Significance
We describe a situation in which bacteria typical of the human oral microbiome are organized spatially by gliding cells, species of Capnocytophaga, that move backward and forward over the substratum. The mobile adhesins that pull the cells over the substratum also attach to cells of nonmotile bacterial species, which are carried up and down the motile cells as cargo. The synchronized transport of nonmotile cargo bacteria helps to shape a polymicrobial community.
Keywords: polymicrobial biofilm, microbiome, swarm, cargo transport, gliding
Abstract
The human microbiome is an assemblage of diverse bacteria that interact with one another to form communities. Bacteria in a given community are arranged in a 3D matrix with many degrees of freedom. Snapshots of the community display well-defined structures, but the steps required for their assembly are not understood. Here, we show that this construction is carried out with the help of gliding bacteria. Gliding is defined as the motion of cells over a solid or semisolid surface without the necessity of growth or the aid of pili or flagella. Genomic analysis suggests that gliding bacteria are present in human microbial communities. We focus on Capnocytophaga gingivalis, which is present in abundance in the human oral microbiome. Tracking of fluorescently labeled single cells and of gas bubbles carried by fluid flow shows that swarms of C. gingivalis are layered, with cells in the upper layers moving more rapidly than those in the lower layers. Thus, cells also glide on top of one another. Cells of nonmotile bacterial species attach to the surface of C. gingivalis and are propelled as cargo. The cargo cell moves along the length of a C. gingivalis cell, looping from one pole to the other. Multicolor fluorescent spectral imaging of cells of different live but nonmotile bacterial species reveals their long-range transport in a polymicrobial community. A swarm of C. gingivalis transports some nonmotile bacterial species more efficiently than others and helps to shape the spatial organization of a polymicrobial community.
Bacteria that attach to and colonize a surface form some of the most stable species of the human microbiome. Rapid availability of nutrition, protection from antibiotics, and long-term colonization are advantages of attaining a specific spatial niche within a microbial community. The mechanisms that drive interactions and guide the architecture of microbial communities are unclear. Active motion via the help of a molecular motor and guidance by a chemotaxis pathway enables some bacteria to sense their environment and find a spatial niche (1, 2). While the chemotaxis ability of a bacterium improves surface colonization (3, 4), motile bacteria do not always dominate a microbiome. For example, of the 13 abundant genera of bacteria that colonize the human gingival region (5), bacteria of three genera—Capnocyophaga (gliding motility) (6), Neisseria (twitching motility) (7), and Lautropia (flagellar motility) (8)—are known to be motile. Nonmotile bacteria can reach a spatial niche via diffusion or fluid flow. However, diffusion is not an efficient means of transport for an object of micrometer size, and fluid flows can be erratic: a sphere of radius 1 μm suspended in water at room temperature takes ∼2.5 h to diffuse a distance of 60 µm (9). Another way by which nonmotile bacteria can be transported is by adhering to motile microorganisms. Only a few examples of this process are known (10–12), and its overall significance is unclear. Here, we describe a mechanism in which motile but nonflagellated bacteria of the human oral microbiome carry nonmotile bacteria as cargo.
Collective motion is observed in biological objects of all sizes, such as bacteria, insects, birds, and fish (13, 14). The transition of an isolated cell to a swarm can be broadly explained by models based on phase transitions (14). Variations in adhesion forces, the mode of motility, and a bias in the direction of motion govern behavior of a particular species. Swarms of flagellated bacteria comprise dense groups of cells that swim in a thin layer of liquid close to a surface without adhering to that surface (15, 16). Bacteria with type 4 pili are propelled as the pili attach, retract, and detach from the surface, performing twitching motility. Bacteria that use mobile cell-surface adhesins are called gliding bacteria. Bacterial gliding occurs when bacteria move in close contact with an external surface, a process that requires continuous expenditure of energy (17–19).
