Abstract
Uridine diphosphate N-acetyl muramic acid (UDP NAM) is a critical intermediate in bacterial peptidoglycan (PG) biosynthesis. As the primary source of muramic acid that shapes the PG backbone, modifications installed at the UDP NAM intermediate can be used to selectively tag and manipulate this polymer via metabolic incorporation. However, synthetic and purification strategies to access large quantities of these PG building blocks, as well as their derivatives, are challenging. A robust chemoenzymatic synthesis was developed using an expanded NAM library to produce a variety of 2-N functionalized UDP-NAMs. In addition, a synthetic strategy to access biorthogonal 3-lactic acid NAM derivatives was developed. The chemoenzymatic UDP synthesis revealed that the bacterial cell wall recycling enzymes MurNAc/GlcNAc anomeric kinase (AmgK) and NAM α-1 phosphate uridylyl transferase (MurU) were permissive to permutations at the two and three positions of the sugar donor. We further explored the utility of these derivatives in the fluorescent labeling of both Gram (−) and Gram (+) PG in whole cells using a variety of bioorthogonal chemistries including the tetrazine ligation. This report allows for rapid and scalable access to a variety of functionalized NAMs and UDP NAMs, which now can be used in tandem with other complementary bioorthogonal labeling strategies to address fundamental questions surrounding PG’s role in immunology and microbiology.
Graphical Abstract
Introduction
Organophosphates are among the essential molecular components for life. In addition to their role in the scaffold of deoxyribonucleic acid (DNA), ribonucleic acid (RNA), energy storage and post-translational modifications of proteins, these phosphate groups hold valuable roles in glycobiology.1–2 From the polymerization of energy storing molecules such as glycogen to glycosylation of lipids on the surface of cells, the addition of a unique sugar molecule onto hetero-atom scaffolds via phosphate chemistry can tune the chemical and physical properties of the biomolecule.3–8 While many phosphate shuttling biomolecules are key intermediates for signaling, they also serve as carriers in many essential biosynthetic pathways.
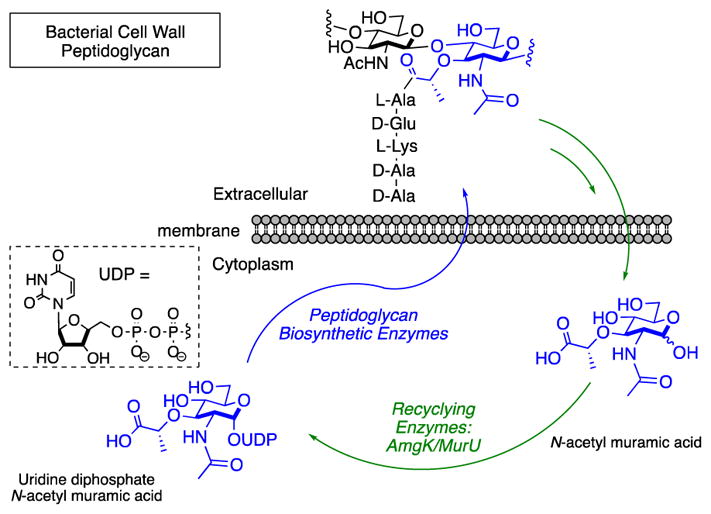
Uridine diphosphates (UDPs) are organophosphate carriers that play a critical role in the biosynthesis of bacterial peptidoglycan (PG), a coat that surrounds each bacterial cell to protect it from environmental changes and stresses.9–10,11 The strength of the bacterial cell wall is directly correlated to its molecular composition; a complex network of peptides and two carbohydrates, N-acetyl glucosamine (NAG) and N-acetyl muramic acid (NAM), which are stitched together through a conserved biosynthetic pathway that utilizes UDP sugar carriers of NAG and NAM carbohydrates (Scheme 1).10
Scheme 1.
Peptidoglycan Recycling and Biosynthetic Pathways
NAM serves as a core structural element exclusively for bacterial cell wall. While bacterial PG is dynamic, constantly changing as bacterial cells grow and divide, the NAM building blocks of this material are conserved across all bacterial species.10 Synthetic and naturally occurring small molecule units derived from bacterial PG with the NAM glycan core are known to activate human innate immune responses.12–13 Interestingly, extending or truncating the peptide portion of these intermediates results in a tunable immune response.14–16
The ability to monitor the natural production and breakdown of this polymer at both the glycan and peptide levels is critical for deciphering how human hosts are recognizing and responding to bacteria at the molecular level.17–20 The development of a variety of small molecule NAM probes as well as methodology to selectively label the NAM core of PG in whole cells could help tease out the molecular fingerprint of these interactions to reveal how different bacteria present their PG to host cells or how host cells process this material.
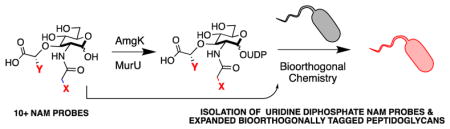
All bacteria naturally build their PG utilizing a series of conserved enzymatic steps beginning with the production of UDP NAM through biosynthetic enzymes MurA and MurB.21 Recently, our laboratory developed methodology to selectively install azide or alkyne bioorthogonal tags onto the NAM backbone of bacterial PG in whole cells for fluorescent visualization via copper catalyzed azide alkyne cycloaddition (CuAAC).22–25 This strategy was achieved by taking advantage of the alternative formation of UDP NAM via the NAM lactol building block and cell wall recycling enzymes MurNAc/GlcNAc anomeric kinase (AmgK) and NAM α-1 phosphate uridylyl transferase (MurU)26 (Scheme 1). Here, we further explored the chemical space of AmgK and MurU substrates to expand both the NAM and UDP NAM tools available to probe bacterial PG biosynthesis and recycling. We developed the necessary synthetic routes to show that the system is amenable to a variety of permutations and demonstrate the utility of the method in labeling bacterial PG.
