Significance
Microtubules consist of parallel protofilament lanes for motor-based intracellular transport. While most motors translocate along individual protofilaments, members of the kinesin-8 family have been reported to sidestep. However, the mechanism of sidestepping is currently not understood. Here, we track the 3D motion of single kinesin-8 motors on freely suspended microtubules. We find that individual motors sidestep with a bias to the left, the probability of which is increased with the time taken per forward step. We relate this behavior to a bifurcation in the step cycle of kinesin-8. Experiments on a kinesin-1 with an elongated neck linker suggest that sidestepping is intrinsic to all processive kinesins possessing long neck linkers, potentially helping them to circumnavigate obstacles on the microtubule.
Keywords: kinesin-8, sidestepping, high processivity, 3D nanometer tracking, step cycle
Abstract
Kinesin-8 motors, which move in a highly processive manner toward microtubule plus ends where they act as depolymerases, are essential regulators of microtubule dynamics in cells. To understand their navigation strategy on the microtubule lattice, we studied the 3D motion of single yeast kinesin-8 motors, Kip3, on freely suspended microtubules in vitro. We observed short-pitch, left-handed helical trajectories indicating that kinesin-8 motors frequently switch protofilaments in a directionally biased manner. Intriguingly, sidestepping was not directly coupled to forward stepping but rather depended on the average dwell time per forward step under limiting ATP concentrations. Based on our experimental findings and numerical simulations we propose that effective sidestepping toward the left is regulated by a bifurcation in the Kip3 step cycle, involving a transition from a two-head–bound to a one-head–bound conformation in the ATP-waiting state. Results from a kinesin-1 mutant with extended neck linker hint toward a generic sidestepping mechanism for processive kinesins, facilitating the circumvention of intracellular obstacles on the microtubule surface.
Cytoskeletal remodeling is essential in eukaryotic cell division and morphogenesis. One of the key factors influencing microtubule dynamics is the motor protein kinesin-8, which is involved in regulation of spindle size, spindle positioning, chromosomal congression (1–3), nuclear positioning (4–6), and tuning ciliary length (7).
Recent in vitro studies, have shown that Saccharomyces cerevisiae kinesin-8 (Kip3) is a slow (∼50 nm/s in comparison with ∼800 nm/s for kinesin-1) plus-end–directed motor protein with high processivity (>11 μm in comparison with ∼1 μm for kinesin-1) (8, 9). At the microtubule plus end Kip3 acts in two ways: It removes tubulin dimers via its motor domains (8–10) and it stabilizes shrinking microtubules via its tail domain (11). The combination of directed motility, depolymerization, and stabilization leads to an elaborate mechanism for microtubule length regulation. In particular, the high processivity of Kip3 funnels almost all motors that land on the microtubule lattice to the microtubule plus end, where the rate of depolymerization is proportional to the flux of incoming motors. Consequently, Kip3 depolymerizes long microtubules faster than short ones (12, 13). This length-dependent depolymerization constitutes a key feature of Kip3 and distinguishes it from other microtubule regulators such as kinesin-13 (14), the Dis1/XMap215 family (15), and EB1 (16). However, in vivo, microtubule-associated proteins, e.g., other kinesins; nonmotor microtubule stabilizing, destabilizing, or cross-linking factors; passenger proteins; and microtubule end-tracking proteins (17), are expected to act as obstacles, blocking the paths of Kip3 motors to the microtubule end. Therefore, to keep its capability to depolymerize microtubules in a length-dependent manner, Kip3 has to be able to circumvent obstacles.
Various studies have explored the motility response of different kinesins when encountering obstacles. While kinesin-1 motors mostly pause and detach (18), motor proteins from the kinesin-2 family were reported to switch to adjacent protofilaments (19, 20). Based on observations in multimotor gliding motility assays, we recently hypothesized that Kip3 can also switch microtubule protofilaments, namely with a bias toward the left (21, 22). We explained these findings by the relatively long neck linker of Kip3 (compared with kinesin-1) and the actual 3D geometry of the motor–microtubule interaction. Other studies based on optical tweezers measurements reported that single Kip3 molecules can (i) enter a diffusive slip state upon longitudinal force application (23) and (ii) switch protofilaments upon sideward force application (24). Interestingly, the latter study hypothesized an intrinsically equal probability for Kip3 to switch protofilaments toward the right or left, being in contrast to our findings of biased sidestepping toward the left (21, 22). To answer whether protofilament switching of single Kip3 motors in the absence of external load is diffusive or directionally biased, we tracked the 3D motion of individual motors on freely suspended microtubules. We observe robust left-handed helical motion, constituting evidence for directionally biased sidestepping. Intriguingly, we observe that leftward-biased sidestepping is not directly coupled to forward stepping but is correlated with the average time it takes the motor to make a forward step (hereafter denoted as average dwell time per forward step). Additionally, we find that the leftward-biased sidestepping probability per forward step increases on (i) prolonging the average dwell time per forward step by limiting the ATP concentration and (ii) weakening the electrostatic interactions of the motor with the microtubule lattice. Based on our observations and numerical simulations, we propose that in the ATP waiting state the motor can switch from a two-head–bound (2HB) to a one-head–bound (1HB) conformation with sidestepping favored if ATP binds in the 1HB conformation. Finally, we show that a kinesin-1 mutant with an extended neck linker can switch protofilaments at rate-limiting ATP conditions, hinting toward a generic side-stepping mechanism for all processive kinesins with long neck linkers.
Results
Single Kip3 Motors Sidestep with a Directional Bias Toward the Left.
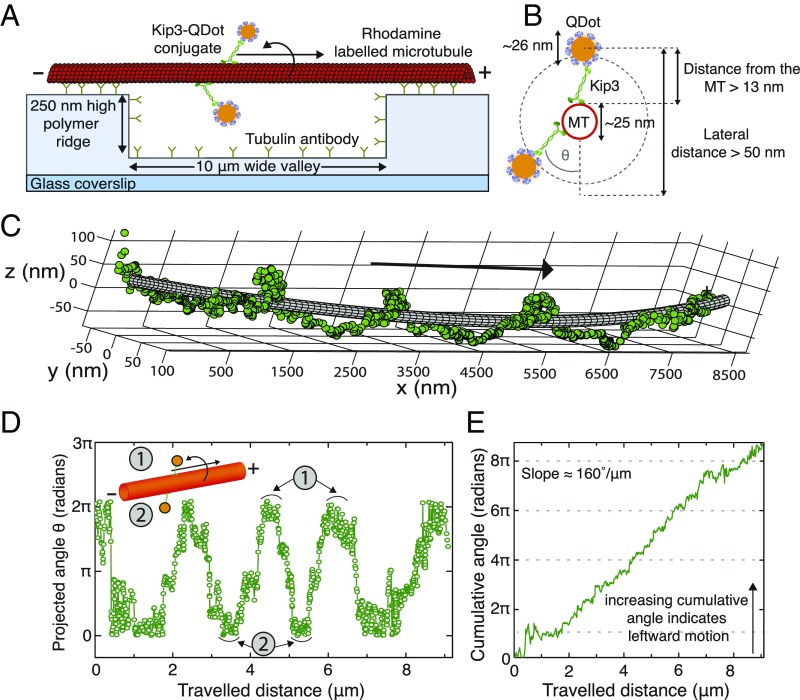
The 3D motion of Kip3 motors along the microtubule lattice was investigated by tracking single Kip3 motors labeled with quantum dots (QDs) at their tails (Kip3-QD), stepping on freely suspended, double-stabilized microtubules. Here, the microtubules were tautly immobilized on 250-nm–high polymer ridges that were separated by 10-μm–wide valleys [Fig. 1A and SI Appendix, Fig. S1; strategy similar to that in Bugiel et al. (25)]. To obtain single-molecule conditions (i.e., not more than one Kip3 motor per Kip3-QD conjugate) a 1:10 ratio of Kip3 motor to QDs was used in the conjugation mixture (SI Appendix, Fig. S2). Kip3-QD conjugates were imaged using highly inclined thin illumination microscopy (HILO) (26) with a custom-built 3D single-molecule tracking setup termed Parallax (27) (SI Appendix, Fig. S3 as well as SI Appendix, 3D Single Molecule Imaging Set-Up, Parallax for more details). The position of individual Kip3-QDs with respect to the microtubule lattice was determined by fitting the 3D Kip3-QD tracks around 25-nm–wide tubes (representing the microtubule), using nonlinear least-squares fitting (SI Appendix, Fitting of Kip3-QDot Tracks). Fig. 1C shows a typical example of a Kip3-QD track, where the motor moved in a left-handed helical trajectory around the longitudinal axis of the microtubule (SI Appendix, Fig. S4 for further example tracks). The mean distance of the Kip3-QD position from the surface of the fitted tube was 24 ± 17 nm (mean ± SD), well within the estimated range (Fig. 1B). To obtain the rotational pitch of the track and the directionality of rotation, the angular information for each Kip3-QD motility event (Fig. 1D) and the cumulative angle (Fig. 1E) were plotted as a function of the traveled distance, respectively. For the example in Fig. 1 the rotational pitch was 2.1 ± 0.2 μm (mean ± SD, n = 3 full rotations), the velocity was 42.6 ± 1.0 nm/s (mean ± SD), and the slope of the cumulative angle was 160°/μm.