Bacteria of the phyla Bacteroidetes are abundant in human gut and oral microbiomes, and many members of this phyla have the ability to navigate surfaces via gliding motility. The Bacteroidetes represent about half of the bacterial population of the human gut microbiome (20, 21), and changes in their concentration correlates with diseases such as obesity, diabetes, and colitis (22–24). The Type IX Protein Secretion System (T9SS) is found in many Bacteroidetes and is required for the secretion of proteins such as cell-surface adhesins, chitinase, cellulase, and proteases. T9SS is found in both motile and nonmotile Bacteroidetes. The core T9SS proteins are SprA, SprE, SprT, and GldK-N. Bacteria with the cell-surface adhesin SprB, motility proteins GldA-B, GldD, and GldF-J, and a functional T9SS are able to move over external surfaces (25).
The human oral microbiome is a catalog of microorganisms from lips, teeth, gingiva, tongue, cheek, palate, and contiguous extensions of the oral cavity (26). Bacteroidetes of the genera Porphyromonas, Prevotella, and Capnocytophaga are found in abundance in the human oral microbiome, and they contain the genes to encode the T9SS. Several reports show the abundance of Capnocytophaga sp. in different sites of the human oral microbiome (5, 27–29). Some Capnocytophaga sp. isolated from the human oral microbiome form spreading colonies (6), and cells lacking the T9SS are deficient in gliding (30). Capnocytophaga sp. isolated from the human oral microbiome possess the genes to encode the T9SS and its associated gliding machinery. This suggests that the mechanism of gliding found in the phyla Bacteroidetes is prevalent in the genus Capnocytophaga. However, no experimental data exist about the mechanism for the motion of single cells of the genus Capnocytophaga, nor is anything known about their collective behavior. Recently, via fluorescent labeling of fixed samples of human oral biofilms, Mark Welch et al. showed the presence of a structured and multigenus consortia within human oral microbial communities that contained bacteria of the genus Capnocytophaga (5). How a nonordered community reaches an ordered state and arranges itself into spatial structures is not clear. Sequence and imaging data showed that bacteria of the Capnocytophaga genus are found in abundance in both human supragingival and subgingival biofilms. Why the members of a genus that represents gliding bacteria are stable and abundant in the oral microbiome of healthy humans is not known. Via spectral imaging of live cells, we study the development of a polymicrobial community that contains eight abundant bacterial species of a human subgingival biofilm. We show that swarming Capnocytophaga shape a polymicrobial community by acting as vehicles of public transport and that patterns within a microbial community arise as a result of this active motion. Over longer timescales, a combination of active motion and cell growth can shape the spatial features of a microbial community.
Results and Discussion
T9SS and Its Associated Rotary Motor Are Functional in Gliding Bacteria of the Human Microbiome.
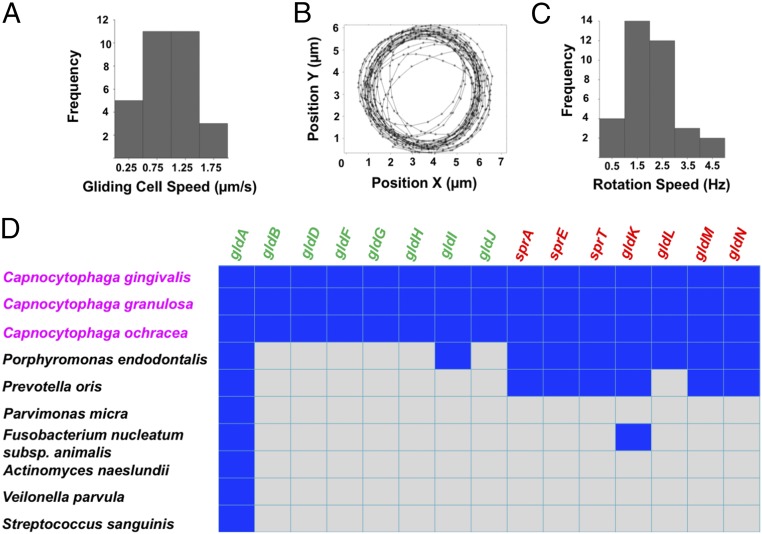
We found that many C. gingivalis cells were able to glide over a glass surface, moving at speeds of about 1 µm/s (Fig. 1A and Movie S1). Also, cells of C. gingivalis could tether to a glass surface and rotate about fixed axes at speeds of about 1.5 Hz (Fig. 1 B and C and Movie S2). This behavior is similar to that of the environmental gliding bacterium Flavobacterium johnsoniae, where rotation of a cell around a fixed axis is attributed to the presence of a rotary motor associated with the T9SS that powers gliding (31). sprA, sprE, sprT, and gldK-N genes encode the proteins that form the core T9SS. gldA-B, gldD, and gldF-J encode proteins that associate with the T9SS to enable gliding motility. The bacterium F. johnsoniae has all 15 genes. T9SS is found in about 62% of the sequenced species of the phyla Bacteroidetes (32). Capnocytophaga species found in the human oral microbiome have all T9SS and gliding motility genes. Of the seven abundant nonmotile bacteria used in this study, Porphyromonas endodontalis and Prevotella oris have T9SS genes, but they lack the gliding motility genes (Fig. 1D). These observations combined with the presence of all T9SS-gliding motility genes and mobile cell-surface adhesins demonstrate that the machinery that enables gliding motility of members of the Bacteroidetes phyla is functional in C. gingivalis.