Results
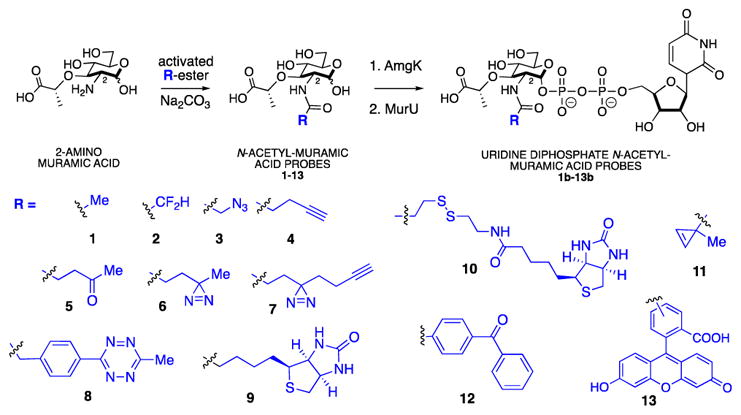
The investigation began with the design and synthesis of an expanded library of NAM derivatives to probe the substrate specificity of AmgK. The modular synthesis of the previously reported N-acetyl azido and alkyne NAM derivatives (3, 4) gave access to large quantities of the precursor, 2-amino muramic acid (Scheme 2). This molecule was poised to functionalize using mild amide bond coupling conditions with a variety of activated ester derivatives in order to generate a library of NAM compounds (Scheme 2, Supporting Information). This library includes multiple bioorthogonal NAMs27 (Scheme 2, Table 1) ranging from ketone condensation intermediates28 (5), to inverse electron demand diels alder (IEDDA) compounds29–32 (8, 11). Photoactivatable intermediates with diazirine33–37 and benzophenone functionality38–40 (6, 7, 12) were also synthesized in addition to di-fluoro methyl41–43 (2) fluorescein (13) and biotin44–46 tags (9, 10). To assess if modifications around the carbohydrate were tolerated, the starting material, 2-amino-muramic acid and a NAM derivative with an azide modification at the 6-position, 6-azido-NAM, were synthesized (Supporting Information).
Scheme 2.
Library of 2-N NAM and UDP NAM probes
Table 1. AmgK and MurU substrate specificity.
Each compound was grouped based on common bioorthogonal reactivity or utility. Turnover of AmgK and MurU are reported with either (Y) representing formation of corresponding product, (N) representing no formation of product and (---) representing untested substrate due to no conversion of AmgK. Product formation was monitored by high-resolution mass spectrometry (HRMS).
| Bioorthogonal Reaction or Chemical Utility | Substrate | AmgK | MurU |
|---|---|---|---|
| Copper Catalyzed Azide-Alkyne Cycloaddition | 3, 4, 14, 6-azido NAM | Y, Y, Y, N | Y, Y, Y, N |
| Ketone Condensation | 5 | Y | Y |
| Photoactivating Crosslinker | 6, 7, 12 | Y, Y, N | Y, Y, --- |
| Inverse Electron-Demand Diel’s-Alder | 8, 11 | Y, N | Y, --- |
| NMR Characterization | 2 | Y | Y |
| Streptavidin Affinity Purification | 9, 10 | Y, Y | N, N |
| Fluorescence Visualization | 13 | N | --- |
| Other | 1, amino | Y, N | Y, --- |
With the library of NAM derivatives in hand, the substrate specificity of the bacterial cell wall recycling enzyme AmgK was probed (Table 1 and 2). Each of the NAM derivatives were treated with purified AmgK26 and product formation was monitored by high resolution mass spectrometry (HRMS) (Supporting Information Table 1). The corresponding mono-phosphate NAM products were observed in almost all of the intermediates with the exception of cyclopropene derivative 11, benzophenone 12 and fluorescein derivative 13 (Table 1, Supporting Information Table 1), suggesting that these modifications were not accepted by the kinase. 6-azido-NAM (Supporting Information, Scheme 3) and 2-amino-muramic acid showed no conversion into the monophosphate, indicating that they are not substrates.
Table 2.
Kinetic Characterization of AmgK with NAM derivatives as substrates.
| Substrate | kcat(s−1) | Km(mM) | kcat/Km (M−1s−1) |
|---|---|---|---|
| 1a | 6.58 ± 0.21 | (3.04 ± 0.58) × 10−2 | (2.16 ± 0.42) × 105 |
| 3a | 14.07 ± 0.72 | 1.12 ± 0.16 | (1.26 ± 0.19) × 104 |
| 4a | 9.74 ± 0.45 | 0.55 ± 0.09 | (1.8 ± 0.3) × 104 |
| 14a | 4.67 ± 0.14 | (3.03 ± 0.54) × 10−2 | (1.54 ± 0.28) × 105 |
| 5b | - | - | 3.45 × 103 |
| 6b | - | - | 1.55 × 103 |
| 7b | - | - | 1.02 × 103 |
| 8b | - | - | 2.25 × 103 |
| 9b | - | - | 2.95 × 103 |
kinetic parameters were calculated by fitting the experimental data into the Michaelis-Menten equation using program GraphPad Prism-6.
Apparent kcat/Km values were calculated by fitting the experimental data to the Lineweaver-Burk equation.
Standard Error (SE) were calculated based on 3 technical replicates of each sample concentration
In order to compare AmgK’s substrate preferences, the kinetic profile of AmgK with the NAM derivatives was measured according to the ATPase enzyme coupled assay used by Mayer and co-workers.47 For 1, apparent Km and kcat were 30.41 ± 5.75 μM and 6.58 ± 0.21 s−1, respectively (Table 2, Supporting Information). These data agree with the values reported by Mayer and coworkers26. For substrates 3 and 4, AmgK showed ~10 fold less efficiency than it had turning over 1. The catalytic efficiencies were ~100 fold less with other derivatives as substrates (Table 2, Supporting Information). Apparent kcat/Km values were acquired for substrates whose rates did not plateau by fitting the experimental kinetic parameters to a double reciprocal plot (Table 2, Supporting Information). 5, 6, 7, 8 and 9 appear to have high Km values based on this analysis, suggesting that the enzyme has low affinity for these larger modifications (Supporting Information).
The monophosphate NAM intermediates were then subjected to uridylyl transferase MurU48 to generate the UDP-sugar (Scheme 2). The production of the UDP NAM derivatives was monitored by HRMS (Table 1, Supporting Information Table 1). MurU converted the monophosphate intermediates of 1–8 to their respective UDP products as observed by HRMS. Furthermore, intermediates 1, 3–5 were converted in milligram quantities (i.e. > 10 mgs) and isolated by preparatory mass directed HPLC for complete NMR characterization (Supporting Information).