Fig. 1.
Single Kip3 motors move with helical trajectories on freely suspended microtubules. (A) Schematic representation of a freely suspended microtubule immobilized on optically transparent polymer ridges by anti–β-tubulin antibodies. Single Kip3-mfGFP motors, labeled with streptavidin-coated QDs at the tail (Kip3-QD), are capable of accessing the entire 3D lattice of the microtubule between two ridges (i.e., over the “valley”). (B) The diameter for the helical path traversed by the Kip3-QD (QD center localized by tracking the QD signal) around the microtubule is greater than 50 nm. (C) The 3D tracking result for an example Kip3-QD event fitted around a 25-nm–wide tube using nonlinear least-squares fitting. (D) Projected angle of the Kip3-QD with respect to the microtubule lattice (indicated as θ in B) shown for every position along the microtubule axis (traveled distance). Here, the maxima (minima) denote the positions of Kip3-QD on top (bottom) of the microtubule. (E) The increase of the cumulative angle with increasing traveled distance indicates the directionality of rotation, with positive values denoting angular changes toward the left (in the direction of motion). The example Kip3-QD moves on the microtubule with a velocity of 43 ± 1 nm/s and has a rotational pitch of 2.1 ± 0.3 µm (both mean ± SD, n = 3 rotations).
For statistical evaluation, we identified 73 Kip3-QD motility events suitable for tube fitting (SI Appendix, Selection Criteria for Further Analysis of Kip3-QDot Tracks). Out of these events, 50 Kip3-QD tracks (in total exhibiting 110 complete rotations) showed clear left-handed helical motion with a rotational pitch of 1.9 ± 0.3 μm and a velocity of 43 ± 5 nm/s (28) (data in SI Appendix, Fig. S5A and Fig. 2A; see SI Appendix, Analysis of Distributions with Error Estimation). Furthermore, the cumulative angles vs. traveled distance for these events showed a clear preference in directionality toward the left (slope = 130 ± 40°/μm; SI Appendix, Fig. S5A). The remaining 23 tracks did not show clear rotations but were rather indicative of erratic transverse motion (SI Appendix, Fig. S5B). However, for these unclear events the average distance from the fitted tube was 14.1 ± 3.2 nm, significantly lower than for the other 50 tracks (17.9 ± 2.9 nm; P = 0.003, Mann–Whitney U test). The unclear tracks have likely arisen from poor tracking, from poor fitting, and/or from motors moving on surface-interacting microtubules not rejected during the preselection procedure (see an example event on a surface interacting microtubule in SI Appendix, Fig. S6). Importantly, none of these tracks displayed any evident right-handed trajectory.
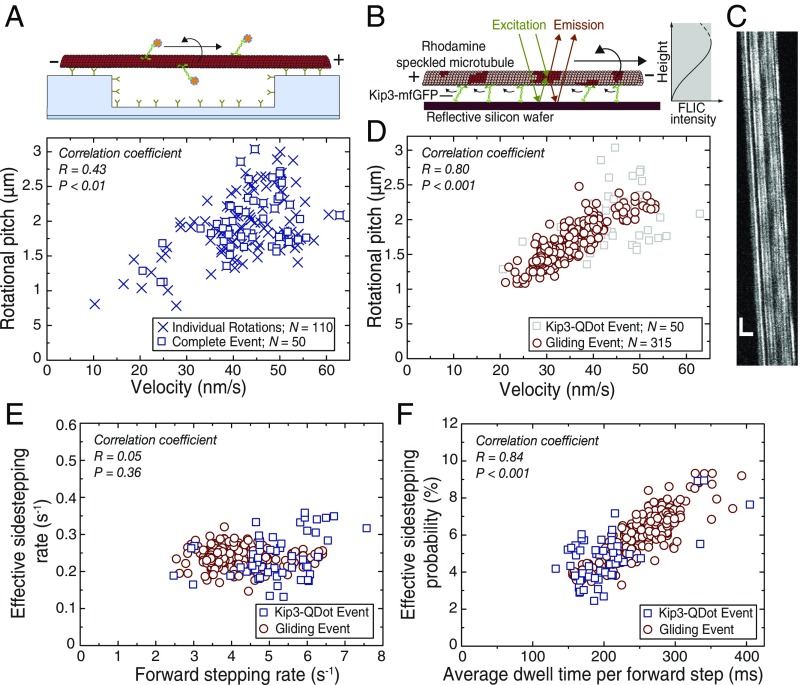
Fig. 2.
Effective sidestepping probability depends on average dwell time per forward step. (A) Rotational pitches of 50 Kip3-QD tracks in dependence of the forward velocity. Each individual rotation is represented by a blue cross while the blue squares represent rotational pitches from complete Kip3-QD tracks. (B) Schematic representation for obtaining rotational information from speckled microbubules gliding on a reflective silicon substrate (22) coated with Kip3-mfGFP motors. Due to FLIC the recorded intensities of the asymmetric speckles increase as a function of height above the substrate. Rotational information of gliding speckled microtubules is interpreted from the variation in the recorded intensity of the speckles on the microtubules. (C) Example kymograph (space–time intensity plot) of a gliding speckled microtubule. Each individual speckle shows periodic intensity fluctuation (every 1.3 µm) corresponding to the rotational pitch of the gliding microtubule. (Vertical scale bar, 20 s; horizontal scale bar, 5 µm.) (D) Rotational pitches of microtubules in Kip3 gliding assays in dependence of the forward velocity. Here, the rotational pitches of 315 microtubules are represented by red circles and single-molecule Kip3-QD data (from A) are shown in gray for reference. There is a significant dependence of the rotational pitch on the forward velocity in both experiments (stepping assay R = 0.43, P < 0.01; gliding assay R = 0.87, P < 0.001; combined data R = 0.80, P < 0.001; Pearson correlation coefficient). (E) Effective sidestepping rate for the data shown in A and B plotted against the forward stepping rate (blue squares, stepping assay; red circles, gliding assay; see SI Appendix, SI Text for details on the calculation). The effective sidestepping rates are similar in both experiments (combined 0.24 ± 0.01 sidesteps per second, n = 365) and no significant dependence of the effective sidestepping rate on the forward stepping rate is observed (combined R = 0.05, P = 0.36). (F) Effective sidestepping probability per forward step in dependence of the average dwell time per forward step (blue squares, stepping assay; red circles, gliding assay; see SI Appendix, SI Text for details on the calculation). The effective sidestepping probability per forward step strongly depends on the average dwell time per forward step (combined R = 0.84, P < 0.001).
Taken together, we observed that Kip3-QD predominantly moved with leftward helical trajectories around microtubules. As the supertwist for double-stabilized microtubules [∼96% 14-protofilament microtubules (29)] is left-handed with a pitch in the range of 7–9 µm (22, 30), the short rotational pitches measured for the single Kip3-QD conjugates implies that the motors must sidestep with a bias toward the left. Currently, we do not know whether sidestepping occurs exclusively to neighboring protofilaments on the left or whether the motors move—although biased toward the left—with a mixture of leftward and rightward sidesteps (Discussion). Consequently, we therefore concentrate on characterizing and modeling the leftward-biased “effective” sidestepping rate and probability, the latter of which amounts to about 4.8% (1 effective sidestep toward the left per 21 forward steps) in the experiment described above.
Leftward-Biased Sidestepping Is Not Directly Coupled to Forward Stepping.
