Significance
Eukaryotic cells depend on precise genome organization within the nucleus to maintain an appropriate gene-expression profile. Critical to this process is the packaging of functional domains of open and closed chromatin to specific regions of the nucleus, but how this is regulated remains unclear. In this study, we show that the zinc finger protein Casz1 regulates higher-order nuclear organization of rod photoreceptors in the mouse retina by repressing nuclear lamina function, which leads to central localization of heterochromatin. Loss of Casz1 in rods leads to an abnormal transcriptional profile followed by degeneration. These results identify Casz1 as a regulator of higher-order genome organization.
Keywords: chromatin, photoreceptors, mouse, retina, neurodegeneration
Abstract
Genome organization plays a fundamental role in the gene-expression programs of numerous cell types, but determinants of higher-order genome organization are poorly understood. In the developing mouse retina, rod photoreceptors represent a good model to study this question. They undergo a process called “chromatin inversion” during differentiation, in which, as opposed to classic nuclear organization, heterochromatin becomes localized to the center of the nucleus and euchromatin is restricted to the periphery. While previous studies showed that the lamin B receptor participates in this process, the molecular mechanisms regulating lamina function during differentiation remain elusive. Here, using conditional genetics, we show that the zinc finger transcription factor Casz1 is required to establish and maintain the inverted chromatin organization of rod photoreceptors and to safeguard their gene-expression profile and long-term survival. At the mechanistic level, we show that Casz1 interacts with the polycomb repressor complex in a splice variant-specific manner and that both are required to suppress the expression of the nuclear envelope intermediate filament lamin A/C in rods. Lamin A is in turn sufficient to regulate heterochromatin organization and nuclear position. Furthermore, we show that Casz1 is sufficient to expand and centralize the heterochromatin of fibroblasts, suggesting a general role for Casz1 in nuclear organization. Together, these data support a model in which Casz1 cooperates with polycomb to control rod genome organization, in part by silencing lamin A/C.
The mammalian nucleus must compartmentalize a genome measuring about 2 m in length into a structure that is less than 600 µm3 in volume (1, 2). To maintain genome organization, chromatin is packaged into functional domains organized according to a variety of mechanisms. At the highest level, the genome is partitioned into active A and inactive B compartments that exhibit segregated higher-order looping interactions that can be further subdivided (3, 4). Topologically associated domains (TADs), similarly divide chromosomes into megabase-scale units with internally restricted chromosome folding (5). TADs are also highly stable among different cell types. Structures such as the nuclear lamina, nucleolus, and nuclear pores additionally associate with chromatin and modify its activity. Epigenetic modifications also partition the genome into a hierarchy of euchromatin and heterochromatin types with differing levels of accessibility and compaction. While it has long been known that these factors cooperate to control the genome, it remains challenging to dissect how cells modify this organization and, in turn, how global changes relate to different transcriptomic states and cellular functions.
The developing mouse retina provides an advantageous model system in which to address these issues. Upon differentiation, rod photoreceptors undergo a process called “chromatin inversion” in which heterochromatin becomes localized to the center of the nucleus and the euchromatin to the periphery (6, 7), which is the opposite of the classic nuclear organization observed in most eukaryotic cells. Chromatin inversion was proposed as an adaptation to nocturnal vision that reduces light scattering in the retina (7). While previous studies have provided a descriptive framework for understanding rod genome organization, determinants of this organization remain largely elusive, with only the lamin B receptor (Lbr) identified so far (8).
Here, we report that Casz1 is a determinant of rod photoreceptor nuclear organization. Casz1 is a zinc finger transcription factor required for both heart and vascular development, and its germline inactivation causes embryonic lethality (9–11). Casz1 is orthologous to the Drosophila melanogaster gene castor (12–14). In flies, castor participates in shaping the lineages of most neuroblasts (stem cells) of the central nervous system (15–19) and appears to act exclusively as a transcriptional repressor (17, 20). Castor is also widely expressed in postmitotic neurons in the fly, but its role in neurons has not been established.
We have previously shown that, similar to castor, Casz1 participates in controlling the temporal output of retinal progenitor cells in the mouse (21). Casz1 expression in retinal progenitor cells increases as development proceeds, and we found that Casz1 has a role in promoting rod production from these progenitors. Intriguingly, Casz1 remains expressed in rods and cones upon differentiation, suggesting that it might have a functional role in photoreceptors. Accordingly, we found that genetic ablation of Casz1 in retinal progenitors led to the formation of retinas that subsequently degenerated over a period of ∼8–12 mo (21), but it remained unclear whether this was due to a role of Casz1 in photoreceptors or was simply a consequence of Casz1 inactivation in progenitors.
To distinguish between these possibilities, we conditionally ablated Casz1 specifically in maturing rod photoreceptors. We show that this leads to a similarly slow retinal degeneration, demonstrating that Casz1 is required to maintain long-term survival of differentiated rods. Importantly, we find that Casz1 is necessary and sufficient to control rod photoreceptor nuclear organization. At the mechanistic level, Casz1 is required to oppose the function of the nuclear lamina and acts, at least in part, by suppressing lamin A/C expression. Our data suggest a role for Casz1 in maintaining the organization of the rod photoreceptor genome, thereby safeguarding the rod transcriptome.
Results
Casz1 Is a Nuclear Protein in Mouse Photoreceptors.
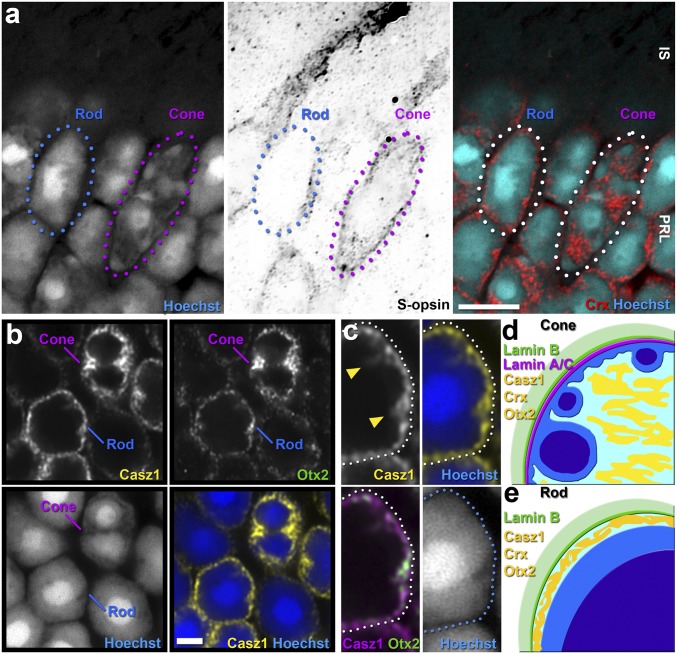
We and others have previously reported that Casz1 mRNA and protein are expressed in differentiating and mature rod and cone photoreceptors within the murine and Xenopus retina (21–23). More recently, it was suggested that Casz1 protein localization in photoreceptors is predominantly cytoplasmic, as the protein was observed to encircle the nuclei of murine rods (24). However, this interpretation did not take into account the unusual inverted chromatin organization of rod photoreceptors found in many nocturnal animals (7), in which the euchromatin is located in a thin ring just beneath the nuclear lamina (Fig. 1A).
Fig. 1.
Casz1 is a nuclear protein in photoreceptors. (A) Airyscan confocal images of mature rod and cone photoreceptors distinguished by staining for the cone photopigment S-opsin. (B) Airyscan confocal images of rods and cones costained for Casz1 or Otx2. (Scale bars: A, 5 μm; B, 2 μm.) (C) Magnified (5,000×) view of a rod photoreceptor shown in B. (D and E) Schematic of cone (conventional) (D) and rod (inverted) (E) nuclear organization. IS, inner segments; PRL, photoreceptor layer. (Scale bars: A, 5 μm; B, 2 μm.)
To determine where Casz1 protein was localized within the photoreceptor nucleus, we performed superresolution confocal microscopy on adult retinas stained using antibodies that we previously validated on Casz1-KO tissue (Fig. 2 A and B) (21). We additionally costained for the homeodomain transcription factor Otx2, which plays a central role in photoreceptor gene expression and localizes to the euchromatin at the nuclear periphery (25, 26). We found that Casz1 colocalized with Otx2 and Hoechst in the nuclear periphery of rods (Fig. 1 B and C). Casz1 was expressed at slightly higher levels in cone photoreceptors, again colocalizing with Otx2 (Fig. 1B). We conclude that Casz1 is enriched in the euchromatic domain of mouse photoreceptor nuclei.