In addition to the 2-N functionalized NAM derivatives, modification at the 3-OH lactic acid portion of the small molecule was desired (Scheme 3). It is known that some bacteria including Mycobacterium tuberculosis (Mtb), naturally modify their bacterial PG to a N-glycolyl moiety at the 2-N position via NamH.49–50 In order to avoid the loss of a 2-N biorthogonal probe through this pathway, we moved to install the functionality at another location of the NAM sugar. As these derivatives have not been made before, a synthesis of lactic acid functionalized NAMs was designed and implemented (Scheme 3, Supporting Information).
Scheme 3.
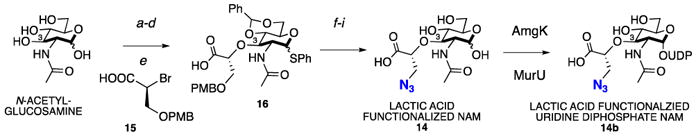
Synthesis of lactic acid N3 functionalized NAM
a. Ac2O, DMAP, pyridine (85%), b. PhSH, SnCl4, DCM, reflux (54%), c. NaOMe, MeOH, r.t. (quant), d. PhCH(OMe)2, TsOH, DMF, 70°C (72%), e. NaH, 15, DMF (57%), f. K2CO3, MeI, DMF, r.t. (80%), g. DDQ, DCM/H2O (quant), h. MsCl, pyridine/DCM (80%), i. 1. NaN3, DMF, 70°C; 2. IRA H+, H2O, 95°C; 3. TCCA (12% over 3 steps).
To efficiently derivatize the 3-OH position, the synthesis of a novel azide modified lactic acid precursor, which would be added to the NAG core, was developed (Scheme 3, Supporting Information). We found that the addition reaction proceeded most efficiently with the brominated lactic acid scaffold 15. The para-methoxybenzyl (PMB) protecting group was selected over other groups due to ease of installation and removal. 15 was readily coupled to the suitably protected NAG 1-thioglycoside to yield the derivatized NAM. Subsequent methyl esterification, PMB deprotection with DDQ revealed the free alcohol, which was activated for azido installation.
To generate the final unprotected NAM lactic acid derivative, a mild deprotection strategy was utilized. Notably, the thiol-protecting group was removed under oxidative conditions with trichloroisocyanuric acid (TCCA)51, an environmentally friendly chemical reagent.. Global deprotection revealed compound 14. 14 was shown to be accepted by recycling enzymes AmgK and MurU to generate the corresponding UDP NAM intermediate 14b by HRLCMS (Table 1 and Supplementary Table 1). Kinetic characterization with AmgK was performed and the data show that the kinase accepted the lactic acid modification with the same efficiency as the natural NAM (Table 2).
With the formation of the UDP-NAM derivatives at the 2 and 3 position of the carbohydrate confirmed, the promiscuity of the bacterial PG biosynthetic enzymes MurC-F was explored (Scheme 1, Supplemental Scheme 1). All of the UDP NAM derivatives subjected to MurC-F were able to be converted into the desired pentapeptide Park’s nucleotide derivatives and were observed by HRMS (Supporting Information Table 1). The relaxed substrate specificity of the PG recycling and biosynthetic enzymes motivated the expansion of the PG glycan labeling method of whole bacterial cells. In order to improve this methodology, three key areas were explored: modification at the 3-postion of the NAM, click-chemistry utility and UDP-NAM-carrier tolerance
First the lactic acid modified NAM derivative 14 was tested for incorporation in whole bacterial cells. E. coli QKU, a MurQ52 knockout E. coli strain that contains the recycling enzymes AmgK and MurU, cells were pulsed with 14 for two doubling times (45 minutes) in the presence of fosfomycin and subsequently treated with CuAAC click chemistry conditions as previously reported22. Cells were visualized with superresolution Structured Illumination Microscopy (SIM)53 (Fig 1, Supplementary Fig 1, Supplemental Material). Fluorescent signal was observed at the septal division ring of the bacterial cells as compared to the positive control cells treated with 3 and the negative control cells treated with 1 in which only background fluorescence was observed (Supplementary Fig. 2 and 3A, respectively). This positive labeling result suggests that NAM derivative 14 can be utilized by the bacterial PG biosynthetic enzymes, which is consistent with the in vitro enzymatic conversion study (Supporting information Table 1).
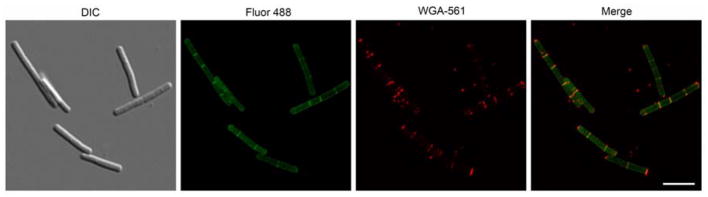
Figure 1. E. coli QKU cells fluorescently modified with the lactic acid NAM derivative 14 and WGA co-staining.
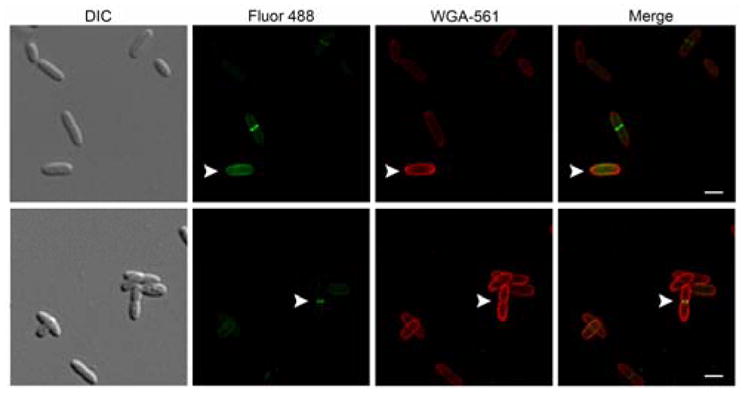
DIC and maximum intensity projection 2D images from super-resolution SIM Z-stacks of E. coli QKU cells treated with 14, fosfomycin, IPTG for 45 min and clicked with Alk488 (green) and then stained with tetramethylrhodamine WGA 561 (red) of whole cell (white arrow, top) and dividing cell (white arrow, bottom) (scale bars, 10 μm). Images are representative of a minimum of three fields viewed per replicate with at least two technical replicates and the experiment was conducted in three biological replicates.