To investigate sidestepping in each of the individual Kip3-QD tracks, we quantified the rotational pitches in dependence of velocity for each of the 50 tracks (Fig. 2A, data for individual rotations also shown). We observed a stochastic variation in Kip3-QD velocities, ranging from 35 nm/s to 55 nm/s, and a significant dependence of the rotational pitch on the velocity (R = 0.43, P < 0.01; Pearson correlation coefficient). When performing Kip3 gliding assays with speckled microtubules using fluorescence interference contrast (FLIC) microscopy [Fig. 2 B and C and Mitra et al. (22)], we obtained a rotational pitch of 1.6 ± 0.1 μm and a velocity of 34 ± 2 nm/s (n = 315 microtubules). Further, we observed a variation in microtubule gliding velocities, ranging from 20 nm/s to 50 nm/s, which was dependent on the density of the motors (SI Appendix, Fig. S7). In agreement with our single Kip3-QD data, the gliding assays showed a similar dependence of the rotational pitch on the velocity (Fig. 2D; R = 0.87, P < 0.001). Combining our data from stepping and gliding assays (correlation between rotational pitch and velocity R = 0.80, P < 0.001), we found the effective sidestepping rate to be constant (0.24 ± 0.01 sidesteps per second; n = 368; correlation between the effective sidestepping rate and the average forward stepping rate R = 0.05, P = 0.36) over a large range of forward stepping rates (2.5–7.5 forward steps per second, Fig. 2E). This indicates that sidestepping is not directly coupled to the forward stepping. Interestingly, the probability to make an effective sidestep toward the left per forward step (denoted as effective sidestepping probability; see SI Appendix, Calculation of Stepping Parameters for details on the calculation) strongly depended on the average dwell time per forward step (Fig. 2F; R = 0.84, P < 0.001).
Effective Sidestepping Probability Increases at Limiting ATP Concentrations.
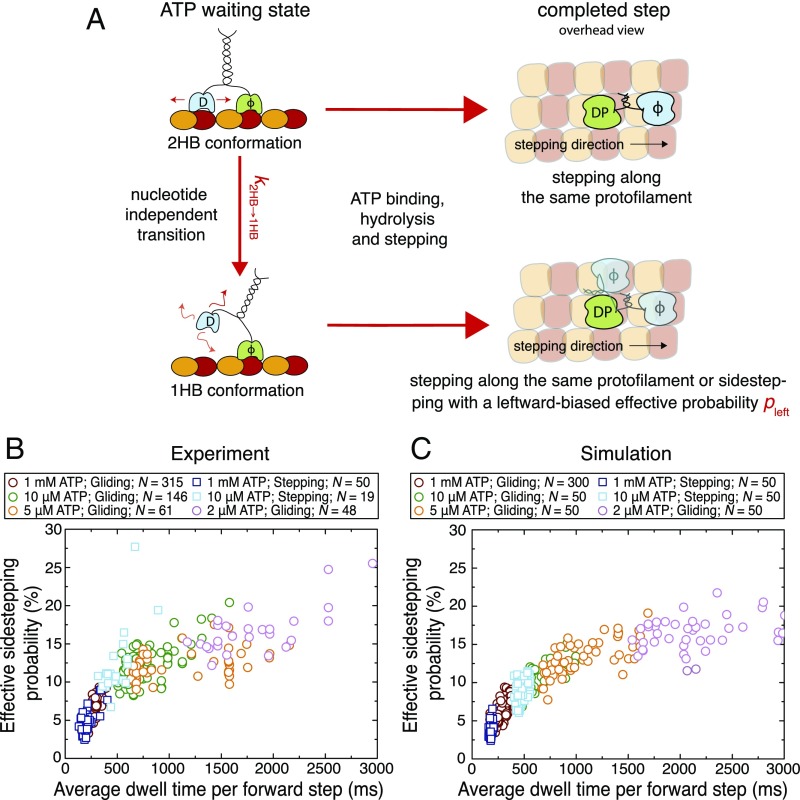
One possibility to explain the increase in the effective sidestepping probability with the average dwell time per forward step is that Kip3 waits for ATP in two different conformations, both of them having different intrinsic sidestepping probabilities. In the case of kinesin-1, it has been shown that the motor can transition from the 2HB conformation to a 1HB conformation while waiting for ATP at limiting ATP concentration (30–35). We suggest a similar transition, which is nucleotide independent but time dependent, occurs for Kip3 even at saturating ATP conditions and propose that sidestepping predominantly occurs from the 1HB conformation (Fig. 3A). Based on these assumptions, we predict an increase in the effective sidestepping probability if the ATP waiting state of Kip3 is prolonged. To test this hypothesis, we performed Kip3-QD stepping assays on suspended microtubules at limiting ATP conditions (10 μM ATP) and observed that most Kip3-QD tracks again showed left-handed helical trajectories on the microtubules. Here, 19 tracks (with 50 complete rotations) yielded a rotational pitch of 0.9 ± 0.2 μm, a velocity of 17 ± 5 nm/s, and a slope of the cumulative angle of 300° ± 100°/μm (SI Appendix, Fig. S5C). Kip3-QD stepping assays at lower ATP concentration (<10 µM ATP) could not be performed reliably because the Kip3-QD tracks were too short (fewer than two rotations before dissociation of Kip3-QDs from the microtubules). We therefore complemented our data by results from Kip3 gliding assays at 10 µM ATP (rotational pitch 0.9 ± 0.1 μm and velocity 12 ± 1 nm/s; n = 146 microtubules), 5 µM ATP (0.9 ± 0.1 μm, 7 ± 5 nm/s; n = 61 microtubules) and 2 µM ATP (0.7 ± 0.1 μm, 5 ± 1 nm/s; n = 48 microtubules). Fig. 3B shows the combined data of the measured effective sidestepping probabilities for each track in dependence of the average dwell time per forward step (alternative representation of the same data in SI Appendix, Fig. S8 A and B). As hypothesized, the effective sidestepping probability increased with longer dwell time per forward step, saturating at 15–20%. This percentage provides an estimate of the maximum effective sidestepping probability () of the motor in the 1HB conformation which is predominantly populated at low ATP concentration (SI Appendix, Fig. S8C).
Fig. 3.
Effective sidestepping probability increases with longer average dwell times per forward step. (A) Illustration of the proposed model to explain the increase in the effective sidestepping probability seen in Fig. 2C. We suggest that Kip3 can undergo a nucleotide-independent transition from a 2HB conformation to a 1HB conformation () in the ATP waiting state. Further, sidestepping is strongly enhanced from the 1HB conformation (with an effective sidestepping probability ). Here, T denotes ATP, D denotes ADP, DP denotes ADP plus phosphate, and ϕ denotes no nucleotide. Loose binding and diffusion are represented by small red arrows. (B) The effective sidestepping probability per forward step is shown in dependence of the average dwell time per forward step for Kip3-QD stepping assays (squares) at 1 mM (blue, n = 50) and 10 μM ATP (cyan, n = 19) as well as for microtubule gliding assays (circles) at 1 mM (red, n = 315), 10 μM (green, n = 146), 5 μM (orange, n = 61), and 2 μM ATP (pink, n = 48). (C) The simulated effective sidestepping probability per forward step is shown in dependence of the average dwell time per forward step for stepping assays (50 single-molecule tracks at 1 mM and 10 μM ATP) as well as for gliding assays (300 microtubules at 1 mM and 50 microtubules each at 10 μM, 5 μM, and 2 μM ATP; same markers and colors as in B). Parameters of the numerical simulation are given in SI Appendix, Table S1. In the simulations, the difference between stepping and gliding assays is only the probability of futile steps (ranging between 0% and 50% for gliding events and between 0% and 20% for stepping events) to account for the experimentally observed velocity variations (SI Appendix, Fig. S7 and Materials and Methods).
To explore other factors that might influence the sidestepping probability, we first changed the dwell time per forward step by temperature in Kip3 gliding assays. We found that the effective sidestepping probability did not vary much between the investigated temperatures of 17 °C, 24 °C, and 32 °C, corresponding to average dwell times per forward step of 120 ms, 280 ms, and 450 ms, respectively (SI Appendix, Fig. S8D). This behavior is in line with the expectation that all rates within the step cycle of Kip3 are similarly affected by temperature. In contrast, at constant temperature the effective sidestepping probabilities varied significantly stronger for the same average dwell times per forward step (adjusted by different ATP concentrations, Fig. 3B). This confirms that sidestepping is linked to a specific state in the step cycle (namely the ATP waiting state) and not to the complete step. Second, we extended the time spent by the motor in the posthydrolysis ADP.Pi state by adding free phosphate (Pi) in Kip3 gliding assays (36). No significant change in the effective sidestepping probability was observed (P = 0.913; SI Appendix, Fig. S8E). Moreover, in control Kip3 stepping assays [using Kip3 multifunctional GFP (Kip3-mfGFP) without QDs] only a marginal increase in the average dwell time per forward step (∼3% increase in the presence of additional Pi) was observed, suggesting that the ADP.Pi state does not play a major role in the sidestepping mechanism. These findings further support our assumption that the ATP waiting state is the crucial state determining the sidestepping probability of Kip3.