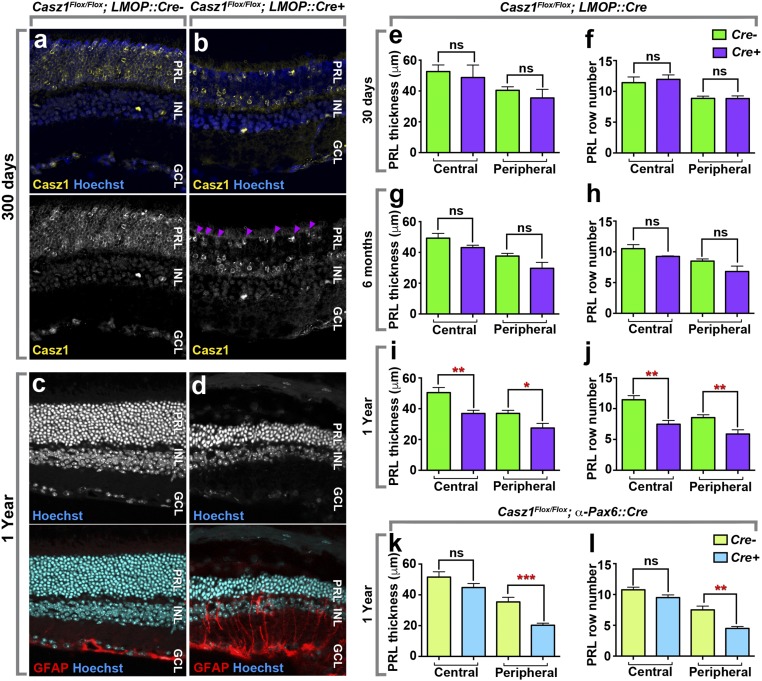
Fig. 2.
Genetic ablation of Casz1 in rod photoreceptors leads to degeneration. (A and B) Casz1 immunohistochemical staining in adult Casz1Flox/Flox retinas (A) or Casz1 Rod-cKO (Casz1Flox/Flox; LMOP::Cre+) retinas (B). Loss of Casz1 protein is observed in most of the cells within the photoreceptor layer (PRL). However, cone photoreceptors retain protein expression as predicted (arrowheads). (C and D) Retinal sections from 1-y-old Casz1 Rod-cKO mice. Photoreceptor degeneration is shown by the thinning of the photoreceptor layer and obvious gliosis detected by the up-regulation of GFAP. (Magnification: A–D, 400×.) (E–L) Quantification of photoreceptor degeneration in Casz1 Rod-cKO or progenitor cKO (Casz1Flox/Flox; αPax6::Cre+) mice. (E–L) Loss of rod cells was measured by quantitating the thickness of the photoreceptor layer (E, G, I, and K) or by counting the number of rods found in columns spanning the apico–basal axis (F, H, J, and L), using defined regions of the central or peripheral retina (E–J). In Casz1 Rod-cKO mice, loss of rod cells was not detectable at 30 d or 6 mo but reached statistical significance at 1 y. As reported previously (21), rod degeneration was also observed in aged Casz1Flox/Flox; α-Pax6::Cre-cKO mice (K and L), in which Cre is expressed specifically in the peripheral retina during retinal development. ns, not significant. *P <0.05, **P < 0.01, ***P < 0.001. The full statistics are presented in SI Appendix, Table S1.
Casz1 Is Required for Long-Term Photoreceptor Survival.
We previously reported that genetic ablation of Casz1 in retinal progenitors led to developmental cell fate-specification defects, followed by photoreceptor degeneration after 8–12 mo (21). Since Casz1 was deleted in the progenitors that give rise to photoreceptors, this degeneration could have been a consequence of aberrant development or could reflect a distinct role for Casz1 in mature photoreceptor survival. To distinguish between these possibilities, we introduced a transgene driving Cre under the control of a Rhodopsin regulatory element (LMOP) (27) into our floxed Casz1 conditional mutant line (21, 28). Using recombination reporter alleles, we previously confirmed that the LMOP::Cre transgene is specifically expressed in rod photoreceptors beginning at around postnatal day (P)9 (27, 29). We harvested control and Casz1 rod-specific conditional knockout (Rod-cKO) retinas and confirmed via immunohistochemistry that Casz1 protein staining was lost specifically in rods (Fig. 2 A and B).
Next, we harvested retinas at various time points and examined them histologically. Similar to our observations after progenitor ablation, loss of Casz1 in differentiated rod photoreceptors caused a slow retinal degeneration characterized by reduced thickness of the photoreceptor layer and gliosis in 1-y-old animals (Fig. 2 C and D). To quantitate these results, we measured the thickness of the photoreceptor layer in microns and additionally counted the number of rows of nuclei within the photoreceptor layer. Both of these measures accurately reflect the overall photoreceptor cell number (30, 31). No significant change in photoreceptor cell layer thickness was detected in 1-mo-old and 6-mo-old animals (Fig. 2 E–H and SI Appendix, Table S1), but 12-mo-old Casz1 Rod-cKO mice exhibited a significant reduction in photoreceptor layer thickness (Fig. 2 I and J) that was quantitatively similar to the degeneration previously observed using the progenitor-specific driver (Fig. 2 K and L).
To understand how Casz1 modifies the transcriptome before degenerative death, we performed RNA sequencing (RNA-seq) on wild-type versus Casz1-mutant retinal cells at P2. The results of this experiment confirmed that photoreceptor gene expression was compromised in the absence of Casz1 before degeneration (SI Appendix, Fig. S1). Taken together, these data demonstrate that Casz1 safeguards photoreceptor cell gene expression and survival.
Casz1 Interacts with Polycomb Repressor Complex Proteins.
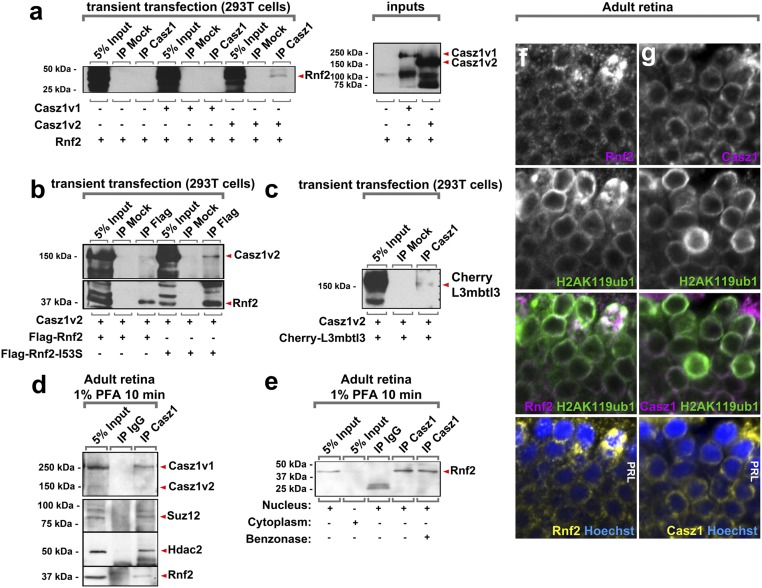
We next attempted to define a molecular pathway responsible for Casz1-dependent effects on cell survival. Previous proteomic work suggested that Casz1 proteins can associate with polycomb proteins, including Rnf2 (Ring1b) (32). Accordingly, we found that Casz1 coimmunoprecipitated with Rnf2 when expressed in 293T cells. However, only the shorter Casz1v2 (Casz1b) splice variant immunoprecipitated Rnf2; the longer Casz1v1 protein did not (Fig. 3A). Likewise, Casz1v2 could be reciprocally immunoprecipitated by Rnf2 (Fig. 3B). L3mbtl3, another protein present in complexes containing Casz1 and Rnf2 (32), was also immunoprecipitated when coexpressed with Casz1v2 (Fig. 3C).
Fig. 3.
Casz1 interacts with the polycomb repressor complex in rod photoreceptors. (A) Casz1 immunoprecipitations (IP) from cotransfected 293 cells contain Rnf2. Only the Casz1v2 immunoprecipitation contains Rnf2 protein. (B) Reverse immunoprecipitation of Casz1v2 using an antibody recognizing Rnf2. (C) Casz1 immunoprecipitations from cotransfected 293 cells contain L3mbtl3. (D) Casz1 immunoprecipitations generated via the RIME workflow (33) from adult retinal nuclear extracts contain the polycomb proteins Suz12 and Rnf2. (E) The interaction between Casz1 and Rnf2 is maintained when genomic DNA is degraded with benzonase. (F and G) Colocalization of Casz1, Rnf2, and the polycomb mark H2AK119ub1 in adult photoreceptors. (Magnification: F and G, 1,890×.)