Fig. 1.
T9SS and its associated rotary motor are functional in gliding bacteria of the human microbiome. (A) The frequency distribution of gliding speeds of a population of C. gingivalis cells peaked around 1 µm/s. (B) The center of mass of a C. gingivalis cell tethered to glass followed a circular trajectory. (C) The frequency distribution of rotation speed of a population of tethered C. gingivalis cells peaked around 1.5 Hz. Experiments in A–C were done at room temperature on glass. (D) A matrix showing the similarity between genes that encode the T9SS and gliding motor proteins of abundant bacteria of the human oral microbiome. The reference gene source was F. johnsoniae. A blue square indicates the presence while a gray square indicates the absence of a gene. BLAST was performed using the Human Oral Microbial Database (26). To eliminate random matches, an e value of e−5 was used as the threshold. The core gliding and core T9SS genes are in green and red, respectively. Bacteria in magenta have all green and red genes and can glide.
Gliding Bacteria from the Human Microbiome Move Collectively.
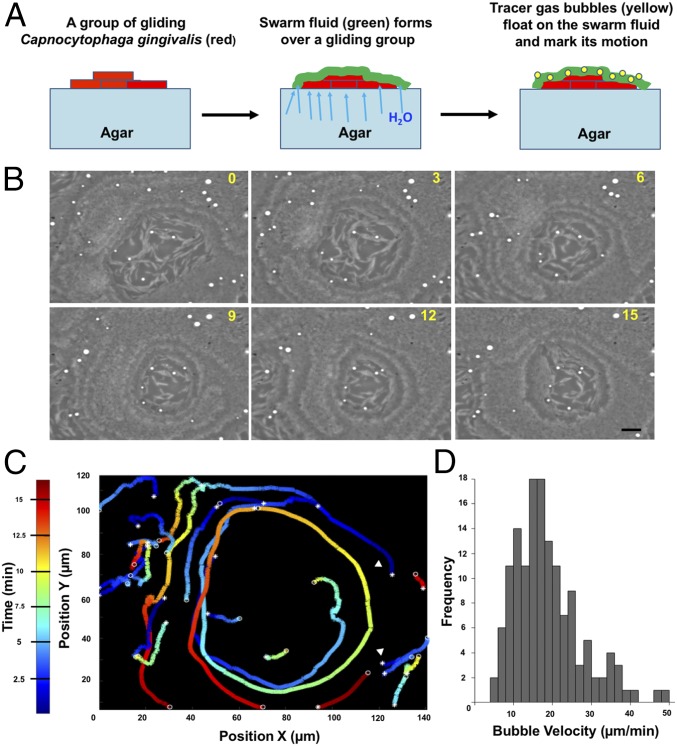
We refer to the collective motion of gliding bacteria as “swarming” and to large groups of moving cells as “swarms.” Compact subgroups that move as a unit are called “slugs.” C. gingivalis cells displayed swarm behavior over the surface of agar. These swarms moved in a synchronized manner, and circular motion of cells was observed near the edge of the swarm. Their motion was tracked with micrometer-sized surfactant-encased gas bubbles. Such bubbles floated on the top layer of the swarm (Fig. 2 A and B and Movie S3). The gas bubbles near the leading edge of a swarm moved in circular trajectories predominantly in the counterclockwise direction (Fig. 2C). The speed of bubbles, which is also the speed of the top layer of the swarm, was around 18 µm/min (Fig. 2D).