We were able to reproduce our experimental findings by numerical simulations based on a bifurcation in the mechanochemical cycle of Kip3 (Fig. 3C; see Materials and Methods and SI Appendix, MATLAB Code for Numerical Simulation of the Stepping Mechanism for the simulation code; parameters are listed in SI Appendix, Table S1). Quantitative agreement between experiment and simulation was obtained by assuming the following: (i) The ATP waiting state is the major rate-limiting step within the mechanochemical cycle of Kip3; i.e., the motor spends most of the 160-ms–long step-cycle time waiting to bind ATP. In the simulation we have therefore chosen However, we note that the model fits our experimental data similarly well for higher ATP waiting times (up to the entire step-cycle time). (ii) The transition rate is approximately fourfold lower than the ATP binding rate at saturating ATP concentration (). (iii) Once the motor has transitioned to the 1HB conformation, the effective sidestepping probability to the neighboring protofilament on the left is For simplicity, we model sidestepping as if it would occur only from the 1HB conformation and exclusively toward the left. Extensions to this model (including Kip3 not exclusively sidestepping toward the left, but toward the left and the right with a leftward bias) are elaborated on in Discussion and in SI Appendix, Fig. S9. The good agreement between the numerical simulation and experimental data shows that the proposed model consistently describes the Kip3 motility at varying ATP concentrations.
Effective Sidestepping Probability Increases with Higher Transition Rates from the 2HB to the 1HB Conformation.
Our model (Fig. 3A) predicts an increase in the effective sidestepping probability if the transition rate from the 2HB to the 1HB conformation () is increased. Here, the transition rate is likely determined by the interaction strength of the loosely bound, trailing motor head (ADP bound) with the microtubule surface (Fig. 4A). As it is known that ADP-bound Kip3 diffusively interacts with microtubules, partially due to electrostatic interactions (37), it should be possible to enhance by removing or screening the microtubule surface charges and thus to increase the effective sidestepping probability.
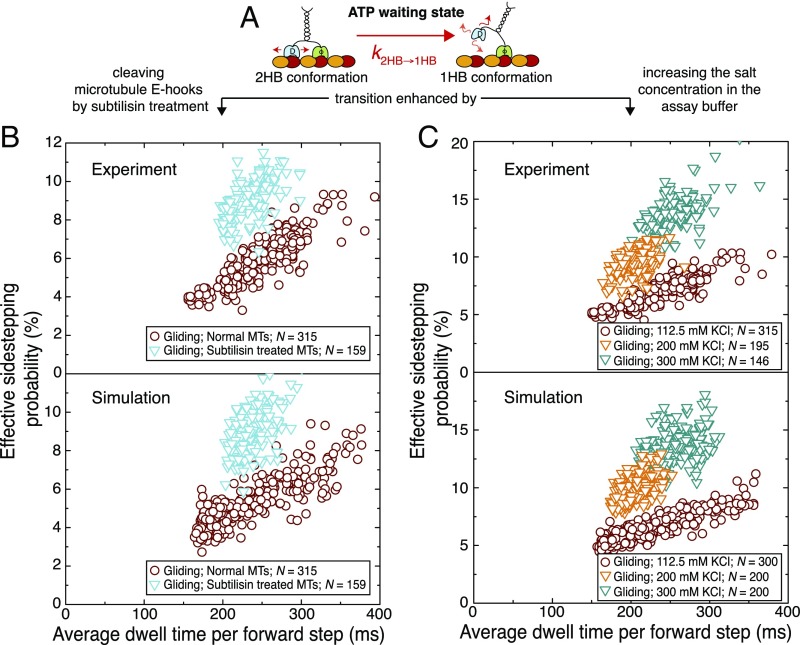
Fig. 4.
Effective sidestepping probability increases upon changing the strength of the motor–microtubule interaction. (A) Illustration of the proposed nucleotide-independent transition from the 2HB to the 1HB conformation in the ATP waiting state. The proposed transition is likely enhanced by cleaving the negatively charged microtubule e-hooks (subtilisin treatment) or by increasing the salt concentration in the buffer (screening of surface charges). (B, Upper) Effective sidestepping probability per forward step in dependence of the average dwell time per forward step for normal (red circles, n = 315) and subtilisin-treated microtubules (cyan triangles, n = 159) in gliding assays (1 mM ATP). (B, Lower) Simulated effective sidestepping probability per forward step in dependence of the average dwell time per forward step for normal microtubules (red circles, n = 300) and subtilisin-treated microtubules (cyan triangles, n = 150) in gliding assays. For subtilisin-treated microtubules, the transition rate was increased by a factor of 2 compared with that for normal microtubules. (C, Upper) Effective sidestepping probability per forward step in dependence of the average dwell time per forward step in gliding assays (1 mM ATP) with 112.5 mM KCl (red circles, n = 315; data corresponding to the red circles in B), 200 mM KCl (orange triangles, n = 195), and 300 mM KCl (green triangles, n = 146) in the assay buffer. (C, Lower) Simulated effective sidestepping probability per forward step in dependence of the average dwell time per forward step for microtubules gliding in assay buffer with 112.5 mM KCl (red circles, n = 300), 200 mM KCl (orange triangles, n = 200), and 300 mM KCl (green triangles, n = 200). To simulate the assays with 200 mM KCl and 300 mM KCl, the transition rate was increased by a factor of 2.5 and 4, respectively, compared with normal assay conditions. Note that the y axis is scaled differently in B and C.
To test this hypothesis, we first performed Kip3 gliding assays with subtilisin-treated microtubules, where the negatively charged e-hooks on the microtubule surface were cleaved off. When performing control Kip3 stepping assays with subtilisin-treated microtubules at 1 mM ADP, we observed an approximately fourfold reduction in the microtubule-association rate and the interaction time (SI Appendix, Fig. S10). Further, control Kip3 stepping assays at 1 mM ATP showed regular motility of Kip3 motors with only ∼10% reduction of the stepping velocity (normal microtubules 54 ± 3 nm/s, subtilisin-treated microtubules 49 ± 4 nm/s; SI Appendix, Fig. S10C). The rotational pitches (1.1 ± 0.1 µm, n = 159 microtubules) for subtilisin-treated microtubules in Kip3 gliding assays at 1 mM ATP were significantly shorter than for normal microtubules while the gliding velocity (35 ± 2 nm/s) was in the same range. Fig. 4B, Upper shows the effective sidestepping probabilities for each individual microtubule in the gliding assays (normal and subtilisin-treated microtubules). Our numerical simulations were in good agreement with the experimental results when we increased the transition rate by a factor of 2 for the Kip3 interaction with subtilisin-treated microtubules (Fig. 4B, Lower).
Second, we performed Kip3 gliding assays at higher salt concentration in the buffer (200 mM KCl and 300 mM KCl) to screen the microtubule surface charges. In control Kip3 stepping assays at 1 mM ATP we observed a significant reduction in the microtubule interaction time (8-fold for 200 mM KCl and 40-fold for 300 mM KCl; SI Appendix, Fig. S10 A and B). Again, the rotational pitches of microtubules in Kip3 gliding assays in assay buffer with 200 mM KCl (1.1 ± 0.1 µm, n = 195 microtubules) and 300 mM KCl (0.8 ± 0.1 µm, n = 146 microtubules) were significantly shorter than those of microtubules in standard buffer (112.5 mM KCl) while the gliding velocity was within the same range (38 ± 1 nm/s for 200 mM KCl and 32 ± 2 nm/s for 300 mM KCl). Fig. 4C, Upper shows the effective sidestepping probabilities for each individual microtubule in the gliding assays at different salt conditions (112.5 mM KCl, 200 mM KCl, and 300 mM KCl). Our numerical simulations were in good agreement with the experimental results when we increased the transition rate by a factor of 2.5 and 4 for the Kip3 interaction with microtubules at 200 mM KCl and 300 mM KCl, respectively (Fig. 4C, Lower).
Taken together, these results confirm that the interaction strength of the trailing motor head with the microtubule lattice (Fig. 4A, in the ATP waiting state) crucially determines the effective sidestepping probability of Kip3.
Extending the Neck Linker of Kinesin-1 Leads to Sidestepping at Limiting ATP Conditions.