To determine whether Casz1 interacts with polycomb proteins in vivo, we next immunoprecipitated Casz1 from adult retinas, where it is expressed almost exclusively in rods (Fig. 2 A and B). Despite robust mRNA expression, we found that full-length Casz1 isoforms could not be detected from retinal lysates using conventional methodology (data not shown), echoing difficulties previously encountered with immunohistochemistry (21). To assist the retrieval of Casz1 protein, we therefore performed cross-linking–assisted immunoprecipitation using the rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) proteomic workflow (33). Using this approach, Rnf2 was very robustly immunoprecipitated from retinal nuclear extracts using Casz1 antibodies (Fig. 3 D and E). Suz12, another polycomb protein associated with polycomb repressive complex 2 (PRC2), was also immunoprecipitated by Casz1 (Fig. 3D). The Mi2/Nurd effector protein Hdac2 had been repeatedly shown to interact with Casz1-containing protein complexes (32, 34), and we accordingly observed that retinal Casz1 immunoprecipitated Hdac2 (Fig. 3D). Additionally, the Casz1/Rnf2 interaction was not altered when DNA was degraded using benzonase, suggesting that the proteins were not tethered together by DNA (Fig. 3E). Finally, we examined the expression of Rnf2 and its target histone modification H2AK119ub1 in adult retinas using antibodies previously validated on mutant cells (35, 36). We found that both Rnf2 and H2AK119ub1 colocalized with Casz1 in the nuclear periphery of rods (Fig. 3 F and G). We conclude that the Casz1v2 isoform forms complexes with transcriptional corepressors, including polycomb.
Casz1 Regulates Nuclear Lamina Function.
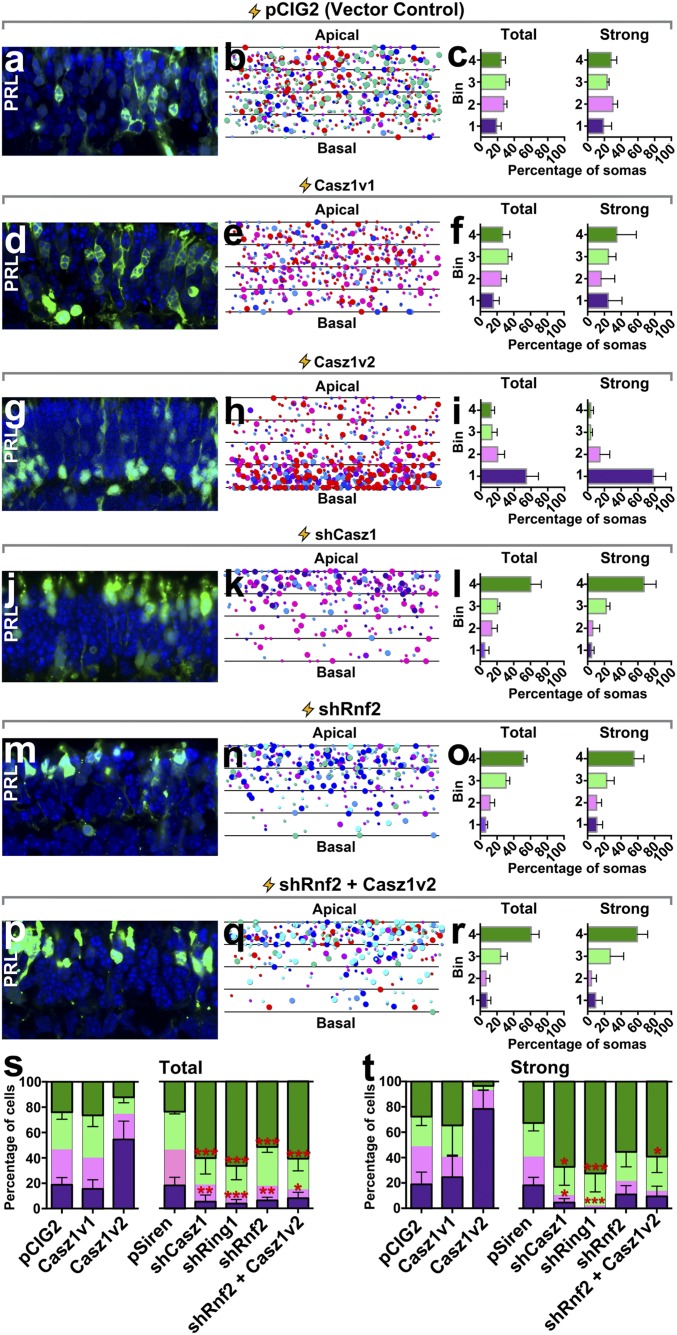
To examine the functional effects of Casz1 on rod photoreceptors, we first overexpressed both Casz1v1 and Casz1v2 splice forms in rods by electroporating postnatal retinal explants with the pCIG2 expression vector, which allows us to mark all transfected cells simultaneously via an IRES2-EGFP cassette. Explants were allowed to develop for 2 wk, and the resultant rods were examined in retinal sections. We noted striking effects of Casz1v2 on rods, with transfected cells exhibiting a clear bias to localize their nuclei to the basal side of the photoreceptor layer, adjacent to the outer plexiform layer (Fig. 4 G–I, S, and T and SI Appendix, Table S2). Opposite effects were observed when RNAi was used to knock down Casz1 or polycomb proteins such as Ring1a or Rnf2, where transfected cell somas migrated to the apical side of the layer (Fig. 4 J–O, S, and T). Moreover, the effect of Casz1v2 overexpression was blocked by knocking down Rnf2, indicating that Casz1v2 depends on Rnf2 to control rod nuclear position (Fig. 4 P–T).
Fig. 4.
Casz1 and polycomb control rod nuclear position. P0/P1 retinal explants were transfected with the indicated constructs and cultured for 2 wk to allow rods to be produced. Explants were then harvested, sectioned, and imaged (A, D, G, J, M, and P; magnification: 630×). Each photoreceptor layer was divided into four equal-sized bins, and the centroid position of each rod nucleus was plotted (B, E, H, K, N, and Q). Rods were considered strongly transfected (strong) if they exhibited pixel saturation under the imaging conditions. These cells are indicated by larger circles in the plots; weaker transfected cells are indicated by smaller circles. Colors correspond to different experimental replicates. The proportion of total or strongly transfected rod nuclei within each of the four bins was then obtained (C, F, I, L, O, and R). (S and T) Comparison of total (S) or strongly transfected (T) rods. The indicated statistical inferences reflect multivariate comparisons via one-way ANOVA and Tukey’s post hoc test for a given bin between all other experimental treatments. Bins 2 and 3 were not analyzed statistically. pCIG2, Casz1v1, and Casz1v2 were not included in the ANOVA and are reproduced here only for visual comparison (Fig. 7). *P < 0.05; **P < 0.01; ***P < 0.001, significantly different versus control.
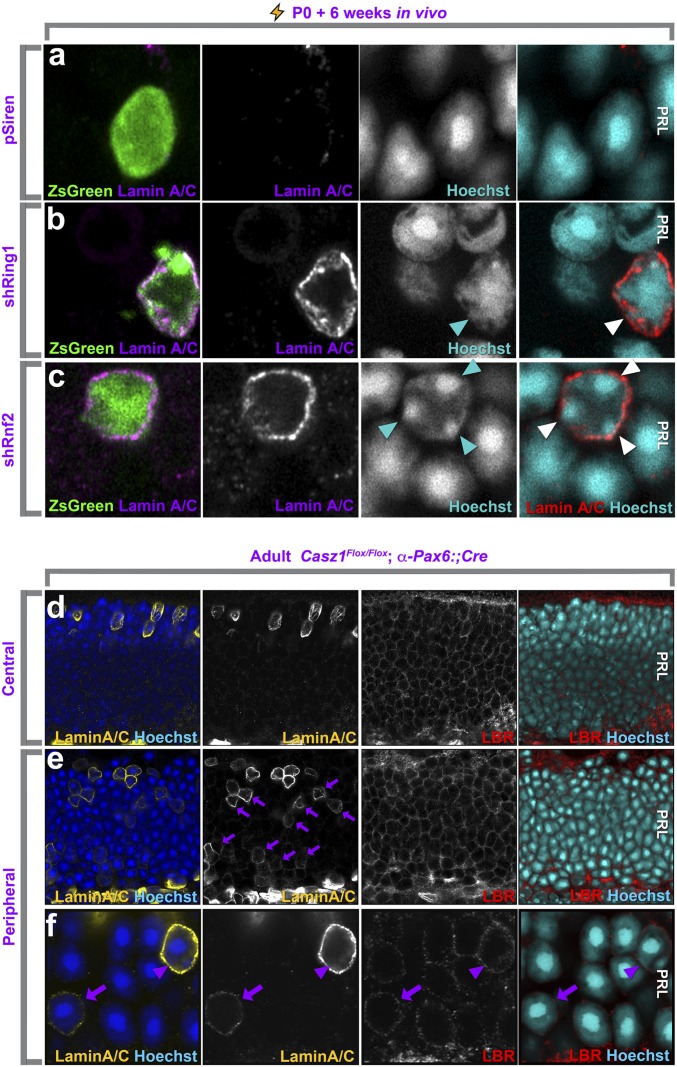
It is well known that the apico–basal nuclear position in the photoreceptor layer is regulated by the nuclear lamina. Basal migration of photoreceptor nuclei is associated with disruption of lamina components such as laminB2, Sun1/2, Syne2, and associated dynein motors (37–42). We thus predicted that Casz1 might affect the expression of lamina proteins. As expected, whereas rods transfected with an empty RNAi control vector exhibited no lamin A/C expression and no alteration in Lbr levels (Fig. 5A and SI Appendix, Fig. S2A), rods transfected with shRNA constructs targeting Ring1, Rnf2, or Casz1 up-regulated lamin A/C (Fig. 5 B and C and SI Appendix, Fig. S2 B and C). Importantly, no cones were targeted by postnatal transfections, in accordance with previous reports, due to the exclusively embryonic temporal window for cone generation (SI Appendix, Fig. S3A) (43, 44).