Other bioorthogonal “click” chemistries were probed for tolerance by the NAM-PG labeling method. Previously, copper catalyzed azide-alkyne cycloaddition (CuAAC) in whole cells was performed. Here the tetrazine-transcyclooctene (TCO) ligation, the fastest click reaction to date was assessed.32, 54 E. coli QKU cells were treated with either 1 or NAM derivative 8 and IEDDA was performed with TCO-TAMRA (Supporting Information). Cells were visualized with superresolution SIM. The positive control cells were labeled with 3 and treated with CuAAc to ensure E. coli QKU labeling competence (Supplementary Fig 4). The optimal probe pulse length of 8 was assessed (Supplementary Fig. 5A) in the presence and absence of fosfomycin. It was determined that 20 min pulse lengths were optimal without fosfomycin (Fig 2). It appeared that fluorescent labeling was only obtained in select cells per field of view when cells were treated with 8 (Fig 2, Supplementary Figure 5B), indicating that the uptake of the tetrazine probe was not uniform throughout cells or that a sub-population of the cells were capable of accepting the tetrazine modification. Cells that were treated with 1, the unlabeled control, showed no fluorescence (Supplementary Fig 3B), suggesting that this limited labeling is not due to the fluorophore non-specifically associating to the bacteria.
Figure 2. E. coli QKU cells modified with 8 for tetrazine-TCO ligation.
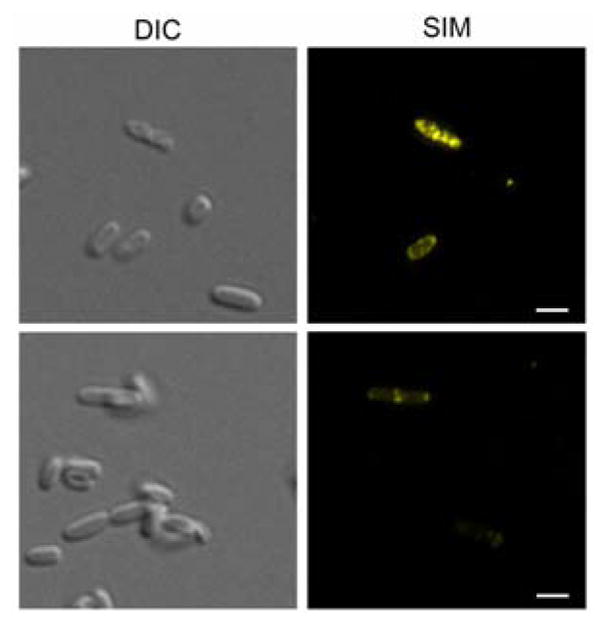
DIC and 2D images from super-resolution SIM Maximum Intensity Projection Images Z-stacks of E. coli QKU cells treated with 8 and IPTG for 20 min and clicked with TCO-TAMRA (yellow) (scale bars, 2 μm). Images are representative of a minimum of three fields viewed per replicate with at least two technical replicates and the experiment was conducted in at least three biological replicates.
Traditionally, we and others have utilized mass spectrometry evidence for probe PG incorporation.22, 55–56 However, due to the low fluorescent probe incorporation with 8 and 14, we desired a method other than mass spectrometry to confirm PG labeling. Wheat germ agglutinin (WGA) co-labeling, a technique commonly used to stain the NAG backbone of the bacterial PG was used.57–58 If co-localization was observed with the TCO-TAMRA probe, then modification of the PG could be considered. Co-localization with tetramethylrhodamine-WGA stain, in E. coli QKU cells treated with 14 was observed suggesting labeling of the PG (Fig. 1, Supplementary Figure 1). No co-localization with 488-WGA stain in E. coli QKU cells treated with 8 was observed (Supplementary Fig 6). Cells that incorporated 8 appeared to have degraded cell morphologies, indicating that tetrazine incorporation is not well tolerated and thus has low WGA signal.
Finally, UDP-NAM modifications were tested for whole cell incorporation. To date, two methods have been used to in-corporate NAM derivatives into the PG of whole cells. One involves the genetic manipulation of the bacterial cell to include the recycling enzymes AmgK and MurU22. The second method involved a laborious 15 step synthesis of uridine diphosphate muramyl tetrapeptide fluorescein derivative59. The latter was a tour de force that demonstrated the unnatural UDP peptide fluorophore derivative could be taken up and embedded into the PG of Gram + lactic acid bacteria. However, the method was limited due to the demanding chemical synthesis of the UDP probe. Therefore, we took advantage of the scalable chemoenzymatic production of the uridine diphosphate sugar derivatives 3b and 4b containing the azide and alkyne functionality, respectively (Scheme 2). This method was amendable to milligram scale synthesis; the phosphate and uridylyl steps were performed subsequently without purification of the phosphate intermediate (Supporting Information). Yields for the multiple enzymatic transformations ranged from 19% to 89%. Whole cell bacterial labeling with these derivatives was explored using compound 3b.
UDP sugars exist as charged species at physiological pH (Scheme 1 and 2), and diffusion through the bacterial cell membrane could be challenging. Therefore, the whole cell studies began with the bacterium Lactobacillus acidophilus, a commensal60 Gram + organism commonly found in yogurt and previously used by Nishimura and coworkers to label bacterial PG using UDP fluorescein lysine derivatives.59 After incubation with L. acidophilus and the azido UDP NAM 3b, SIM imaging revealed fluorescent labeling as compared to the control cells which were incubated with the natural UDP NAM compound 1b (Supplementary Fig. 7 A and B). L. acidophilus cells that were treated with 3b and CuAAC were then stained with WGA-561 and PG co-localization was observed (Fig. 3). These data imply that the other UDP derivatives isolated on scale (4b–6b) could be incorporated. We note that bacterial labeling was only achieved while bacteria were actively growing (Supplementary Fig 8A,B).
Figure 3. L. acidophilus labeling with azido UDP NAM 3b and WGA costaining.
DIC and 2D Maximum Intensity Projection images from super-resolution SIM Z-stacks L. acidophilus cells treated with 3b, clicked with Alk488 (green) and treated with WGA-561 (red) (scale bars, 10 μm). Images are representative of a minimum of three fields viewed per replicate with at least two technical replicates and the experiment was conducted in at three biological replicates.