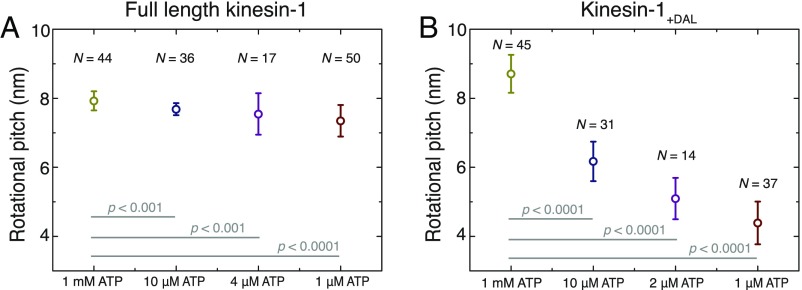
To investigate whether the proposed sidestepping mechanism of Kip3 can be generalized, we performed kinesin-1 gliding assays at varying ATP concentrations (1 mM, 10 µM, 4 µM, and 1 µM), prolonging the ATP waiting state to the same range as for Kip3 at 1 mM ATP. We found that the rotational pitches decreased only slightly from 7.9 ± 0.3 μm at 1 mM ATP (n = 44) to 7.3 ± 0.5 μm at 1 µM ATP (n = 50), even though the average dwell time per forward step increased significantly from 10 ms to more than 900 ms, sixfold longer than for Kip3 at 1 mM ATP (Fig. 5A). The observed decrease in the rotational pitch corresponds to an effective sidestepping probability of ∼0.1%, effectively only one sidestep per 1,000 forward steps, consistent with earlier studies that kinesin-1 strictly follows individual microtubule protofilaments (22, 30, 38).
Fig. 5.
Kinesin-1 with an extended neck linker switches protofilaments at limiting ATP concentration. (A) Rotational pitch of microtubules in gliding assays with wild-type kinesin-1 at various ATP concentrations. The rotational pitch decreased only slightly from 7.9 ± 0.3 μm at 1 mM ATP (n = 44) to 7.3 ± 0.5 μm at 1 µM ATP (n = 50), even though the average dwell time per forward step increased significantly from 10 ms (gliding velocity 817 ± 5 nm/s) to more than 900 ms (gliding velocity 9 ± 1 nm/s). (B) Rotational pitch of microtubules in gliding assays with the kinesin-1+DAL mutant (neck linker extended by 3-aa DAL at the C-terminal direction) at various ATP concentrations. The rotational pitch decreased roughly twofold from 8.7 ± 0.6 μm at 1 mM ATP (n = 45) to 4.4 ± 0.6 μm at 1 µM ATP (n = 37), while the average dwell time per forward step increased from 17 ms (gliding velocity 485 ± 6 nm/s) to 1,600 ms (gliding velocity 5 ± 1 nm/s).
Recently, it has been suggested that motors from the kinesin-2 and the kinesin-8 families can switch protofilaments due to an intrinsically longer neck linker than kinesin-1 (17 aa in comparison with 14 aa) (19, 21). Likewise, it was found that kinesin-1 mutants with very long neck-linker inserts stochastically sidestep (39) and kinesin-1 mutants with short neck-linker inserts (3 additional amino acids) sidestep in the presence of obstacles on the microtubule lattice (40). We therefore asked whether a short neck-linker insert would enable kinesin-1 to sidestep even in the absence of obstacles and whether, similar to Kip3, the sidestepping was dependent on the ATP waiting time. In gliding assays with the kinesin-1+DAL mutant [neck linker extended by the 3-aa DAL in the C-terminal direction (41)] at 1 mM ATP we observed a rotational pitch of 8.7 ± 0.6 µm (n = 45), indicating the absence of sidestepping. However, on performing gliding assays in limiting ATP concentrations (10 µM, 2 µM, and 1 µM) the rotational pitch decreased by up to ∼50% at 1 µM ATP (4.4 ± 0.6 µm; n = 37), while the average dwell time per forward step increased from 17 ms at 1 mM ATP to 1,600 ms at 1 µM ATP (Fig. 5B). This indicates that the neck-linker extension enables the sidestepping of kinesin-1+DAL under conditions where the ATP waiting state is prolonged (limiting ATP conditions). At 1 µM ATP the effective sidestepping probability of kinesin-1+DAL was ∼1.3% (1 sidestep per 77 forward steps) but higher effective sidestepping probabilities may be possible on further prolonging the ATP waiting state.
Discussion
Single Kip3 Motors Sidestep in a Directionally Biased Manner.
Using an assay that allows for tracking the 3D motion of Kip3-QD conjugates on freely suspended microtubules in the absence of external load, we observed that single Kip3 motor proteins moved in left-handed helical trajectories around the longitudinal axis of microtubules (Fig. 1). The rotational pitch was significantly shorter than the supertwist of the employed microtubules, implying 1 effective directional sidestep per 21 forward steps. Thus, our results validate the earlier hypothesis that Kip3 does not exclusively move along a single microtubule protofilament, but also sidesteps to neighboring protofilaments with a bias to the left (21). Recently, Bugiel et al. (24) verified sidestepping of single Kip3 motors in optical tweezers assays but suggested that protofilament switching occurred intrinsically unbiased toward both directions. However, in contrast to our experiments, in that study motility was observed under axial and/or longitudinal load with 590 nm silica microspheres attached to the motors. This experimental design may potentially have influenced the stepping mechanism of Kip3, e.g., by increased transition rates into the weakly bound slip state (23).
Proposed Model for the Kip3 Sidestepping Mechanism.
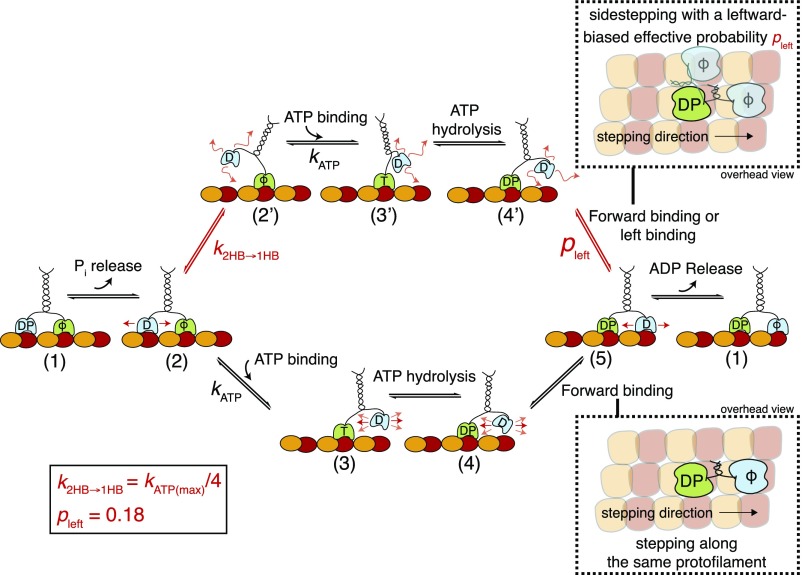
We observed that Kip3 sidestepping was not directly coupled to forward stepping. Rather, the effective sidestepping probability increased with the average dwell time per forward step in experiments performed at physiological ATP concentration [millimolar concentrations present in cells (42)] or at limiting ATP concentrations (Figs. 2 and 3). In contrast, when modifying the dwell time per forward step by varying the temperature, we observed only a marginal change in the effective sidestepping probability over a large range of average dwell times per forward step (SI Appendix, Fig. S8D). This implies that sidestepping is not linked to the complete step (as all rates are assumed to be similarly affected by temperature) but rather to a specific state in the step cycle. Increasing the dwell time of the motor in the posthydrolysis ADP.Pi state by slowing down phosphate release (via applying additional Pi in imaging solution) also did not significantly affect the effective sidestepping probability (SI Appendix, Fig. S8E). Taken together, these findings suggest that sidestepping is linked to the ATP waiting state. We therefore propose a model (Fig. 6) based on the following: (i) The ATP waiting state (state 2) is one of the rate-limiting steps in the Kip3 step cycle (), (ii) the transition from the 2HB conformation to the 1HB conformation occurs in the ATP waiting state (state 2 to state 2′), and (iii) the sidestepping of Kip3 is strongly enhanced in the 1HB conformation (state 4′ to state 5).
Fig. 6.