Fig. 5.
Up-regulation of lamin A/C is associated with polycomb or Casz1 loss of function. (A–C) Airyscan confocal images of retinal explants transfected with control constructs or shRNA constructs targeting Ring1 or Rnf2. Arrowheads mark heterochromatin contacting the nuclear lamina specifically in transfected cells. (D–F) Airyscan confocal analysis of lamin A/C or Lbr expression in Casz1-cKO retinas generated using the α-Pax6::Cre driver. The central retina does not express Cre during development and serves as an internal control (D). (E and F) Arrows mark lamin A/C up-regulation in basally located rod nuclei. The arrowhead in F marks an apically localized cone nucleus, which normally expresses lamin A/C. (Magnification: A–C, 6,300×; D and E, 945×; F, 3,150×.)
To confirm that the observed up-regulation of lamin A/C was due to specific knockdown of Ring1 or Rnf2 rather than off-target effects, we overexpressed Mysm1, which deubiquitinates H2AK119 in opposition to Ring1 and Rnf2 (45). Accordingly, Mysm1 likewise led to the up-regulation of lamin A/C expression (SI Appendix, Fig. S2D). We also confirmed that the apical nuclear position phenotype triggered by shRnf2 could be rescued by coexpressing it with a human Rnf2 construct that was mismatched with the shRNA hairpin due to evolutionary divergence (SI Appendix, Fig. S4).
We next examined Casz1-cKO rods generated using the retinal progenitor α–Pax6::Cre driver (46). Within the central retina, where the Cre driver is generally not expressed during development, we found that, as previously reported (8, 47), lamin A/C expression was confined to cones, which localize apically in the photoreceptor layer (Fig. 5D and SI Appendix, Fig. S5). Conversely, many more photoreceptors expressed lamin A/C in the peripheral retina (Fig. 5 E and F), where the Cre driver is most highly expressed during development (46). Many of these cells were very clearly rods, based on their nuclear morphology and more basal nuclear position (Fig. 5 E and F). Lamin A/C+ rods also lacked expression of cone markers (SI Appendix, Fig. S3 B and C). Thus, loss of either Casz1 or its polycomb cofactors results in the ectopic up-regulation of lamin A/C.
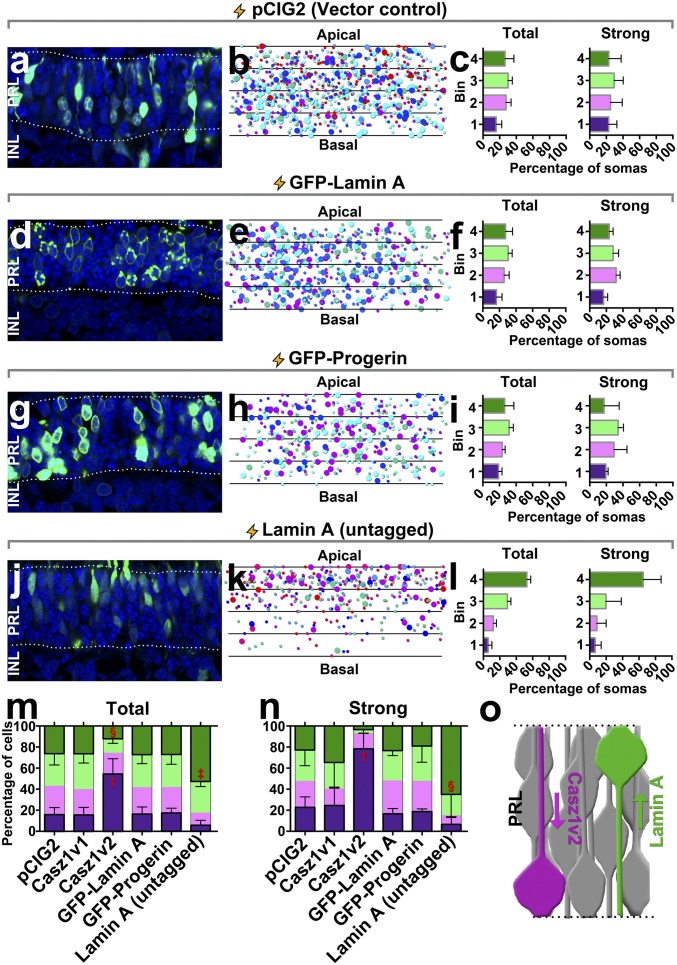
To determine whether lamin A/C was sufficient to explain the observed nuclear position effects, we overexpressed N-terminal GFP-fusion proteins encoding lamin A (prelamin A) and progerin, a lamin variant harboring a C-terminal truncation, but found that these constructs had no effect on nuclear position (Fig. 6 A–N and SI Appendix, Table S3). We additionally overexpressed a construct encoding an untagged version of lamin A in photoreceptors using explant electroporation. Surprisingly, we found that untagged lamin A was sufficient to force localization of photoreceptor soma to the apical side of the neural retina (Fig. 6 J–N). These results suggested that the N-terminal GFP tag was function blocking. Both tagged and untagged lamin A proteins localized correctly to the nuclear periphery, although the GFP-lamin A construct generated protein inclusions that were not apparent in cells transfected with the untagged version (SI Appendix, Fig. S6). We conclude that lamin A is sufficient to promote apical migration of photoreceptor nuclei.
Fig. 6.
Lamin A controls rod nuclear position. (A–L) P0 Retinal explants were transfected with the indicated constructs and cultured for 2 wk to allow rods to be produced. Explants were then harvested, sectioned, and imaged. Note that the EGFP signal in D and G is fused directly to lamin A, whereas the EGFP signal in A and J is coexpressed from an IRES cassette. Each photoreceptor layer was divided into four equal-sized bins, and the centroid position of each rod nucleus was plotted as detailed in Fig. 4. (Magnification: A, D, G, and J, 630×.) (M and N) Comparison of total (M) or strongly transfected (N) rods. The indicated statistical inferences reflect multivariate comparisons via one-way ANOVA and Tukey’s post hoc test for a given bin versus all other experimental treatments. Bins 2 and 3 were not analyzed statistically. †P < 0.0001, significantly different from all other treatments; ‡P < 0.001, significantly different from all other treatments; §P < 0.05, significantly different from all other treatments. (O) Model for the effects of Casz1 and lamin A on nuclear position. Casz1v2 opposes the function of the nuclear lamina, leading to basal localization of the rod nucleus. Lamin A promotes apical nuclear localization and is dominant over Casz1v2.
In addition to nuclear position, the lamina also regulates heterochromatin tethering (8). In the absence of functional lamina tethers such as Lbr, cells undergo chromatin inversion in which chromocenters relocalize to the center of the nucleus instead of maintaining their default localization at the nuclear envelope (7, 8). Previous work using transgenic mice had shown that lamin C was not sufficient to affect chromatin inversion, but the role of lamin A was not explored.
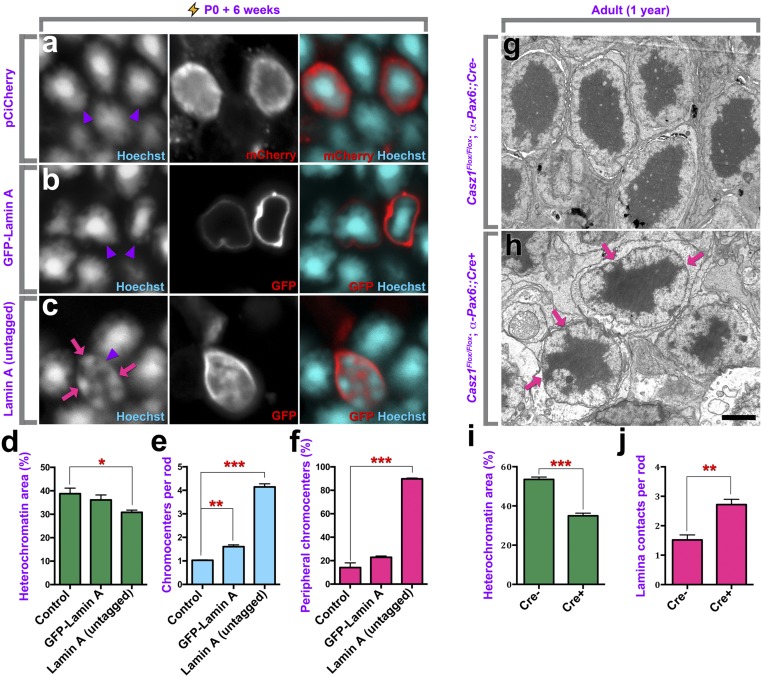
To assess the effects of lamin A on chromatin inversion, we electroporated retinas of P0 mice in vivo and harvested the tissues 6 wk later, when rod chromatin inversion is complete. Whereas control or GFP-Lamin A overexpression had little to no effect on chromatin inversion (Fig. 7 A–F), a striking blockade of chromatin inversion was observed in rods overexpressing untagged lamin A (Fig. 7 C–F). While the proportion of the nucleus occupied by heterochromatin was reduced by only ∼20% (Fig. 7D and SI Appendix, Table S4), both the number of chromocenters and the proportion of chromocenters contacting the nuclear lamina were significantly increased compared with control or GFP-lamin A (Fig. 7 E and F). These data show that lamin A is a powerful suppressor of chromatin inversion.