As the growth conditions are challenging for L. acidophilus, other bacteria species were assayed for UDP NAM 2b labeling. S. aureus, B. subtilis and two strains of E. coli: E. coli DH5a and E. coli RMF795, an E. coli mutant strain with a semi-permeable outer membrane.61 S. aureus, B. subtilis, and the E. coli strains were treated with the azido UDP NAM 3b. SIM imaging revealed no fluorescent labeling after click incorporation of fluorophore as compared to the control cells which were incubated with the natural UDP NAM 1b (Supplementary Figure 9A–D). We hypothesize that a subset of species may be capable of UDP NAM uptake or tolerant to modification on NAM residues. These probes could help define which bacteria have this capability.
Discussion
Traditionally, the synthesis of UDP and its sugar cargos has been extremely demanding, and therefore underexplored. As UDP sugars are critical for nearly every step of the biosynthesis of bacterial PG, access to these phosphate intermediates is valuable to understanding the vitality of this polymer. The modular synthesis of 2-N functionalized NAMs allowed for the development of a library of NAM derivatives to include substrates beyond the azide and alkyne compounds (3, 4). We chose to construct the library at the 2-N-postion (Scheme 1) as modification at the 6 position was not tolerated by AmgK (Table 1, entry 1). We have expanded the NAM probes to include derivatives with capabilities of Staudinger ligations, IEDDA, photo activating, affinity tagging, as well as fluorine NMR spectroscopy analysis (Scheme 2). With these derivatives in hand, we were able to assess the substrate specificity and kinetic profile of the recycling enzyme AmgK, a key enzyme in the production line of uridine diphosphate NAM sugars (Table 1 and Table 2). AmgK appears to tolerate modifications at the 2 and 3 positions of the carbohydrate, converting modifications such as levulinic acid, biotin, tetrazine, and diazirine to the respective phosphor-sugars. Compounds that lacked the acetyl (Table 1, entry 8) or displayed a quaternary center alpha to the carbonyl of the amide (11–13) were not accepted by the enzyme (Table 1). As there is no crystal structure for this kinase, we propose that AmgK “reads” the carbonyl state of the NAM. Although AmgK was able to turnover substrates containing atomistically large modifications at the 2-position, kinetic characterization revealed that not all modifications were well tolerated. Bulky modification to the 2-position resulted in a ~100-fold decrease in the catalytic efficiency of the enzyme (5–9, Table 2). Yet, modification at the 3-position was comparable to the natural substrate (Table 2), suggesting that the enzyme is more tolerant to modification at this position. Further experiments are necessary to test the steric limitations of this position on the carbohydrate.
We further explored the chemoenzymatic synthesis of UDP NAM derivatives, of which until this work were previously limited through synthetic methodology and no UDP NAM derivatives with modifications on the carbohydrate have been reported. We optimized the purification and compound characterization of these UDP derivatives and further explored their utility in the conversion to the pentapeptide intermediates of PG biosynthesis known as Park’s nucleotide (Scheme 1). Substrates that AmgK converted to the mono-phosphate were next used to assess substrate specificity for the other PG recycling enzyme, MurU, and the four PG biosynthetic enzymes (MurC-F) (Scheme 1). The data show that all modifications tolerated by AmgK were accepted by the subsequent enzymes, except the biotin modifications, 9 and 10, which was not turned over by MurU (Table 1, Supporting Information Table 1). This is the first report of a large collection of Park’s nucleotide derivatives and will be valuable probes in studying bacterial cell wall biosynthesis in a variety of organisms.
Other types of bio-orthogonal chemistry were assayed; the tetrazine NAM derivative 8 was subjected to the whole cell labeling methodology in which genetically engineered E. coli cells containing the recycling enzymes AmgK and MurU were subjected to IEDDA with TCO TAMRA. While few cells were labeled using this methodology, the ones that were fluorescent have different morphologies (Fig 2) with what appears to be a thinner cell wall compared to the healthier cells in the field of view. While it appears that the smaller bioorthonoal functionality is more desirable to study global whole cell NAM labeling, the tetrazine modification upon optimization could prove useful in studying live-cell UDP NAM production and shuttling.
In order to test if the PG labeling method was amenable to modification at the 3-postion of the NAM, a new synthesis of a modified 3-muramic acid was developed. The synthesis of the 3-azido NAM derivative 14 was designed and optimized to correctly set the muramic acid stereochemistry while incorporating the unnatural bioorthogonal azide handle (Scheme 3). Biochemical assays indicated that both the recycling and early PG biosynthetic enzymes are permissive to the azide modification (Table 1, 2 and Supporting Table 1). When 14 was used in the whole cell labeling of E. coli QKU cells, labeling was concentrated to the PG septal division ring (Figure 1, Supplementary Fig 1). A small population of cells appears to be labeled beyond the septal division ring (Figure 1, top row white arrow). This preliminary observation suggests that the modification at the lactic acid is incorporated into lipid linked PG intermediates62 and/or new PG but may fail to incorporate effectively into the growing PG polymer, suggesting that different transglycosylases have different substrate preferences (Fig 1 and Supplemental Fig 1). Further detailed mass spectrometry analysis is necessary to quantify the pools of PG that are modified. 14 and derivatives thereof will be extremely valuable in studying the PG of bacteria that are known to modify the 2-position, such as Mtb, which is known to N-glycolylate this position to evade an immune response49, 63–66. NAM derivatives containing modification at the 3-position could be used to probe this resistance mechanism.
This work expands and improves the methodology to label bacterial PG at the NAM residue. Previously, this methodology was limited to the genetic manipulation of organisms to include the recycling enzymes amgK and murU as well as lengthy synthesis of UDP peptide probes. In order to eliminate this genetic editing step and make the synthesis scalable and accessible, we took advantage of the chemoenzymatic production of the azide UDP NAM derivatives and developed the methodology to produce these analogues. PG co-labeling was only achieved in L. acidophilus as observed by superresolution microscopy (Figure 3). The result complements the previous work of Nishimura and coworkers where fluorescein was synthetically attached to the lysine of UDP-NAM pentapeptide to label the PG of L. acidophilus.59 With the application of superresolution microscopy, new details about the localization of the NAM residues in L. acidophilus along the PG-division ring and side-wall have been observed (Fig. 3).