Fig. 2.
Synchronized swarm behavior of bacteria from the human microbiome. (A) A graphical representation of the use of gas bubbles to trace fluid flow patterns of a swarm. (B) Time-lapse images of a swarm with gas bubbles (white) moving on top. The time lapse is in minutes. (Scale bar: 10 µm.) (C) Trajectories of the gas bubbles moving in a circular pattern on a swarm. Color map depicts time. (D) A frequency distribution of the speed of gas bubbles. Experiments in B–D were done at 37 °C on an agar pad.
Speeds of Layers of a Swarm Are Additive.
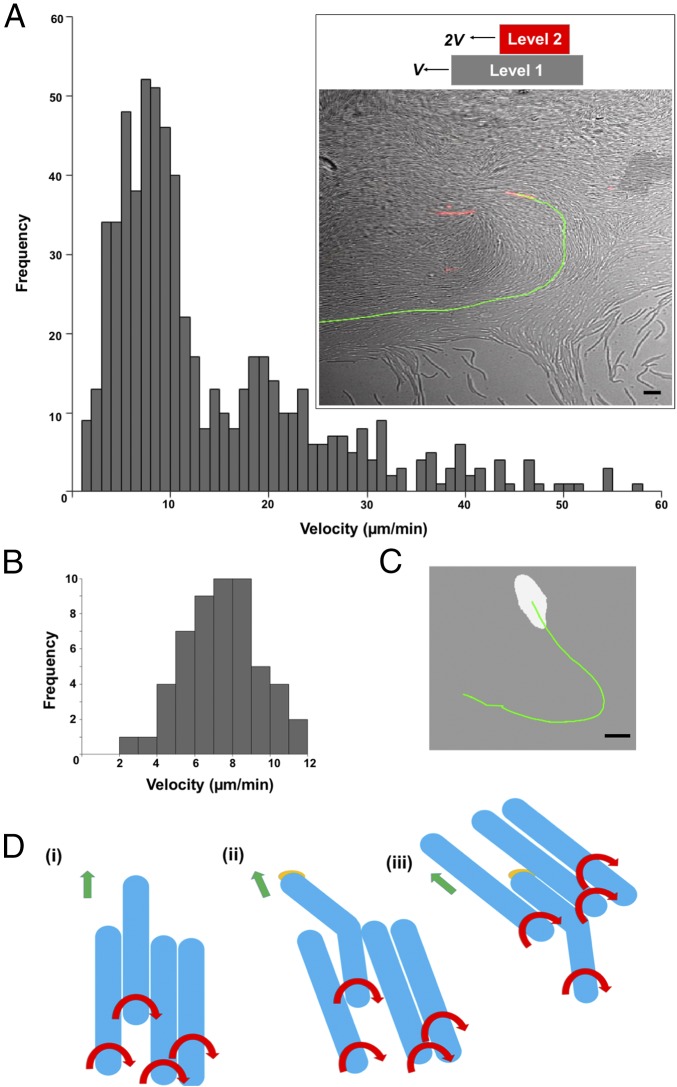
The swarms of C. gingivalis formed layers that were superimposed on top of one other. If layers move in similar directions and cells in the bottom layer move with a speed v, then cells in the second layer would move with a speed 2v. To test this hypothesis, a small percentage of fluorescently labeled C. gingivalis cells were mixed with unlabeled C. gingivalis cells. Fluorescently labeled cells were tracked in a swarm containing both labeled and unlabeled cells (Fig. 3A, Inset, and Movie S4). The speed distribution of cells was bimodal with peaks around both 9 and 18 µm/min (Fig. 3A). Slugs that appeared near the edge of a swarm were tracked, and their speed was around 9 µm/min (Fig. 3 B and C and Movie S5). This suggests that the speed of a swarming cell and its synchronization correlate with its location in a 3D spatial domain.
Fig. 3.