Fig. 7.
Lamin A/C is sufficient to block the inversion of rod nuclear architecture. (A–C) Retinas were transfected in vivo at P0, and rods were allowed to differentiate and mature over 6 wk. Retinas were then harvested, and nuclei were visualized via Hoechst staining. Arrowheads mark transfected cells. Note that GFP in B is fused directly to lamin A, whereas the mCherry/GFP signal in A and C is coexpressed from an IRES cassette. (Magnification: A–C, 1,890×.) (D–F) Quantitation of heterochromatin area (D), the number of chromocenters present per rod nucleus (E), and the percentage of chromocenters contacting the nuclear lamina (F). (G and H) Transmission electron micrographs of 1-y-old Casz1-cKO rods. Arrows mark aberrant lamina/heterochromatin contacts. (Scale bar, 2 μm.) (I and J) Quantitation of the percentage of the nuclear area occupied by heterochromatin (I) and the number of lamina/heterochromatin contacts per nucleus (J). *P < 0.05, **P < 0.01, ***P < 0.001, significantly different versus control; n = 3.
The strong effects elicited by lamin A overexpression led us to revisit Casz1-cKO rods, which up-regulate lamin A/C (Fig. 5 E and F and SI Appendix, Fig. S3 B and C). While increased chromocenter/lamina contacts had been observed in Casz1/polycomb RNAi electroporations (Fig. 5 B and C and SI Appendix, Figs. S2 and S3A), defects in Casz1-mutant rods were not easy to detect at adult stages in vivo using confocal or even Airyscan superresolution microscopy. The only discernable effect was that some mutant rod nuclei appeared to be slightly more disordered than in controls. However, this defect was much more apparent when rods were visualized with transmission electron microscopy (TEM). We examined Casz1-cKO rods from the peripheral retinas of Casz1Flox/Flox; α-Pax6::Cre-cKO animals, where Cre is expressed (46), and compared these with rods from the peripheral retinas of Cre− animals. We found that Casz1-cKO rods exhibited highly eccentric heterochromatin in comparison with littermate controls (Fig. 7 G and H). This manifested as a significant reduction in the overall proportion of the nucleus occupied by heterochromatin (Fig. 7I and SI Appendix, Table S4). Chromocenter/lamina contacts were also increased by approximately twofold in the Casz1-KO animals (Fig. 7J and SI Appendix, Table S4). Taken together, these results indicate that Casz1/polycomb function is required for lamin A/C repression in rods, thereby ensuring normal nuclear positioning and chromatin inversion.
Casz1 Is Sufficient to Induce Aspects of Photoreceptor Nuclear Organization in Fibroblasts.
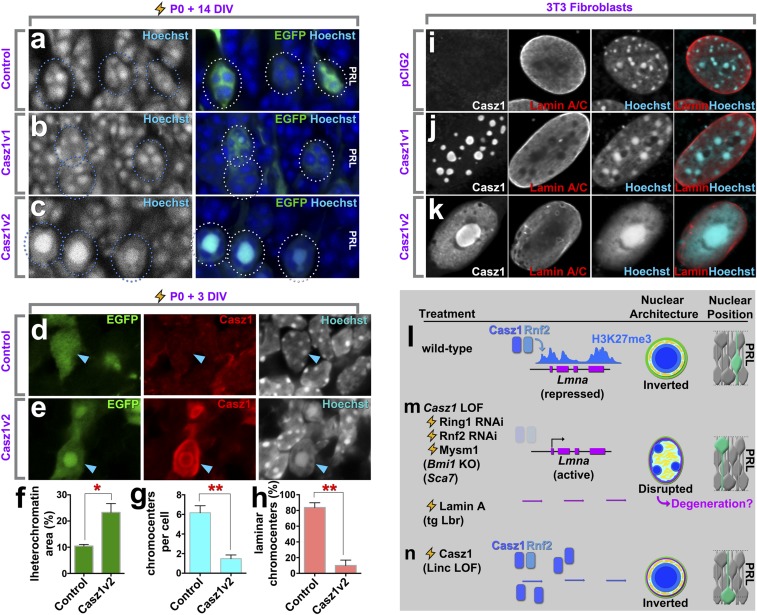
We next investigated whether Casz1 might contribute directly to chromatin inversion. We first misexpressed control, Casz1v1, or Casz1v2 in immature rods via explant electroporation at P0 and studied nuclear organization at P14, when all rods have been born but their chromatin inversion is not yet completed. While control and Casz1v1 transfections had no effect on rod nuclei organization, Casz1v2 dramatically accelerated chromatin inversion (Fig. 8 A–C). Next, we wondered whether Casz1 would be capable of reorganizing heterochromatin in nonphotoreceptor cells. We analyzed progenitor cells, which constitute the majority of transfected cells, 3 d after electroporation at P0. Progenitors overexpressing Casz1v2 exhibited centralized and enlarged chromocenters similar to those found in mature rods, whereas control transfections had no effect (Fig. 8 D and E). To determine whether these large central chromocenters represented the merging of preexisting heterochromatin or instead reflected de novo production, we performed quantitative microscopy. We found that progenitors overexpressing Casz1v2 had a decreased mean number of chromocenters per nucleus and a significantly larger nuclear area than control cells (Fig. 8 F and G and SI Appendix, Table S5). Finally, the percentage of chromocenters touching the nuclear periphery was significantly reduced in cells transfected with Casz1v2 compared with controls (Fig. 8H).
Fig. 8.
Casz1 is sufficient for aspects of inverted nuclear organization. Retinal explants were transfected at P0 and cultured for 14 d (A–C) or 3 d (D and E) in vitro with the indicated constructs. Transfected cells were identified by EGFP expression and analyzed for the expression of Casz1. DNA was stained with Hoechst dye. There is little endogenous Casz1 expression in retinal progenitors at ∼P4, as we previously reported (21). (F–H) Hoechst staining in transfected cells was quantitatively analyzed for the percentage of the nuclear area occupied by chromocenters (F), the average number of chromocenters per nucleus (G), and the percentage of chromocenters contacting the nuclear margin (H). *P < 0.05; **P < 0.001; n = 3 independent experiments. (I–K) 3T3 fibroblasts were transfected with the indicated constructs and harvested after 72 h. Cells were subjected to immunocytochemistry for Casz1 and lamin A/C, and nuclei were visualized Hoechst staining. (L–N) Summary of data and working model. (Magnification: A–E and I–K, 1890×.) (L) In rods, the Lmna gene is coated by the repressive polycomb mark H3K27me3 and is transcriptionally silent (SI Appendix, Fig. S4) (66, 67). Casz1 interacts with the polycomb protein Rnf2 and contributes to Lmna repression and thereby chromatin inversion. Nuclear position within the photoreceptor layer is randomized. (M) Casz1 or polycomb cKO or RNAi leads to the up-regulation of lamin A/C expression. Disruption of rod nuclear organization and degeneration also has been reported in mutants for the polycomb gene Bmi1 and for Sca7 (61, 73), although lamin A/C up-regulation has been reported only for the latter (8). Reintroduction of heterochromatic tethers such as lamin A or Lbr (8) disrupts the inverted nuclear architecture of rods. In the photoreceptor layer, rod nuclei relocalize to the apical side. (N) Casz1 overexpression is sufficient to reorganize the nuclei of heterologous cells even in the presence of lamin A/C. In the photoreceptor layer, rod nuclei localize to the basal side, although lamin A/C is not expressed in rods.
To determine whether Casz1 could reorganize chromatin in nonretinal cells, we overexpressed Casz1 constructs in NIH 3T3 fibroblasts. Overexpression of control or Casz1v1 constructs had little effect on nuclear organization, whereas Casz1v2 transfection led to an aggregation and expansion of chromocenters (Fig. 8 I–K). A proportion of cells exhibited nuclei that strikingly resembled those of mature rod nuclei, with a single chromocenter in the middle of the nuclei. In these cells, euchromatin became redistributed to the nuclear periphery, although euchromatic marks often remained depleted at the lamina (SI Appendix, Fig. S7). As lamin A/C is highly expressed in fibroblasts, we examined cells transfected with control or Casz1v2 constructs for the expression lamin A/C to ask whether nuclear reorganization correlated with effects on lamin A/C levels. We found that lamin A/C remained expressed in NIH 3T3 fibroblasts (Fig. 8K). We surmise that while Casz1 is required to silence lamin A/C expression in the retina, Casz1v2 is additionally sufficient to oppose lamina function independently of lamin A/C or Lbr transcriptional repression.