It is worth noting, that the growth conditions of the bacterial cells resulted in different labeling efficiencies for L. acidophilus (Supplemental Figure 8A,B). L. acidophilus is a facultative anaerobe and is notoriously difficult to grow under laboratory conditions. We qualitatively note that when cells were actively growing, labeling efficiency of the PG appeared to be higher than when cells were dormant (Supporting Information, Supplementary Figure 8A,B), indicating that the cells may have been under different stresses, and those that were more actively growing and dividing are more likely to uptake the UDP NAM derivative. We note that these NAM probes may be used to explore bacterial nucleotide sugar transportation. To date, there are not any known UDP-NAM transporters that could allow for this event to occur. However, transporters for UDP galactose (UGT), UDP NAG, GDP fucose (GFT) and CMP sialic acid (CST) exist.67 The identification of UDP NAM sugar transporters could be potential sources for selective antibiotic targets.
In conclusion, we have developed two critical synthetic methods: chemoenzymatic synthesis of UDP-NAM sugar derivatives and modification chemistry for the 3-position of the NAM lactic acid. These syntheses were used to explore the substrate specificity of the PG recycling and biosynthetic enzymes and revealed relaxed specificity at the 2 and 3 positions of the NAM. These probes allowed for new biological applications of the PG-glycan-labeling method to be developed: bioorthogonal tetrazine ligation and lactic acid modification. The robust chemoenzymatic synthesis of the 2-azido UDP NAM derivative allowed a Gram-positive commensal to be labeled on the sugar backbone at unprecedented resolution. The NAM-PG labeling method and the expanded substrate scope will be used in tandem with other complementary bioorthogonal labeling strategies55–56, 59, 68–78 to illuminate fundamental aspects of PG biosynthesis and immune processing. These tools will allow the study of NAM production and breakdown of a variety of pathogen and commensal bacteria to reveal novel antibiotic targets and immune recognition elements.
Supplementary Material
Acknowledgments
We thank Professor Joseph Fox and the Fox Laboratory, specifically Yi Li, the Boyd Laboratory at the University of Delaware, specifically Nathan McDonald. We thank Professor Burnaby Munsen at the University of Delaware. We thank Dr. Papa Nii Asare-Okai, the director of the MS facility as well as Dr. Yang in the Chemistry and Biochemistry Department at the University of Delaware, with assistance in MS. We thank Dr. Jeffrey Caplan, the Director of Bioimaging and the Delaware Biotechnology Institute for assistance with superresolution microscopy. For financial support, this project was supported by the NIH U01 Common Fund program with grant number U01CA221230-01; Delaware COBRE program, with a grant from the National Institute of General Medical Sciences-NIGMS P20GM104316-01A1. C.L.G. is a Pew Biomedical Scholar and Cottrell Scholar, and thanks the Pew Foundation and the Research Corporation for Science Advancement. K.E.D. and M.R.J thank the NIH for support through a CBI training grant: 5T32GM008550. For instrumentation support, the Delaware COBRE and INBRE programs supported this project with a grant from the National Institute of General Medical Sciences–NIGMS (5 P30 GM110758-02, P20GM104316-01A1 and P20 GM103446) from the National Institutes of Health.
ABBREVIATIONS
- AmgK
MurNAc/GlcNAc anomeric kinase
- CMP
cyotidine monophosphate
- CST
CMP sialic acid transporter
- CuAAC
copper catalyzed azide alkyne cycloaddition
- DNA
Deoxyribonucleic acid
- GDP
guanosine diphosphate
- GFT
GDP fucose transporter
- HPLC
High-Performance Liquid Chromatography
- HRMS
High resolution mass spectrometry
- IEDDA
Inverse electron demand diels al-der
- Mtb
Mycobacterium tuberculosis
- MurU
NAM α-1 phosphate uridylyl
- NAG
N-acetylglucosamine
- NAM
N-acetylmuramic acid
- PG
Peptidoglycan
- PMB
paramethoxybenzyl
- RNA
Ribonucleic acid
- SIM
Structured illumination microscopy
- TCO
trans-cyclooctene
- TCCA
trichloroisocyanuric acid
- TLC
thin layer chromatography
- UDP
Uridine diphosphate
- UGT
UDP galactose transporter
- WGA
Wheat Germ Agglutinin
Footnotes
The authors declare no competing financial interest.
AUTHOR CONTRIBUTION
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website. Supplementary Information includes supplementary figures and tables, biochemical methods, synthetic procedures and compound characterization and is available as a single PDF file.