Speeds of layers of a swarm are additive. (A) A bimodal frequency distribution of the speed of fluorescently labeled cells moving within a swarm. (Inset) A swarm with a fluorescently labeled cell (red) and the trajectory of that cell (green). (Scale bar: 5 µm.) (B) The frequency distribution of the speed of slugs (a small group of mobile cells). (C) A slug (white) with the trajectory of its motion in green. (Scale bar: 5 µm.) Experiments in A–C were done at 37 °C on an agar pad. (D) An explanation for how a group of cells moves counterclockwise. (i) A group of flexible cells (blue) moving as right-handed screws. Red arrows show the direction of rotation of the cell body, and green arrows show the direction of motion of a group. (ii) The leading end of one cell sticks to an external surface via multiple adhesins (orange) while the rest of the cell body keeps twisting. This produces a torsion that bends the cell toward the left of the direction of motion of the group. (iii) Cells in a group orient themselves along the cell body of the bent cell, and the group moves counterclockwise.
Counterclockwise Motion of a Swarm Can Be Explained from the Right-Handed Motion of Single Cells.
Single gliding cells move in a manner similar to a right-handed screw. If the front end of a flexible gliding cell moving as a right-handed screw sticks to an external surface, the cell body bends, resulting in the front end of the cell pointing toward the left (Fig. 3D and SI Appendix, Fig. S2). Models that explain group behavior suggest that, in a group, the direction of motion of an individual cell depends on the direction of motion of its neighbors, and perturbations in the noise of the system result in varying patterns of a swarm (14). Cells that are in the vicinity of a bent cell move along the direction of the bend, and this direction of motion gets propagated in a swarm, resulting in its counterclockwise motion (Fig. 3D).
A Cargo-Transporter Relationship Exists Between Nonmotile and Gliding Bacteria of the Human Microbiome.
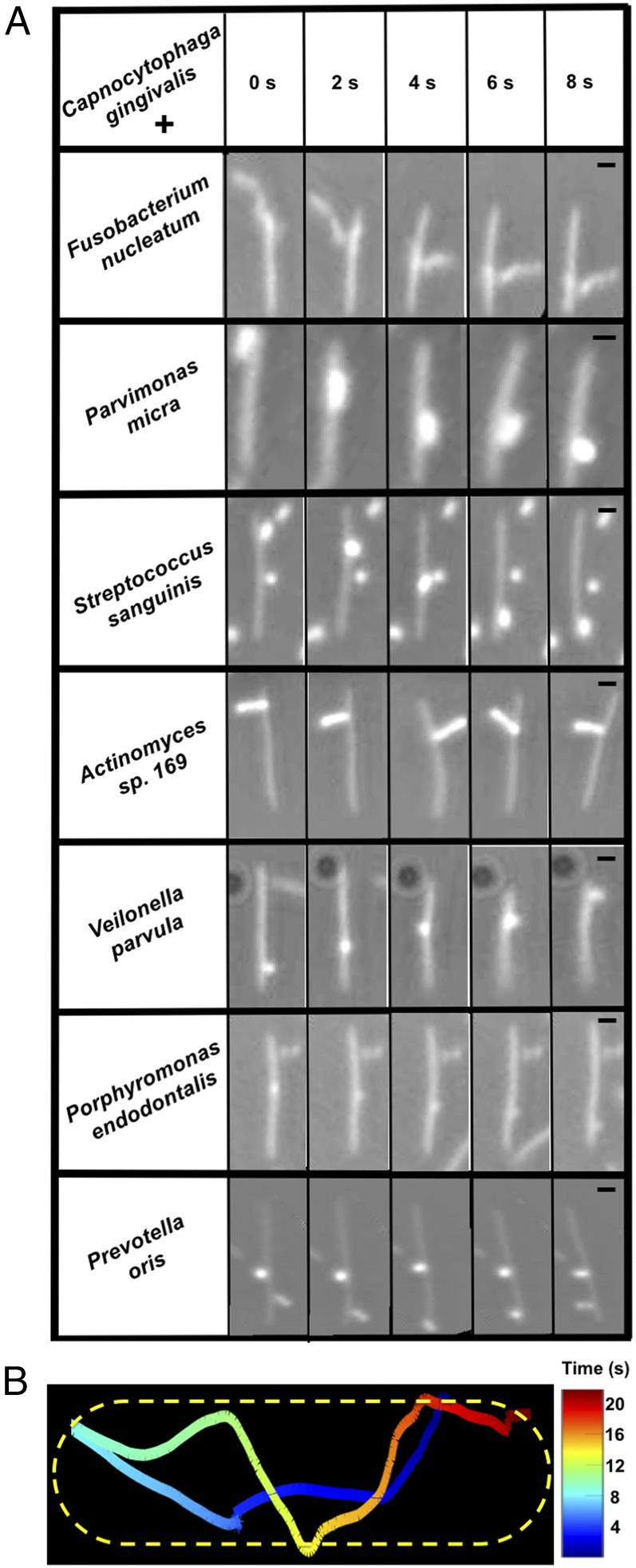