Discussion
Photoreceptor cells are among the most metabolically active cells in the body (48). Probably for this reason, rods and cones are highly sensitive to transcriptomic perturbations. Deficiency in the transcription factors required for activating the rod gene-expression program, such as Crx, Mef2c/d, Neurod1, and Otx2, is associated with photoreceptor cell death and retinal degenerative diseases such as retinitis pigmentosa and cone–rod dystrophy (49–54). Reduction of rod gene expression is associated with cell death in many experimental models, since as many as 50 photoreceptor genes must be expressed at high levels to maintain survival (55, 56).
Murine rod photoreceptors also maintain a highly specialized inverted nuclear organization in which euchromatin is located adjacent to the nuclear lamina and heterochromatin is greatly expanded and centralized (6, 7, 57, 58). Intriguingly, Crx and Nrl are also essential for establishing higher-order chromosome-looping configurations in both rods and cones (59), and mutants exhibit macroscopic nuclear disorganization in rods (60–63). While these transcription factors thus control higher-order genome organization, it seems unlikely that they act directly to control the striking reorganization (7) and expansion (57, 58) of heterochromatin observed during rod maturation, since both are prominently associated with euchromatin. Our data suggest instead that Casz1 contributes to heterochromatin organization and expansion in rod photoreceptors (Fig. 8 L–N).
Casz1 Safeguards Rod Gene Expression.
While a previous study had concluded that Casz1 protein was localized within the cytoplasm of photoreceptors (24), we suggest that this finding failed to take into account the unusual nuclear organization of rod photoreceptors. Examination of Casz1 immunohistochemistry using Airyscan confocal microscopy allowed us to resolve Casz1 localization below the Rayleigh diffraction limit, which revealed clear nuclear staining. Moreover, we showed independently that Casz1 protein was associated with transcriptional modifiers such as the polycomb repressor complex and that these cofactors were essential for Casz1-dependent functions. Finally, transcriptome analysis of Casz1-cKO cells confirms that rod gene expression is reduced as early as P2 (SI Appendix, Fig. S1), which likely contributes to the eventual photoreceptor degeneration observed in old retinas. Casz1 was localized to the euchromatin of rods and cones but is prominently localized to the margins of chromocenters in retinal progenitors (21) or in transfected 3T3 fibroblasts. Further experiments will be required to understand what regulates this shift in nuclear localization.
Lamin A Is Sufficient to Reorganize and Reposition the Rod Photoreceptor Nucleus.
To understand the effects of Casz1 on the rod photoreceptor nucleus, we examined known determinants of the nuclear lamina. We initially focused on Lbr, since it was the only determinant previously shown to be sufficient to reverse rod chromatin inversion (8). To our surprise, we observed no obvious effect of Casz1 on Lbr expression in rods; moreover, we found that weak Lbr expression persisted in adult stages. Indeed, the latter observation was recently reported by Hughes et al. (64), who performed genomic and transcriptomic analysis of sorted rods and cones. They suggested instead that lamin A/C might be the critical determinant of rod chromatin inversion, since the Lmna gene is completely silent and becomes inaccessible in differentiated rods. Our reanalysis of published RNA-seq data and our own expression profiling (SI Appendix, Fig. S5) confirm that Lbr levels fall in accordance with Solovei et al. (8), at the level of both transcript and protein, and that these levels never reach zero, in accordance with Hughes et al. (64). We submit that the very low levels of Lbr expressed by adult rods likely fall below a functional threshold.
The clear up-regulation of lamin A/C in both Casz1-KO and polycomb RNAi was nevertheless perplexing, as lamin C had previously been shown to have no effect on chromatin inversion (8). Moreover, it had been suggested that lamin A is not expressed in neurons (65). However, when examining published RNA-seq data (66, 67), we observed that lamin A was quantitatively the dominant transcript in cone photoreceptors (SI Appendix, Fig. S5). Accordingly, when we expressed lamin A in rods, we found that it was sufficient to reorganize rod chromatin architecture and reposition the nucleus to the apical side of the photoreceptor layer. That these activities are specific to lamin A and not lamin C is in agreement with human mutations associated with Hutchinson–Guilford progeria syndrome, which abolish the splicing and processing of the lamin A C terminus and concomitantly abrogate heterochromatin tethering (68–70).
While the mechanistic basis for effects on nuclear position remains to be clarified, we note that the apical localization of cone photoreceptors requires linkers of the nucleoskeleton to the cytoskeleton (LINC) proteins (37–39, 41, 42), suggesting that in rods lamin A/C might provide an anchor for the LINC proteins, allowing them to transmit forces to the nucleus.
While Casz1 regulates the nuclear organization of rods, its role in cone photoreceptors remains to be determined. We hypothesize that Casz1 might also be required for cone nuclear organization, but the more conventional nuclear architecture of cones is not as simple to analyze using microscopy. Moreover, lamin A/C remains expressed in cones, albeit at low levels, whereas it is undetected in rods. It is therefore possible that Casz1/polycomb simply reduces lamin A/C expression levels in cones. Another possibility is that rod- and cone-specific transcription factors alter the function of Casz1. Rod-specific polycomb complexes have recently been shown to repress cone gene expression (71), suggesting that Casz1 might conversely function to suppress rod-specific genes in cones. Importantly, we carefully examined rods manipulated via Casz1 cKO or polycomb RNAi to confirm that lamin A/C up-regulation and nuclear reorganization occur independently of a rod vs. cone fate switch (SI Appendix, Fig. S3).
Degeneration, lamin A/C up-regulation, and nuclear reorganization appear to be a slow process in Casz1-KO retinas, whereas these processes appeared much faster when using RNAi to knockdown Casz1 or polycomb. We suggest that this is probably due to the slow kinetics of Cre-mediated exon excision, as both Cre drivers lead to mosaic recombination. Indeed, we observe many “escaper” cells in the retinas of Rod-cKO mutants as late as P300. RNAi manipulations may also lead to lamin A/C up-regulation before rod maturation, whereas the LMOP::Cre driver, which is active only from P7 onward (29, 72), may lead to Casz1 inactivation after rods have completed chromatin inversion. Preventing chromatin inversion might thus be more efficient than reversing it.
Cooperation Between Casz1v2 and Polycomb in Regulating Rod Lamina Function.
We had previously shown that only the Casz1v2 isoform promotes rod production when misexpressed in retinal progenitors (21). Intriguingly, we find that Casz1v2, but not Casz1v1, associates with polycomb proteins, suggesting a mechanistic basis for the functional differences elicited by the two splice forms. However, this finding is difficult to explain at the structural level, since the amino acid sequence of Casz1v2 is completely represented within the longer Casz1v1 transcript, except for the single amino acid residue at the C terminus of Casz1v2. One possibility is that the longer C-terminal domain of Casz1v1 sterically hinders the interaction with Rnf2.
Polycomb recruitment to the genome is a mysterious and poorly understood subject, and much work remains to be done before concluding that Casz1 regulates this process directly. However, our functional data strongly suggest that Casz1 and polycomb cooperate to regulate the function of the nuclear lamina, acting to suppress the expression of lamin A/C. Accordingly, rods mutant for the polycomb gene Bmi1 have been shown to exhibit heterochromatin decondensation (73), which closely resembles the phenotype we found in Casz1-KO rods, and polycomb marks are observed on the Lmna promoter and gene body in rod-specific ChIP-seq datasets (SI Appendix, Fig. S5) (67). Like Bmi1, Casz1 associates with polycomb repressive complex 1 (PRC1), which has numerous variants. Interestingly, Samd7, another PRC1-associated protein, was recently shown to suppress cone gene expression in rod photoreceptors (71). Although lamin A/C is expressed in cones and not rods, we do not see up-regulation of other cone markers in Casz1-cKO rods (SI Appendix, Fig. S3) or in our RNA-seq dataset. Moreover, both Casz1 and other polycomb mutants degenerate (73–75), whereas Samd7 mutants do not. These data suggest the existence of multiple PRC1 variants with nonoverlapping functions.
