Acad Sci USA. 1998;95:7502–7507. doi: 10.1073/pnas.95.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satsangi J, Parkes M, Louis E, Hashimoto L, Kata N, Welsh K, Terwilliger J D, Lathrop G M, Bell J I, Jewell D P. Nat Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 11.Hampe J, Schreiber S, Shaw S H, Lau K F, Bridger S, Macpherson A J, Cardon L R, Sakul H, Harris T J, Buckler A, et al. Am J Hum Genet. 1999;64:808–816. doi: 10.1086/302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elson C O, McCabe R, Cong Y, Brandwein S, Weaver C, Leiter E H, Sundberg J P, McGhee J R. Inflammatory Bowel Diseases: From Bench to Bedside. 1997. , eds. T. Andus, H. Goebell, P. Layer, & J. Scholmerich (Kluwer, Dordrecht, The Netherlands), pp. 21–26. [Google Scholar]
- 13.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 14.Berg D J, Davidson N, Kühn R, Müller W, Menon S, Holland G, Thompson-Snipes L, Leach M W, Rennick D. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bristol I J, Cong Y, Farmer M A, Zheng X X, Strom T B, Elson C O, Sundberg J P, Leiter E H. Inflamm Bowel Dis. 2000;6:290–302. doi: 10.1002/ibd.3780060407. [DOI] [PubMed] [Google Scholar]
- 16.Cong Y, Brandwein S L, McCabe R P, Lazenby A, Birkenmeier E H, Sundberg J P, Elson C O. J Exp Med. 1998;187:855–864. doi: 10.1084/jem.187.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mähler M, Bristol I J, Leiter E H, Birkenmeier E H, Elson C O, Sundberg J P. Am J Physiol. 1998;274:G544–G551. doi: 10.1152/ajpgi.1998.274.3.G544. [DOI] [PubMed] [Google Scholar]
- 18.Mähler M, Bristol I J, Sundberg J P, Churchill G, Birkenmeier E H, Elson C O, Leiter E H. Genomics. 1999;55:147–156. doi: 10.1006/geno.1998.5636. [DOI] [PubMed] [Google Scholar]
- 19.Asseman C, Mauze S, Leach M W, Coffman R L, Powrie F. J Exp Med. 1999;190:995–1004. doi: 10.1084/jem.190.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandwein S L, McCabe R P, Cong Y, Waites K B, Ridwan B U, Dean P A, Ohkusa T, Birkenmeier E H, Sundberg J P, Elson C O. J Immunol. 1997;159:44–52. [PubMed] [Google Scholar]
- 21.Sugiyama F, Churchill G A, Higgins D C, Johns C, Makaritsis K P, Gavras H, Paigen B. Genomics. 2001;71:70–77. doi: 10.1006/geno.2000.6401. [DOI] [PubMed] [Google Scholar]
- 22.Sen S, Churchill G A. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Procaccino F, Reinshagen M, Hoffmann P, Zeeh J M, Lakshmanan J, McRoberts J A, Patel A, French S, Eysselein V E. Gastroenterology. 1994;107:12–17. doi: 10.1016/0016-5085(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 24.Guglietta A, Sullivan P B. Eur J Gastroenterol Hepatol. 1995;7:945–950. doi: 10.1097/00042737-199510000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Schulze-Osthoff K, Los M, Baeuerle P A. Biochem Pharmacol. 1995;50:735–741. doi: 10.1016/0006-2952(95)02011-z. [DOI] [PubMed] [Google Scholar]
- 26.Satriano J, Schlondorff D. J Clin Invest. 1994;94:1629–1636. doi: 10.1172/JCI117505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogura Y, Bonen D K, Inohara N, Nicolae D L, Chen F F, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr R H, et al. Nature (London) 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 28.Beutler B, Poltorak A. Drug Metab Dispos. 2001;29:474–478. [PubMed] [Google Scholar]
- 29.Poltorak A, Merlin T, Nielsen P J, Sandra O, Smirnova I, Schupp I, Boehm T, Galanos C, Freudenberg M A. J Immunol. 2001;167:2106–2111. doi: 10.4049/jimmunol.167.4.2106. [DOI] [PubMed] [Google Scholar]
- 30.Cormier R T, Hong K H, Halberg R B, Hawkins T L, Richardson P, Mulherkar R, Dove W F, Lander E S. Nat Genet. 1997;17:88–91. doi: 10.1038/ng0997-88. [DOI] [PubMed] [Google Scholar]
- 31.Cormier R T, Bilger A, Lillich A J, Halberg R B, Hong K H, Gould K A, Borenstein N, Lander E S, Dove W F. Oncogene. 2000;19:3182–3192. doi: 10.1038/sj.onc.1203646. [DOI] [PubMed] [Google Scholar]
- 32.Engle S J, Hoying J B, Boivin G P, Ormsby I, Gartside P S, Doetschman T. Cancer Res. 1999;59:3379–3386. [PubMed] [Google Scholar]
- 33.Rioux J D, Silverberg M S, Daly M J, Steinhart A H, McLeod R S, Griffiths A M, Green T, Brettin T S, Stone V, Bull S B, et al. Am J Hum Genet. 2000;66:1863–1870. doi: 10.1086/302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fearon E R, Cho K R, Nigro J M, Kern S E, Simons J W, Ruppert J M, Hamilton S R, Preisinger A C, Thomas G, Kinzler K W, et al. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 35.Jacob C O, Lee S K, Strassmann G. J Immunol. 1996;156:3043–3050. [PubMed] [Google Scholar]
- 36.Das G, Gould D S, Augustine M M, Fragoso G, Scitto E, Stroynowski I, Van Kaer L, Schust D J, Ploegh H, Janeway C A., Jr J Exp Med. 2000;192:1521–1528. doi: 10.1084/jem.192.10.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darvasi A. Mamm Genome. 1997;8:163–116. doi: 10.1007/s003359900382. [DOI] [PubMed] [Google Scholar]