Genes that encode the bacterial T9SS and the cell-surface adhesin SprB are present in C. gingivalis, which is abundant in human oral biofilms. C. gingivalis belongs to the phylum Bacteroidetes and is related to the model gliding bacterium F. johnsoniae. Addition of an antibody raised against the SprB of F. johnsoniae to a culture of C. gingivalis caused cell aggregation (SI Appendix, Fig. S1), suggesting that C. gingivalis carries SprB on its surface. SprB is known to bind to polysaccharides, which are abundant on most cell walls. To test the hypothesis that this adhesin might bind to the cell surfaces of nonmotile bacteria, seven abundant and nonmotile bacterial species found in human subgingival biofilms were cocultured with gliding C. gingivalis. The nonmotile bacteria were P. endodontalis, P. oris, Parvimonas micra, Actinomyces sp. oral taxon-169, Fusobacterium nucleatum, Streptococcus sanguinis, and Veilonella parvula. Single cells of these nonmotile species attached to the cell surface of C. gingivalis and were transported as cargo along a C. gingivalis cell from one pole to the other (Fig. 4A and Movies S6–S12). The adhesin SprB of F. johnsoniae moves in a spiral fashion on the cell-surface (17). A nonmotile bacterium on the surface of a C. gingivalis cell moves in a similar way (Fig. 4B).
Fig. 4.
A cargo-transporter relationship exists between nonmotile and gliding bacteria of the human microbiome. (A) Seven different species of nonmotile bacteria were carried along the surface of C. gingivalis cells, moving from one end of the cell to the other. The images have been aligned to reveal the motion of the cargo relative to the C. gingivalis cell. The raw data are shown in Movies S6–S12. (Scale bar: 1 µm.) The eight bacteria shown in the matrix are some of the abundant microbial species found in a human subgingival biofilm. (B) The trajectory of the nonmotile bacterium P. oris transported along the length of a C. gingivalis cell shown as a detailed example of the cargo-transporter relationship described above. P. oris moved from one pole to the other and looped back after it reached a pole. The C. gingivalis cell is outlined by a dotted line; the color map indicates time. Experiments were done at room temperature on glass.
Long-Range Cargo Transport Shapes a Microbial Community.