Whether alterations in lamina function can explain the cell death observed here in Casz1 Rod-cKO mutants and in retinal polycomb mutants (73, 75, 76), the exact molecular mechanism remains to be formally determined. We note, however, that lamin A expression alters the nuclear organization of rods at the macroscopic level. Disorganized nuclear organization was previously shown to lead to defective partitioning of euchromatic genes into heterochromatin in degenerating Sca7 transgenic rods (61), where lamin A/C is also up-regulated (8). Additionally, murine rods normally maintain their euchromatic transcriptional apparatus at the nuclear periphery and perhaps at the nuclear lamina itself (77). We hypothesize that the conversion of this territory to a suppressive transcriptional environment by lamin A disrupts photoreceptor gene expression, as supported by our RNA-seq analysis of Casz1-cKO retinas. This hypothesis is countered by results from Lbr transgenic mice, which do not appear to degenerate, suggesting that nuclear reorganization and degeneration are separable (8). However, it remains unclear whether the nuclear reorganization triggered by Lbr versus lamin A/C is equivalent. Indeed, Lbr is expressed naturally in rods, whereas lamin A/C is never normally expressed (SI Appendix, Fig. S5). More work will be required to determine whether and how gene expression and nuclear organization are linked in Casz1-KO rods.
Our results suggest that Casz1 and lamina proteins contribute oppositely to chromatin inversion and that the specific outcome observed depends on protein dosage. For example, overexpressing Casz1v2 led to the precocious inversion and expansion of heterochromatin, both in Lbr+ retinal progenitors and immature rods. Casz1v2 misexpression similarly promoted the basal localization of Lbr+ rod photoreceptor nuclei in transfected explants. Moreover, overexpressing Casz1 in Lbr+ retinal progenitors or lamin A/C+ 3T3 fibroblasts likewise led to the expansion and/or aggregation of chromocenters. These data suggest that Casz1 can bypass the effects of lamina tethers to promote aspects of chromatin inversion. On the other hand, overexpression of lamin A reduced heterochromatin area in Casz1+ rod photoreceptors and blocked the ability of Casz1 to localize rod nuclei to the basal side of the photoreceptor layer, even when Casz1 was cotransfected. Finally, heterochromatin is maintained in normal configuration in retinal progenitor cells, which express both Casz1 (21) and Lbr (8). We suggest that the increasing expression levels of Casz1v2 observed during retinal development (21) progressively contribute to chromatin inversion. In the future, it will be of interest to define how exactly Casz1 and lamin A/C expression are opposed at the posttranscriptional level.
In conclusion, this study uncovers Casz1 as a regulator of nuclear architecture essential for cell survival. Since Casz1 is expressed in other types of neurons as well as tissues such as muscle and skin (21–23), we propose that it might function similarly to control nuclear organization in many different cell types.
Methods
A detailed description of the materials and methods is presented in SI Appendix, Methods.
Animal Care.
All animal work was performed in accordance with guidelines from the Institut de Recherches Cliniques de Montréal (IRCM) animal care committee and the Canadian Council on Animal Care. CD1 mice were obtained from Taconic. Casz1Flox/Flox (21), α–Pax6::Cre (46), LMOP::Cre (72), IKCre-A (78), and R26-Stop-EYFP (79) alleles and genotyping protocols were previously described. (See also SI Appendix, Methods and Table S1.) No differences were observed between Cre− and Cre+ heterozygote animals, which therefore are referred to as “control” throughout the text and figures. Animals of either sex were used.
Biochemistry.
Immunoprecipitation and Western blotting from cultured cell lines were performed as described previously, with minor modifications (29). Cross-linking–assisted immunoprecipitation was performed as described (33). Antibodies and concentrations are presented in SI Appendix, Methods and Table S2.
Histology.
In vivo and ex vivo retinal transfection, tissue fixation, and immunohistochemistry were performed essentially as described (21, 29). Antibodies and concentrations are presented in SI Appendix, Methods and Table S2. For TEM, 1-y-old animals were perfused with fixative composed of 2.5% glutaraldehyde, 2% paraformaldehyde in 100 mM cacodylate buffer (pH 7.0), and 2 mM CaCl2. Retinal tissue was passed through an acetone dehydration buffer series, sectioned to 90–100 nm, and imaged using an FEI Tecnai 12 BioTwin instrument. Further details are presented in SI Appendix, Methods.
Quantitation and Statistical Analysis.
Statistical operations were conducted using Microsoft Excel and GraphPad Prism 6 software. To measure nuclear position, the photoreceptor layer of transfected retinal regions was divided into equal-sized bins, each covering 25% of the layer. The nuclear position of transfected cells was then marked, and the proportions of nuclei in each bin were quantitated. To measure heterochromatin and euchromatin areas, regions of the nucleus were traced manually in TEM images or confocal z-planes using the Freehand Selection Tool in the ImageJ (NIH) software package. Measured areas were related to the total area of the nucleus. The n values refer to independent experiments or animals as per the text.
RNA-Seq Analysis.
RNA-seq methods and bioinformatics are described in SI Appendix, Methods. Data have been deposited in the GEO database (accession no. GSE115778).
Supplementary Material
Acknowledgments
We thank Irina Solovei for generously sharing reagents and advice; Harald Herrmann and Monika Zwerger for antibodies; Cheryl Arrowsmith, Harohiko Koseki, Heinrich Leonhardt, and Bas Van Steensel for sharing DNA constructs; Christine Jolicoeur for invaluable technical support; Jessica Barthe and Marie-Claude Lavallée for animal husbandry; Julie Lord and Éric Massicote for help with flow cytometry; Odile Neyret-Djossou and Alexis Blanchet-Cohen for help with RNA-seq experiments; and members of the M.C. laboratory and the IRCM Neuroscience laboratories for insightful discussions. This work was supported by the Foundation for Fighting Blindness Canada and the Canadian Institutes of Health Research (CIHR) Operating Grant PJT-148553. P.M. was supported by a CIHR postdoctoral fellowship and currently holds the Gladys and Lorna J. Wood Chair for Research in Vision. M.C. holds the Gaëtane and Roland Pillenière Chair in Retina Biology and is a Research Scholar Emeritus of the Fonds de Recherche du Québec–Santé.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE115778).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803069115/-/DCSupplemental.
References
- 1.Misteli T. Physiological importance of RNA and protein mobility in the cell nucleus. Histochem Cell Biol. 2008;129:5–11. doi: 10.1007/s00418-007-0355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monier K, Armas JC, Etteldorf S, Ghazal P, Sullivan KF. Annexation of the interchromosomal space during viral infection. Nat Cell Biol. 2000;2:661–665. doi: 10.1038/35023615. [DOI] [PubMed] [Google Scholar]
- 3.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberhart A, et al. Epigenetics of eu- and heterochromatin in inverted and conventional nuclei from mouse retina. Chromosome Res. 2013;21:535–554. doi: 10.1007/s10577-013-9375-7. [DOI] [PubMed] [Google Scholar]
- 7.Solovei I, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 8.Solovei I, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Charpentier MS, et al. CASZ1 promotes vascular assembly and morphogenesis through the direct regulation of an EGFL7/RhoA-mediated pathway. Dev Cell. 2013;25:132–143. doi: 10.1016/j.devcel.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christine KS, Conlon FL. Vertebrate CASTOR is required for differentiation of cardiac precursor cells at the ventral midline. Dev Cell. 2008;14:616–623. doi: 10.1016/j.devcel.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, et al. Essential role of the zinc finger transcription factor Casz1 for mammalian cardiac morphogenesis and development. J Biol Chem. 2014;289:29801–29816. doi: 10.1074/jbc.M114.570416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui X, Doe CQ. ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development. 1992;116:943–952. doi: 10.1242/dev.116.4.943. [DOI] [PubMed] [Google Scholar]
- 13.Mellerick DM, Kassis JA, Zhang SD, Odenwald WF. Castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron. 1992;9:789–803. doi: 10.1016/0896-6273(92)90234-5. [DOI] [PubMed] [Google Scholar]
- 14.Vacalla CM, Theil T. Cst, a novel mouse gene related to Drosophila castor, exhibits dynamic expression patterns during neurogenesis and heart development. Mech Dev. 2002;118:265–268. doi: 10.1016/s0925-4773(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 15.Baumgardt M, Karlsson D, Terriente J, Díaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 2006;20:2618–2627. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kambadur R, et al. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 19.Tran KD, Doe CQ. Pdm and castor close successive temporal identity windows in the NB3-1 lineage. Development. 2008;135:3491–3499. doi: 10.1242/dev.024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stampfel G, et al. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature. 2015;528:147–151. doi: 10.1038/nature15545. [DOI] [PubMed] [Google Scholar]
- 21.Mattar P, Ericson J, Blackshaw S, Cayouette M. A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron. 2015;85:497–504. doi: 10.1016/j.neuron.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin NM, Gibbs D, Conlon FL. Differential regulation of CASZ1 protein expression during cardiac and skeletal muscle development. Dev Dyn. 2014;243:948–956. doi: 10.1002/dvdy.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackshaw S, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, et al. Identification of CASZ1 NES reveals potential mechanisms for loss of CASZ1 tumor suppressor activity in neuroblastoma. Oncogene. 2017;36:97–109. doi: 10.1038/onc.2016.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omori Y, et al. Analysis of transcriptional regulatory pathways of photoreceptor genes by expression profiling of the Otx2-deficient retina. PLoS One. 2011;6:e19685. doi: 10.1371/journal.pone.0019685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuel A, Housset M, Fant B, Lamonerie T. Otx2 ChIP-seq reveals unique and redundant functions in the mature mouse retina. PLoS One. 2014;9:e89110. doi: 10.1371/journal.pone.0089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le YZ, et al. Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol Vis. 2004;10:1011–1018. [PubMed] [Google Scholar]