Model of the Kip3 sidestepping mechanism. After completing a step, the motor has released ADP from the leading head and has both heads tightly bound to the microtubule (1). After Pi release from the trailing head, the interaction of this head with the microtubule becomes weaker, but both heads are still bound to the microtubule (2). While waiting for ATP in this 2HB conformation, a bifurcation in the pathway occurs: The leading head (lower path) binds ATP () with the motor in the 2HB conformation or the trailing head (upper path) unbinds from the microtubule before ATP binding and the motor transitions to a 1HB conformation (2′). This transition () is nucleotide independent and can explain different sidestepping probabilities per forward step: If the leading head (lower path) binds ATP with the motor in the 2HB conformation (2), the trailing head moves forward (3), and ATP hydrolysis () induces the binding of this head (4 → 5). Here, the neck linker is restricted and binding is limited to the tubulin dimer directly in front of the bound head (forward stepping on the same protofilament). If the leading head (upper path) binds ATP with the motor in the 1HB conformation (2′), the trailing head is moved forward (3′), and ATP hydrolysis also induces binding (4′ → 5). However, here the neck linker is less restricted and the unbound head can access more binding sites (e.g., forward and leftward). If we assume that the most probable binding site alternative to the one in the front is directly to the left of the bound head (21), we can add a probability pleft (= 0.18) for switching to the protofilament on the left. At physiological ATP conditions (1 mM), both pathways are possible but the effective sidestepping probability is limited because the transition from the 2HB to the 1HB conformation is less likely than ATP binding to the motor in the 2HB conformation (). In contrast, at low ATP conditions, the 1HB conformation becomes more likely, leading to an increased sidestepping probability. Consequently, the effective sidestepping probability saturates (approaching ) at limiting ATP concentration where the 1HB conformation (2′) is predominantly occupied. Here, T denotes ATP, D denotes ADP, DP denotes ADP plus phosphate, and ϕ denotes no nucleotide. Loose binding and diffusion are represented by small red arrows.
First, we hypothesize that the ATP waiting state is one of the rate-limiting steps in the Kip3 step cycle at physiological ATP concentration. Here, we assume that Kip3 has a similar stepping mechanism to that of kinesin-1 and therefore we use the step cycle of kinesin-1 as in ref. 43. However, in contrast to kinesin-1 [∼10–15 ms average dwell time per forward step (35, 44)], Kip3 spends 10-fold more time per forward step (140–170 ms), which means that at least one of the substeps needs to be prolonged. While the posthydrolysis ADP.Pi state is the rate-limiting step for kinesin-1 (36), we show that for Kip3 the longer average dwell time per step cannot be attributed to the ADP.Pi state (Fig. 6, state 1) because only a minor increase (∼3%) in the average dwell time per forward step was observed upon adding Pi into the imaging solution. Further, we believe that ATP hydrolysis (Fig. 6, state 3), SI Appendix, Fig. S8F), is as efficient as in kinesin-1 (thydrolysis ms) (35) and should not increase the dwell time per forward step. Finally, while we cannot rule out a prolonged dwell time in state 4, an elongation of this state would make the motor vulnerable to premature Pi release followed by microtubule unbinding (as shown for kinesin-1) (45). Hence, we propose that ATP binding is rate limiting. Possible explanations for the slow kinetics of ATP binding—but efficient ATP hydrolysis—involve the presence of structural elements in the motor domain that block the ATP binding site (similar to kinesin-5) (46) and/or front-head gating, where rearward strain slows nucleotide binding to the front head (47, 48). Without further biochemical data, a more detailed description is not possible at this point and for simplicity we assume that ATP binding can be estimated with the measured Michaelis–Menten constant from control Kip3 stepping assays (SI Appendix, Fig. S8F).
Second, we suggest that Kip3 binds ATP in either a 2HB or a 1HB conformation with the leading head (without nucleotide) tightly bound to the microtubule lattice. Here, the trailing head (ADP bound) is either weakly interacting with the microtubule (state 2) or remains freely diffusive (within a space confined by the motor geometry; state 2′). The suggested nucleotide-independent transition from a 2HB conformation to a 1HB conformation has been previously reported for kinesin-1 (31–35, 43) if the ATP waiting state was substantially prolonged by performing experiments at limiting ATP concentrations. Because we assume that Kip3 spends most of the step cycle in the ATP waiting state (∼140–170 ms), which is more than 20-fold longer than for kinesin-1 (<8 ms) (35), the transition from the 2HB to the 1HB conformation would occur not only at limiting ATP concentrations but already at physiological ATP concentrations. Therefore, ATP can bind in both the 2HB and the 1HB conformation at physiological conditions, but the likelihood of ATP binding in the 1HB conformation increases at limiting ATP concentrations.
Third, we propose an enhanced sidestepping probability if Kip3 binds ATP in the 1HB conformation. While we can only speculate about the molecular origin of such a behavior, it is conceivable that both the length and the docking conformation of the neck linker are of importance: (i) The accessible tubulin binding sites depend on the reach of the unbound motor head, which strongly relates to the structure of the neck linker. Kip3 has a longer neck linker (17 aa) than kinesin-1 (14 aa) (21) and the first heptad of the neck coiled coil is predicted to be loosely coiled (49). This gives the unbound Kip3 motor head the capacity to reach tubulin binding sites on neighboring protofilaments. (ii) The reach of the unbound motor head (post-ATP binding and hydrolysis; state 4 or 4′ in Fig. 6) is determined by the interaction of the neck linker with the motor domain (49, 50). For example, partial or complete neck-linker docking in the 2HB conformation (while waiting for ATP; state 2) can potentially restrict the actual reach of the motor head during the diffusive search for the next tubulin binding site (post-ATP binding and hydrolysis; states 3 and 4). In contrast, the transition from the 2HB to the 1HB conformation (in the ATP waiting state; state 2′) may lead to the complete undocking of the neck linker from the motor domain and therefore allow—upon ATP binding and hydrolysis (states 3′ and 4′)—the motor to access more microtubule binding sites than just the forward binding site. In our numerical model, we currently assume that Kip3 can sidestep only when binding ATP in the 1HB conformation and that the only possible binding site (alternative to the forward binding site) is situated directly to the left of the bound head (21).
While the described model reproduces our experimental results very well, we cannot rule out a number of additional scenarios: (i) Kip3 also sidesteps with a basal probability when binding ATP in the 2HB conformation. In our numerical model, this scenario can be accounted for by introducing an additional sidestepping probability pleft(2HB). However, to reproduce our experimental results (in particular the initial slope of the data in Fig. 3B), pleft(2HB) would need to be at least an order of magnitude smaller than pleft, indicating that the 1HB conformation is the main state from which the motor can sidestep (SI Appendix, Fig. S9A). (ii) Kip3 does not exclusively sidestep toward the left, but toward the left and the right with a leftward bias. This scenario can be accounted for in our model by incorporating an additional probability for sidestepping toward the right and adjusting the other simulation parameters accordingly. This addition does not change the experimentally observed increase in the effective sidestepping probability as a function of average dwell time per forward step but rather increases the variance of the effective sidestepping probability for the individual motors (SI Appendix, Fig. S9B). Comparing this variance between experimental and numerical results suggests that the ratio of rightward steps to leftward steps must be small, strongly favoring sidestepping toward the left over sidestepping toward the right. This suggestion is in accordance with the asymmetric geometry of the motor–neck-linker complex when bound to the microtubule (21). (iii) In addition to the leftward sidesteps as described in the model, Kip3 switches protofilaments in an unbiased manner when switching out of the step cycle to a diffusive slip state that is suggested to occur upon Pi release from the microtubule-bound motor head in state 4 (Fig. 6) (23). This might explain the observations made in Bugiel et al. (24) where it was shown that Kip3 switches protofilaments in both directions upon sideward force application [note that the likelihood to switch to the slip state increases exponentially with force (23)]. In fact, while active leftward sidestepping incorporated within the step cycle may allow the motor to reach the microtubule plus end efficiently, unbiased sidestepping in a diffusive state may allow more efficient circumnavigation of roadblocks under high traffic (51). Kip3 might have evolved a refined dual-mode sidestepping strategy to tackle variable traffic situations. The exact sidestepping mechanism employed by Kip3 can most likely be determined in future work only by resolving individual sidesteps using high-precision and high-speed 3D tracking of Kip3 motor heads on suspended microtubules without external load.
Effective Sidestepping Probability Can Be Regulated by Microtubule and Buffer Modification.
Irrespective of the above discussed scenarios, our model suggests that the interaction strength of the trailing head with the microtubule surface in the ATP waiting state determines the effective sidestepping probability of Kip3. Reducing the interaction strength and thereby increasing the transition rate from the 2HB to the 1HB conformation () in the ATP waiting state should increase the effective sidestepping probability. Indeed, shorter rotational pitches were observed in gliding rotation assays with subtilisin-treated microtubules (Fig. 4B) or with higher salt concentration in the buffer (Fig. 4C). There, the digestion (subtilisin treatment) or screening (increased salt) of the negatively charged microtubule e-hooks leads to a reduced electrostatic interaction between structural elements in the kinesin-8 motor domain [loop 2 and loop 12 (52, 53)] and the microtubule lattice, in the ADP state (37). Our observations suggest that posttranslational modifications of microtubules could affect the sidestepping mechanism of Kip3 without the need for motor regulatory factors.
General Strategy for Kinesin Motors to Circumvent Obstacles.