References
- 1.Westheimer FH. Why nature chose phosphates. Science. 1987;235(4793):1173–8. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci. 2012;367(1602):2513–6. doi: 10.1098/rstb.2012.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou WK, Hinderlich S, Reutter W, Tanner ME. Sialic acid biosynthesis: stereochemistry and mechanism of the reaction catalyzed by the mammalian UDP-N-acetylglucosamine 2-epimerase. J Am Chem Soc. 2003;125(9):2455–61. doi: 10.1021/ja021309g. [DOI] [PubMed] [Google Scholar]
- 4.Cori GT, Cori CF. Glucose-6-phosphatase of the liver in glycogen storage disease. J Biol Chem. 1952;199(2):661–7. [PubMed] [Google Scholar]
- 5.Lomako J, Lomako WM, Kirkman BR, Whelan WJ. The role of phosphate in muscle glycogen. Biofactors. 1994;4(3–4):167–71. [PubMed] [Google Scholar]
- 6.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259(5):3308–17. [PubMed] [Google Scholar]
- 7.Warren L, Felsenfeld H. N-Acetylmannosamine-6-phosphate and N-acetylneuraminic acid-9-phosphate as intermediates in sialic acid biosynthesis. Biochem Biophys Res Commun. 1961;5:185–90. doi: 10.1016/0006-291x(61)90107-3. [DOI] [PubMed] [Google Scholar]
- 8.Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291(5512):2376–8. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 9.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62(1):181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strominger JL, Park JT, Thompson RE. Composition of the cell wall of Staphylococcus aureus: its relation to the mechanism of action of penicillin. J Biol Chem. 1959;234:3263–8. [PubMed] [Google Scholar]
- 11.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36(4):407–77. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strominger JL. Bacterial cell walls, innate immunity and immunoadjuvants. Nat Immunol. 2007;8(12):1269–71. doi: 10.1038/ni1207-1269. [DOI] [PubMed] [Google Scholar]
- 13.Rangan KJ, Pedicord VA, Wang YC, Kim B, Lu Y, Shaham S, Mucida D, Hang HC. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science. 2016;353(6306):1434–1437. doi: 10.1126/science.aaf3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–72. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 15.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278(8):5509–12. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 16.Wolf AJ, Underhill DM. Peptidoglycan recognition by the innate immune system. Nat Rev Immunol. 2018;18:243–254. doi: 10.1038/nri.2017.136. [DOI] [PubMed] [Google Scholar]
- 17.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535(7610):65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa M, Yang K, Hashimoto M, Park JH, Kim YG, Fujimoto Y, Nunez G, Fukase K, Inohara N. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem. 2006;281(39):29054–63. doi: 10.1074/jbc.M602638200. [DOI] [PubMed] [Google Scholar]
- 19.Humann J, Lenz LL. Bacterial peptidoglycan degrading enzymes and their impact on host muropeptide detection. J Innate Immun. 2009;1(2):88–97. doi: 10.1159/000181181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dziarski R. Recognition of bacterial peptidoglycan by the innate immune system. Cell Mol Life Sci. 2003;60(9):1793–804. doi: 10.1007/s00018-003-3019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep. 2001;18(5):503–19. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 22.Liang H, DeMeester KE, Hou CW, Parent MA, Caplan JL, Grimes CL. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat Commun. 2017;8:15015. doi: 10.1038/ncomms15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41(14):2596–9. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67(9):3057–64. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 26.Gisin J, Schneider A, Nagele B, Borisova M, Mayer C. A cell wall recycling shortcut that bypasses peptidoglycan de novo biosynthesis. Nat Chem Biol. 2013;9(8):491–3. doi: 10.1038/nchembio.1289. [DOI] [PubMed] [Google Scholar]
- 27.Patterson DM, Nazarova LA, Prescher JA. Finding the right (bioorthogonal) chemistry. ACS Chem Biol. 2014;9(3):592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 28.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287(5460):2007–10. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 29.Patterson DM, Nazarova LA, Xie B, Kamber DN, Prescher JA. Functionalized cyclopropenes as bioorthogonal chemical reporters. J Am Chem Soc. 2012;134(45):18638–43. doi: 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Seckute J, Cole CM, Devaraj NK. Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew Chem Int Ed Engl. 2012;51(30):7476–9. doi: 10.1002/anie.201202122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z, Xie X, Fox JM. Diastereoselective synthesis of methylenecyclopropanes from chiral cyclopropene derivatives. Angew Chem Int Ed Engl. 2006;45(24):3960–2. doi: 10.1002/anie.200600531. [DOI] [PubMed] [Google Scholar]
- 32.Blackman ML, Royzen M, Fox JM. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J Am Chem Soc. 2008;130(41):13518–9. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat Chem Biol. 2005;1(2):93–7. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka Y, Kohler JJ. Photoactivatable crosslinking sugars for capturing glycoprotein interactions. J Am Chem Soc. 2008;130(11):3278–9. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- 35.Feng L, Hong S, Rong J, You Q, Dai P, Huang R, Tan Y, Hong W, Xie C, Zhao J, Chen X. Bifunctional unnatural sialic acids for dual metabolic labeling of cell-surface sialylated glycans. J Am Chem Soc. 2013;135(25):9244–7. doi: 10.1021/ja402326z. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka Y, Bond MR, Kohler JJ. Photocrosslinkers illuminate interactions in living cells. Mol Biosyst. 2008;4(6):473–80. doi: 10.1039/b803218a. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Hao P, Li L, Tan CY, Cheng X, Chen GY, Sze SK, Shen HM, Yao SQ. Design and synthesis of minimalist terminal alkyne-containing diazirine photo-crosslinkers and their incorporation into kinase inhibitors for cell- and tissue-based proteome profiling. Angew Chem Int Ed Engl. 2013;52(33):8551–6. doi: 10.1002/anie.201300683. [DOI] [PubMed] [Google Scholar]
- 38.Kumasaka R, Kikuchi A, Yagi M. Photoexcited states of UV absorbers, benzophenone derivatives. Photochem Photobiol. 2014;90(4):727–33. doi: 10.1111/php.12257. [DOI] [PubMed] [Google Scholar]
- 39.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods. 2005;2(5):377–84. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 40.Morin B, Cadet J. Type I benzophenone-mediated nucleophilic reaction of 5′-amino-2′,5′-dideoxyguanosine. A model system for the investigation of photosensitized formation of DNA-protein cross-links. Chem Res Toxicol. 1995;8(5):792–9. doi: 10.1021/tx00047a020. [DOI] [PubMed] [Google Scholar]
- 41.Okaru AO, Brunner TS, Ackermann SM, Kuballa T, Walch SG, Kohl-Himmelseher M, Lachenmeier DW. Application of (19)F NMR Spectroscopy for Content Determination of Fluorinated Pharmaceuticals. J Anal Methods Chem. 2017;2017:9206297. doi: 10.1155/2017/9206297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martino R, Gilard V, Desmoulin F, Malet-Martino M. Interest of fluorine-19 nuclear magnetic resonance spectroscopy in the detection, identification and quantification of metabolites of anticancer and antifungal fluoropyrimidine drugs in human biofluids. Chemotherapy. 2006;52(5):215–9. doi: 10.1159/000094744. [DOI] [PubMed] [Google Scholar]