- 28.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramamurthy V, et al. Numb regulates the polarized delivery of cyclic nucleotide-gated ion channels in rod photoreceptor cilia. J Neurosci. 2014;34:13976–13987. doi: 10.1523/JNEUROSCI.1938-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cayouette M, Behn D, Sendtner M, Lachapelle P, Gravel C. Intraocular gene transfer of ciliary neurotrophic factor prevents death and increases responsiveness of rod photoreceptors in the retinal degeneration slow mouse. J Neurosci. 1998;18:9282–9293. doi: 10.1523/JNEUROSCI.18-22-09282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cayouette M, Smith SB, Becerra SP, Gravel C. Pigment epithelium-derived factor delays the death of photoreceptors in mouse models of inherited retinal degenerations. Neurobiol Dis. 1999;6:523–532. doi: 10.1006/nbdi.1999.0263. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, et al. SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev. 2013;27:749–766. doi: 10.1101/gad.210963.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammed H, et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11:316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z, Lam N, Thiele CJ. Zinc finger transcription factor CASZ1 interacts with histones, DNA repair proteins and recruits NuRD complex to regulate gene transcription. Oncotarget. 2015;6:27628–27640. doi: 10.18632/oncotarget.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morimoto-Suzki N, et al. The polycomb component Ring1B regulates the timed termination of subcerebral projection neuron production during mouse neocortical development. Development. 2014;141:4343–4353. doi: 10.1242/dev.112276. [DOI] [PubMed] [Google Scholar]
- 36.Tardat M, et al. Cbx2 targets PRC1 to constitutive heterochromatin in mouse zygotes in a parent-of-origin-dependent manner. Mol Cell. 2015;58:157–171. doi: 10.1016/j.molcel.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Insinna C, Baye LM, Amsterdam A, Besharse JC, Link BA. Analysis of a zebrafish dync1h1 mutant reveals multiple functions for cytoplasmic dynein 1 during retinal photoreceptor development. Neural Dev. 2010;5:12. doi: 10.1186/1749-8104-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maddox DM, et al. A mutation in Syne2 causes early retinal defects in photoreceptors, secondary neurons, and Müller glia. Invest Ophthalmol Vis Sci. 2015;56:3776–3787. doi: 10.1167/iovs.14-16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razafsky D, Blecher N, Markov A, Stewart-Hutchinson PJ, Hodzic D. LINC complexes mediate the positioning of cone photoreceptor nuclei in mouse retina. PLoS One. 2012;7:e47180. doi: 10.1371/journal.pone.0047180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Razafsky D, et al. Lamin B1 and lamin B2 are long-lived proteins with distinct functions in retinal development. Mol Biol Cell. 2016;27:1928–1937. doi: 10.1091/mbc.E16-03-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsujikawa M, Omori Y, Biyanwila J, Malicki J. Mechanism of positioning the cell nucleus in vertebrate photoreceptors. Proc Natl Acad Sci USA. 2007;104:14819–14824. doi: 10.1073/pnas.0700178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu J, et al. KASH protein Syne-2/nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Melo J, Peng GH, Chen S, Blackshaw S. The Spalt family transcription factor Sall3 regulates the development of cone photoreceptors and retinal horizontal interneurons. Development. 2011;138:2325–2336. doi: 10.1242/dev.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emerson MM, Surzenko N, Goetz JJ, Trimarchi J, Cepko CL. Otx2 and Onecut1 promote the fates of cone photoreceptors and horizontal cells and repress rod photoreceptors. Dev Cell. 2013;26:59–72. doi: 10.1016/j.devcel.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nijnik A, et al. Sanger Institute Microarray Facility; Sanger Mouse Genetics Project The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood. 2012;119:1370–1379. doi: 10.1182/blood-2011-05-352666. [DOI] [PubMed] [Google Scholar]
- 46.Marquardt T, et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 47.Razafsky DS, Ward CL, Kolb T, Hodzic D. Developmental regulation of linkers of the nucleoskeleton to the cytoskeleton during mouse postnatal retinogenesis. Nucleus. 2013;4:399–409. doi: 10.4161/nucl.26244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong-Riley MT. Energy metabolism of the visual system. Eye Brain. 2010;2:99–116. doi: 10.2147/EB.S9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andzelm MM, et al. MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron. 2015;86:247–263. doi: 10.1016/j.neuron.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Béby F, et al. Otx2 gene deletion in adult mouse retina induces rapid RPE dystrophy and slow photoreceptor degeneration. PLoS One. 2010;5:e11673. doi: 10.1371/journal.pone.0011673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freund CL, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 52.Koike C, et al. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007;27:8318–8329. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- 54.Omori Y, et al. Mef2d is essential for the maturation and integrity of retinal photoreceptor and bipolar cells. Genes Cells. 2015;20:408–426. doi: 10.1111/gtc.12233. [DOI] [PubMed] [Google Scholar]
- 55.Hsiau TH, et al. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One. 2007;2:e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegert S, et al. Genetic address book for retinal cell types. Nat Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- 57.Kizilyaprak C, Spehner D, Devys D, Schultz P. In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS One. 2010;5:e11039. doi: 10.1371/journal.pone.0011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Popova EY, et al. Developmentally regulated linker histone H1c promotes heterochromatin condensation and mediates structural integrity of rod photoreceptors in mouse retina. J Biol Chem. 2013;288:17895–17907. doi: 10.1074/jbc.M113.452144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng GH, Chen S. Active opsin loci adopt intrachromosomal loops that depend on the photoreceptor transcription factor network. Proc Natl Acad Sci USA. 2011;108:17821–17826. doi: 10.1073/pnas.1109209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1:e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helmlinger D, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hennig AK, Peng GH, Chen S. Transcription coactivators p300 and CBP are necessary for photoreceptor-specific chromatin organization and gene expression. PLoS One. 2013;8:e69721. doi: 10.1371/journal.pone.0069721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tran NM, et al. Mechanistically distinct mouse models for CRX-associated retinopathy. PLoS Genet. 2014;10:e1004111. doi: 10.1371/journal.pgen.1004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hughes AE, Enright JM, Myers CA, Shen SQ, Corbo JC. Cell type-specific epigenomic analysis reveals a uniquely closed chromatin architecture in mouse rod photoreceptors. Sci Rep. 2017;7:43184. doi: 10.1038/srep43184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jung HJ, et al. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc Natl Acad Sci USA. 2012;109:E423–E431. doi: 10.1073/pnas.1111780109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim JW, et al. Recruitment of rod photoreceptors from short-wavelength-sensitive cones during the evolution of nocturnal vision in mammals. Dev Cell. 2016;37:520–532. doi: 10.1016/j.devcel.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mo A, et al. Epigenomic landscapes of retinal rods and cones. eLife. 2016;5:e11613. doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCord RP, et al. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23:260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Omori Y, et al. Samd7 is a cell type-specific PRC1 component essential for establishing retinal rod photoreceptor identity. Proc Natl Acad Sci USA. 2017;114:E8264–E8273. doi: 10.1073/pnas.1707021114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le YZ, et al. Mouse opsin promoter-directed Cre recombinase expression in transgenic mice. Mol Vis. 2006;12:389–398. [PubMed] [Google Scholar]
- 73.Barabino A, et al. Loss of Bmi1 causes anomalies in retinal development and degeneration of cone photoreceptors. Development. 2016;143:1571–1584. doi: 10.1242/dev.125351. [DOI] [PubMed] [Google Scholar]
- 74.Iida A, et al. Roles of histone H3K27 trimethylase Ezh2 in retinal proliferation and differentiation. Dev Neurobiol. 2015;75:947–960. doi: 10.1002/dneu.22261. [DOI] [PubMed] [Google Scholar]
- 75.Yan N, et al. Postnatal onset of retinal degeneration by loss of embryonic Ezh2 repression of Six1. Sci Rep. 2016;6:33887. doi: 10.1038/srep33887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ueno K, et al. Transition of differential histone H3 methylation in photoreceptors and other retinal cells during retinal differentiation. Sci Rep. 2016;6:29264. doi: 10.1038/srep29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X, et al. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem. 2002;277:43288–43300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
- 78.Tarchini B, Jolicoeur C, Cayouette M. In vivo evidence for unbiased Ikaros retinal lineages using an Ikaros-Cre mouse line driving clonal recombination. Dev Dyn. 2012;241:1973–1985. doi: 10.1002/dvdy.23881. [DOI] [PubMed] [Google Scholar]
- 79.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.