Most rotational studies on plus-end–directed kinesins (20, 21, 54, 55) including a kinesin-1 mutant with a large flexible insert at the neck linker (20) revealed left-handed trajectories around microtubules. Since all these motors have longer neck linkers than wild-type kinesin-1 (21, 56), the sidestepping probability could be dependent on the neck-linker length. In addition, while the transition from the 2HB to the 1HB conformation () could be similar between various kinesin motors (35), the ATP waiting state could vary and the transition to the 1HB conformation is not likely if the ATP binding rate is high (). Therefore, both of these factors have to coincide to observe sidestepping. This can be seen in the experimental data from our gliding rotation assays at varying ATP concentrations with wild-type kinesin-1 and the kinesin-1+DAL mutant (neck linker extended by 3 aa). At 1 µM ATP, virtually no sidestepping was observed for wild-type kinesin-1, whereas significant sidestepping was observed for kinesin-1+DAL (1 sidestep per 77 forward steps). If we assume that at this ATP concentration both motor constructs bind ATP only in the 1HB conformation, the data suggest a probability of less than 0.001 for wild-type kinesin-1 and 0.013 for kinesin-1+DAL. Based on this observation we propose that all processive kinesins may have an alternative pathway in the step cycle where they could transition to a 1HB conformation. We believe this transition occurs stochastically if the ATP waiting state becomes rate limiting or can be enforced if the motors encounter an obstacle at the forward binding site. Directly in front of an obstacle, motors are expected to wait in the 1HB conformation, most likely after ATP hydrolysis, but the unbound head cannot attach to the forward binding site. Because kinesin motors with short neck linkers cannot reach any other binding site, they would pause and detach at the obstacles (18, 57–59) while other kinesins with longer neck linkers could switch the protofilament to circumvent the obstacles (kinesin-2) (19, 40).
In conclusion, we suggest that Kip3 has evolved an elaborate sidestepping mechanism to switch protofilaments to avoid obstacles along the microtubule lattice and potentially also to navigate traffic jams at microtubule ends (60). Directed sidestepping incorporated within the motor step cycle enables the motor to reach the end of crowded microtubules in a single processive run, i.e., without obstacle-forced detachment.
Materials and Methods
Protein Expression and Purification.
Kip3-mfGFP.
The Kip3 motors under investigation are based on a construct with a C-terminally fused mfGFP tag, where a loop in the GFP sequence contains an octo-histidine, a streptavidin-binding peptide (SBP), and a c-Myc tag in tandem (61). Details of the expression and purification are provided in SI Appendix, Kip3-mfGFP Expression and Purification.
Wild-type kinesin-1 and kinesin-1 neck-linker mutant kinesin-1+DAL.
Full-length Drosophila melanogaster kinesin-1 heavy chain (DmKHC) with a C-terminal histidine tag was expressed and purified as described in Korten et al. (62). Kinesin-1+DAL, a kinesin-1 mutant construct truncated at residue 559 (with a C-terminal histidine tag), having an extension of 3-aa residues (DAL) at the C-terminal direction of the neck linker, was a generous gift from the laboratory of William O. Hancock, Department of Biomedical Engineering, Pennsylvania State University, University Park, PA. The construct was expressed and purified as described in Shastry and Hancock (63).
Fabrication of Polymer Structures on Glass.
The patterned glass coverslips were fabricated using UV Nanoimprint lithography (UV-NIL) (64). The detailed protocol used for fabrication is provided in SI Appendix, Fabrication of Polymer Structures on Glass.
In Vitro Motility Assays.
We have described the Parallax 3D single-molecule imaging setup (27) in SI Appendix, 3D Single Molecule Imaging Set-Up, Parallax and provided details on the Kip3-QD stepping assay on suspended microtubules; gliding assays with Kip3, kinesin-1, and kinesin-1+DAL; and other control assays in SI Appendix, In Vitro Motility Assays. Further, we have described the image acquisition and FIESTA based image analysis protocols used for the different motility assays (65) (SI Appendix, Image Acquisition and SI Appendix, Image Analysis). Finally, the details on fitting the Kip3-QDot tracks are provided in SI Appendix, Fitting of Kip3-QDot Tracks.
Calculation of Effective Sidestepping Probability and Rate.
Double-stabilized microtubules used in the experiment have a left-handed supertwist with a pitch of (22), a tubulin interdimer distance of (66), and 14 protofilaments. Using this information, the forward stepping rate, average dwell time per forward step, effective sidestepping probability per forward step, and the effective sidestepping rate were calculated for all of the Kip3-QD events and gliding events, as detailed in SI Appendix, Calculation of Stepping Parameters.
Numerical Simulations.
Kip3 gliding and stepping events were simulated as single Poisson steppers, using Monte Carlo simulations on a MATLAB-based computer program. Here, we assume three states: (i) a 2HB ATP waiting state (2HB-ATP; Fig. 6, state 2); (ii) a 1HB ATP waiting state (1HB-ATP; state 2′); and (iii) a hydrolysis state that includes ATP hydrolysis, motor head attachment, and ADP as well as Pi release (remainder of the cycle). The simplification for the states was based on the hypothesis that there are two different motor conformations when Kip3 waits for ATP (therefore ATP dependent), while the rest of the step cycle is independent of the ATP concentration. Furthermore, the binding rates for ATP as well as the remaining transition rates were the same in both conformations. The transition from the 2HB-ATP to 1HB-ATP () is nucleotide independent and was estimated from the effective sidestepping probability at physiological ATP conditions. To account for the experimentally observed velocity variations (at the same ATP concentrations) a probability of futile steps pfutile was added, which introduces a heterogeneity in the ATP binding rate for each simulated motor. The parameters and were determined experimentally and the parameters and were approximated from the best fit of the model to the data. Averaged dwell times per forward step and effective sidestepping probabilities for each simulated event were obtained by averaging over two rotations (14 protofilament switches constitute one rotation) for stepping events and by averaging over six rotations for gliding events. The simulation code is provided in SI Appendix, MATLAB Code for Numerical Simulation of the Stepping Mechanism and the parameters for the simulation are listed in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank Marcus Braun, Hauke Drechsler, Veikko Geyer, Till Korten, and Zdenek Lansky for comments on the manuscript; all members of the S.D. laboratory as well as Michael Bugiel and Erik Schäffer for fruitful discussions; Wilhelm Walter for expression of the Kip3-mfGFP construct; William Hancock for providing the Kinesin-1+DAL construct and related discussions; Friedrich Schwarz for help in building the Parallax setup; the Microstructure Facility of Center for Molecular and Cellular Bioengineering, in part funded by the State of Saxony and the European Fund for Regional Development — EFRE, for the production of the patterned coverslips; and Corina Bräuer for technical support. We acknowledge financial support from the European Research Council (Starting Grant 242933), the Deutsche Forschungsgemeinschaft through the Sonderforschungsbereich 1027 (Project A8) and the Center for Advancing Electronics Dresden, and the Technische Universität Dresden.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801820115/-/DCSupplemental.