- 43.Dalvit C, Fagerness PE, Hadden DT, Sarver RW, Stockman BJ. Fluorine-NMR experiments for high-throughput screening: theoretical aspects, practical considerations, and range of applicability. J Am Chem Soc. 2003;125(25):7696–703. doi: 10.1021/ja034646d. [DOI] [PubMed] [Google Scholar]
- 44.Sano T, Vajda S, Cantor CR. Genetic engineering of streptavidin, a versatile affinity tag. J Chromatogr B Biomed Sci Appl. 1998;715(1):85–91. doi: 10.1016/s0378-4347(98)00316-8. [DOI] [PubMed] [Google Scholar]
- 45.Stayton PS, Freitag S, Klumb LA, Chilkoti A, Chu V, Penzotti JE, To R, Hyre D, Le Trong I, Lybrand TP, Stenkamp RE. Streptavidin-biotin binding energetics. Biomol Eng. 1999;16(1–4):39–44. doi: 10.1016/s1050-3862(99)00042-x. [DOI] [PubMed] [Google Scholar]
- 46.Laitinen OH, Hytonen VP, Nordlund HR, Kulomaa MS. Genetically engineered avidins and streptavidins. Cell Mol Life Sci. 2006;63(24):2992–3017. doi: 10.1007/s00018-006-6288-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reith J, Berking A, Mayer C. Characterization of an N-acetylmuramic acid/N-acetylglucosamine kinase of Clostridium acetobutylicum. J Bacteriol. 2011;193(19):5386–92. doi: 10.1128/JB.05514-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renner-Schneck M, Hinderberger I, Gisin J, Exner T, Mayer C, Stehle T. Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens. J Biol Chem. 2015;290(17):10804–13. doi: 10.1074/jbc.M114.620989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raymond JB, Mahapatra S, Crick DC, Pavelka MS., Jr Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J Biol Chem. 2005;280(1):326–33. doi: 10.1074/jbc.M411006200. [DOI] [PubMed] [Google Scholar]
- 50.Hansen JM, Golchin SA, Veyrier FJ, Domenech P, Boneca IG, Azad AK, Rajaram MV, Schlesinger LS, Divangahi M, Reed MB, Behr MA. N-glycolylated peptidoglycan contributes to the immunogenicity but not pathogenicity of Mycobacterium tuberculosis. J Infect Dis. 2014;209(7):1045–54. doi: 10.1093/infdis/jit622. [DOI] [PubMed] [Google Scholar]
- 51.Basu N, Maity SK, Chaudhury A, Ghosh R. Trichloroisocyanuric acid (TCCA): an efficient green reagent for activation of thioglycosides toward hydrolysis. Carbohydr Res. 2013;369:10–3. doi: 10.1016/j.carres.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Uehara T, Suefuji K, Jaeger T, Mayer C, Park JT. MurQ Etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. J Bacteriol. 2006;188(4):1660–2. doi: 10.1128/JB.188.4.1660-1662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94(12):4957–70. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devaraj NK, Weissleder R, Hilderbrand SA. Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjug Chem. 2008;19(12):2297–9. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. 2014;506(7489):507–10. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013;8(3):500–5. doi: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida M, Stadler J, Bertholdt G, Gerisch G. Wheat germ agglutinin binds to the contact site A glycoprotein of Dictyostelium discoideum and inhibits EDTA-stable cell adhesion. EMBO J. 1984;3(11):2663–70. doi: 10.1002/j.1460-2075.1984.tb02191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, Huang KC. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci U S A. 2014;111(11):E1025–34. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sadamoto R, Niikura K, Sears PS, Liu H, Wong CH, Suksomcheep A, Tomita F, Monde K, Nishimura S. Cell-wall engineering of living bacteria. J Am Chem Soc. 2002;124(31):9018–9. doi: 10.1021/ja026133x. [DOI] [PubMed] [Google Scholar]
- 60.Jiang Y, Lu X, Man C, Han L, Shan Y, Qu X, Liu Y, Yang S, Xue Y, Zhang Y. Lactobacillus acidophilus induces cytokine and chemokine production via NF-kappaB and p38 mitogen-activated protein kinase signaling pathways in intestinal epithelial cells. Clin Vaccine Immunol. 2012;19(4):603–8. doi: 10.1128/CVI.05617-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122(3):491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiao Y, Srisuknimit V, Rubino F, Schaefer K, Ruiz N, Walker S, Kahne D. Lipid II overproduction allows direct assay of transpeptidase inhibition by beta-lactams. Nat Chem Biol. 2017;13(7):793–798. doi: 10.1038/nchembio.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sansonetti PJ, Di Santo JP. Debugging how bacteria manipulate the immune response. Immunity. 2007;26(2):149–61. doi: 10.1016/j.immuni.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 64.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55(3):778–87. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 65.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32(2):287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 66.Coulombe F, Divangahi M, Veyrier F, de Leseleuc L, Gleason JL, Yang Y, Kelliher MA, Pandey AK, Sassetti CM, Reed MB, Behr MA. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J Exp Med. 2009;206(8):1709–16. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hadley B, Maggioni A, Ashikov A, Day CJ, Haselhorst T, Tiralongo J. Structure and function of nucleotide sugar transporters: Current progress. Comput Struct Biotechnol J. 2014;10(16):23–32. doi: 10.1016/j.csbj.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113(6):767–76. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 69.Gale RT, Brown ED. New chemical tools to probe cell wall biosynthesis in bacteria. Curr Opin Microbiol. 2015;27:69–77. doi: 10.1016/j.mib.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 70.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333(6039):222–5. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kocaoglu O, Calvo RA, Sham LT, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem Biol. 2012;7(10):1746–53. doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51(50):12519–23. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc. 2015;10(1):33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nelson JW, Chamessian AG, McEnaney PJ, Murelli RP, Kazmierczak BI, Spiegel DA. A biosynthetic strategy for re-engineering the Staphylococcus aureus cell wall with non-native small molecules. ACS Chem Biol. 2010;5(12):1147–55. doi: 10.1021/cb100195d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sadamoto R, Matsubayashi T, Shimizu M, Ueda T, Koshida S, Koda T, Nishimura S. Bacterial surface engineering utilizing glucosamine phosphate derivatives as cell wall precursor surrogates. Chemistry. 2008;14(33):10192–5. doi: 10.1002/chem.200801734. [DOI] [PubMed] [Google Scholar]
- 76.Shieh P, Siegrist MS, Cullen AJ, Bertozzi CR. Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc Natl Acad Sci U S A. 2014;111(15):5456–61. doi: 10.1073/pnas.1322727111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc Natl Acad Sci U S A. 2006;103(29):11033–8. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lebar MD, May JM, Meeske AJ, Leiman SA, Lupoli TJ, Tsukamoto H, Losick R, Rudner DZ, Walker S, Kahne D. Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. J Am Chem Soc. 2014;136(31):10874–7. doi: 10.1021/ja505668f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.