References
- 1.DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin SG. Microtubule-dependent cell morphogenesis in the fission yeast. Trends Cell Biol. 2009;19:447–454. doi: 10.1016/j.tcb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Chang F, Nurse P. How fission yeast fission in the middle. Cell. 1996;84:191–194. doi: 10.1016/s0092-8674(00)80973-3. [DOI] [PubMed] [Google Scholar]
- 6.Tran PT, Marsh L, Doye V, Inoué S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niwa S, et al. KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev Cell. 2012;23:1167–1175. doi: 10.1016/j.devcel.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Varga V, et al. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 9.Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 10.Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 11.Su X, et al. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reese L, Melbinger A, Frey E. Crowding of molecular motors determines microtubule depolymerization. Biophys J. 2011;101:2190–2200. doi: 10.1016/j.bpj.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melbinger A, Reese L, Frey E. Microtubule length regulation by molecular motors. Phys Rev Lett. 2012;108:258104. doi: 10.1103/PhysRevLett.108.258104. [DOI] [PubMed] [Google Scholar]
- 14.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 15.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Rolls MM, Hancock WO. An EB1-kinesin complex is sufficient to steer microtubule growth in vitro. Curr Biol. 2014;24:316–321. doi: 10.1016/j.cub.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amos LA, Schlieper D. Microtubules and maps. Adv Protein Chem. 2005;71:257–298. doi: 10.1016/S0065-3233(04)71007-4. [DOI] [PubMed] [Google Scholar]
- 18.Schneider R, Korten T, Walter WJ, Diez S. Kinesin-1 motors can circumvent permanent roadblocks by side-shifting to neighboring protofilaments. Biophys J. 2015;108:2249–2257. doi: 10.1016/j.bpj.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoeprich GJ, Thompson AR, McVicker DP, Hancock WO, Berger CL. Kinesin’s neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys J. 2014;106:1691–1700. doi: 10.1016/j.bpj.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunnbauer M, et al. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol Cell. 2012;46:147–158. doi: 10.1016/j.molcel.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Bormuth V, et al. The highly processive kinesin-8, Kip3, switches microtubule protofilaments with a bias toward the left. Biophys J. 2012;103:L4–L6. doi: 10.1016/j.bpj.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitra A, Ruhnow F, Nitzsche B, Diez S. Impact-free measurement of microtubule rotations on kinesin and cytoplasmic-dynein coated surfaces. PLoS One. 2015;10:e0136920. doi: 10.1371/journal.pone.0136920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jannasch A, Bormuth V, Storch M, Howard J, Schäffer E. Kinesin-8 is a low-force motor protein with a weakly bound slip state. Biophys J. 2013;104:2456–2464. doi: 10.1016/j.bpj.2013.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bugiel M, Böhl E, Schäffer E. The kinesin-8 Kip3 switches protofilaments in a sideward random walk asymmetrically biased by force. Biophys J. 2015;108:2019–2027. doi: 10.1016/j.bpj.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bugiel M, Mitra A, Girardo S, Diez S, Schäffer E. Measuring microtubule supertwist and defects by three-dimensional-force-clamp tracking of single kinesin-1 motors. Nano Lett. 2018;18:1290–1295. doi: 10.1021/acs.nanolett.7b04971. [DOI] [PubMed] [Google Scholar]
- 26.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, McKenna JD, Murray JM, Ostap EM, Goldman YE. Parallax: High accuracy three-dimensional single molecule tracking using split images. Nano Lett. 2009;9:2676–2682. doi: 10.1021/nl901129j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruhnow F, Kloβ L, Diez S. Challenges in estimating the motility parameters of single processive motor proteins. Biophys J. 2017;113:2433–2443. doi: 10.1016/j.bpj.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyman AA, Chrétien D, Arnal I, Wade RH. Structural changes accompanying GTP hydrolysis in microtubules: Information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J Cell Biol. 1995;128:117–125. doi: 10.1083/jcb.128.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nitzsche B, Ruhnow F, Diez S. Quantum-dot-assisted characterization of microtubule rotations during cargo transport. Nat Nanotechnol. 2008;3:552–556. doi: 10.1038/nnano.2008.216. [DOI] [PubMed] [Google Scholar]
- 31.Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 32.Asenjo AB, Sosa H. A mobile kinesin-head intermediate during the ATP-waiting state. Proc Natl Acad Sci USA. 2009;106:5657–5662. doi: 10.1073/pnas.0808355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toprak E, Yildiz A, Hoffman MT, Rosenfeld SS, Selvin PR. Why kinesin is so processive. Proc Natl Acad Sci USA. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verbrugge S, Lansky Z, Peterman EJG. Kinesin’s step dissected with single-motor FRET. Proc Natl Acad Sci USA. 2009;106:17741–17746. doi: 10.1073/pnas.0905177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mickolajczyk KJ, et al. Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proc Natl Acad Sci USA. 2015;112:E7186–E7193. doi: 10.1073/pnas.1517638112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milic B, Andreasson JOL, Hancock WO, Block SM. Kinesin processivity is gated by phosphate release. Proc Natl Acad Sci USA. 2014;111:14136–14140. doi: 10.1073/pnas.1410943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bormuth V, Varga V, Howard J, Schäffer E. Protein friction limits diffusive and directed movements of kinesin motors on microtubules. Science. 2009;325:870–873. doi: 10.1126/science.1174923. [DOI] [PubMed] [Google Scholar]
- 38.Ray S, Meyhöfer E, Milligan RA, Howard J. Kinesin follows the microtubule’s protofilament axis. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoeprich GJ, Mickolajczyk KJ, Nelson SR, Hancock WO, Berger CL. The axonal transport motor kinesin-2 navigates microtubule obstacles via protofilament switching. Traffic. 2017;18:304–314. doi: 10.1111/tra.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muthukrishnan G, Zhang Y, Shastry S, Hancock WO. The processivity of kinesin-2 motors suggests diminished front-head gating. Curr Biol. 2009;19:442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel A, et al. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 43.Hancock WO. The kinesin-1 chemomechanical cycle: Stepping toward a consensus. Biophys J. 2016;110:1216–1225. doi: 10.1016/j.bpj.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coy DL, Wagenbach M, Howard J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem. 1999;274:3667–3671. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- 45.Mickolajczyk KJ, Hancock WO. Kinesin processivity is determined by a kinetic race from a vulnerable one-head-bound state. Biophys J. 2017;112:2615–2623. doi: 10.1016/j.bpj.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muretta JM, et al. The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc Natl Acad Sci USA. 2015;112:E6606–E6613. doi: 10.1073/pnas.1512305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen GY, Mickolajczyk KJ, Hancock WO. The kinesin-5 chemomechanical cycle is dominated by a two-heads-bound state. J Biol Chem. 2016;291:20283–20294. doi: 10.1074/jbc.M116.730697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Thirumalai D. Dissecting the kinematics of the kinesin step. Structure. 2012;20:628–640. doi: 10.1016/j.str.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Goldtzvik Y, Thirumalai D. Parsing the roles of neck-linker docking and tethered head diffusion in the stepping dynamics of kinesin. Proc Natl Acad Sci USA. 2017;114:E9838–E9845. doi: 10.1073/pnas.1706014114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertalan Z, Budrikis Z, La Porta CAM, Zapperi S. Navigation strategies of motor proteins on decorated tracks. PLoS One. 2015;10:e0136945. doi: 10.1371/journal.pone.0136945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, et al. Motility and microtubule depolymerization mechanisms of the kinesin-8 motor, KIF19A. eLife. 2016;5:e18101. doi: 10.7554/eLife.18101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arellano-Santoyo H, et al. A tubulin binding switch underlies Kip3/kinesin-8 depolymerase activity. Dev Cell. 2017;42:37–51.e8. doi: 10.1016/j.devcel.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yajima J, Mizutani K, Nishizaka T. A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat Struct Mol Biol. 2008;15:1119–1121. doi: 10.1038/nsmb.1491. [DOI] [PubMed] [Google Scholar]
- 55.Pan X, Acar S, Scholey JM. Torque generation by one of the motor subunits of heterotrimeric kinesin-2. Biochem Biophys Res Commun. 2010;401:53–57. doi: 10.1016/j.bbrc.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hariharan V, Hancock WO. Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell Mol Bioeng. 2009;2:177–189. doi: 10.1007/s12195-009-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korten T, Diez S. Setting up roadblocks for kinesin-1: Mechanism for the selective speed control of cargo carrying microtubules. Lab Chip. 2008;8:1441–1447. doi: 10.1039/b803585g. [DOI] [PubMed] [Google Scholar]
- 58.Tarhan MC, Orazov Y, Yokokawa R, Karsten SL, Fujita H. Biosensing MAPs as “roadblocks”: Kinesin-based functional analysis of tau protein isoforms and mutants using suspended microtubules (sMTs) Lab Chip. 2013;13:3217–3224. doi: 10.1039/c3lc50151e. [DOI] [PubMed] [Google Scholar]
- 59.Telley IA, Bieling P, Surrey T. Obstacles on the microtubule reduce the processivity of kinesin-1 in a minimal in vitro system and in cell extract. Biophys J. 2009;96:3341–3353. doi: 10.1016/j.bpj.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leduc C, et al. Molecular crowding creates traffic jams of kinesin motors on microtubules. Proc Natl Acad Sci USA. 2012;109:6100–6105. doi: 10.1073/pnas.1107281109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobayashi T, et al. Engineering a novel multifunctional green fluorescent protein tag for a wide variety of protein research. PLoS One. 2008;3:e3822. doi: 10.1371/journal.pone.0003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korten T, Chaudhuri S, Tavkin E, Braun M, Diez S. Kinesin-1 expressed in insect cells improves microtubule in vitro gliding performance, long-term stability and guiding efficiency in nanostructures. IEEE Trans Nanobioscience. 2016;15:62–69. doi: 10.1109/TNB.2016.2520832. [DOI] [PubMed] [Google Scholar]
- 63.Shastry S, Hancock WO. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilles S, et al. UV nanoimprint lithography with rigid polymer molds. Microelectron Eng. 2009;86:661–664. [Google Scholar]
- 65.Ruhnow F, Zwicker D, Diez S. Tracking single particles and elongated filaments with nanometer precision. Biophys J. 2011;100:2820–2828. doi: 10.1016/j.bpj.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.