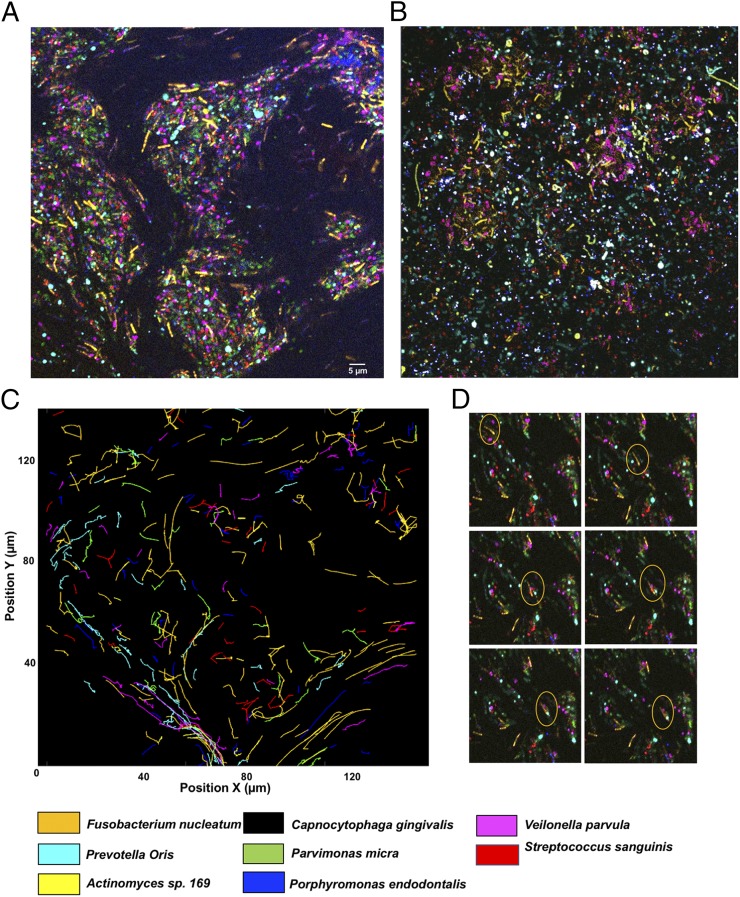
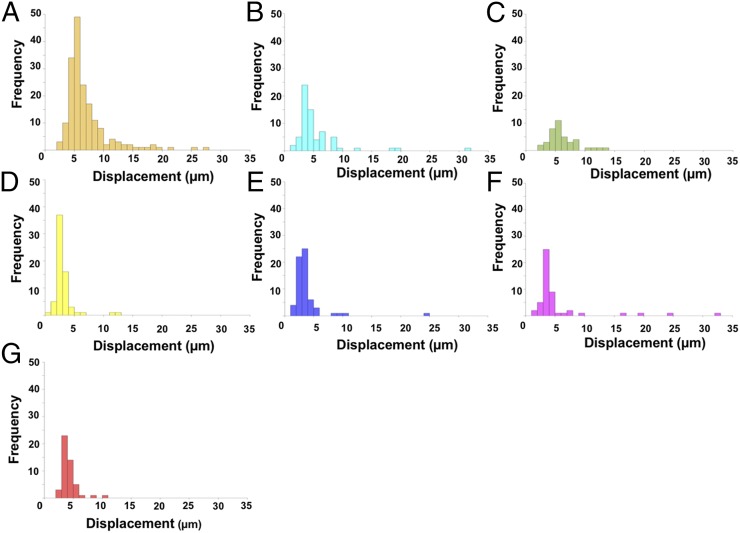
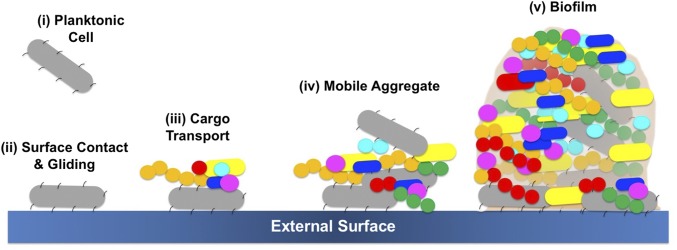
The spatial localization of bacteria was studied in a polymicrobial community comprising C. gingivalis and the seven nonmotile bacteria used for the experiments of Fig. 4A. To explore if C. gingivalis swarms had the capability to transport the nonmotile bacteria over long distances, the cell surfaces of the nonmotile bacteria were labeled using seven different amino-specific succinimidyl ester Alexa Fluor fluorescent dyes. Physiological features, such as the motility of labeled C. gingivalis, remained unaltered, thus providing a powerful tool for live-cell imaging. A polymicrobial community containing seven fluorescently labeled nonmotile bacteria and gliding C. gingivalis was imaged, and spectral imaging was used to separate the fluorescent signals. Long-range transport of nonmotile bacteria by swarming C. gingivalis was observed in a polymicrobial community (Fig. 5A and Movie S13). Within 20 min, the polymicrobial community evolved in such a way that many nonmotile bacteria were arranged as islands surrounded by swarms of C. gingivalis and some nonmotile bacteria (Fig. 5A). A control, in which C. gingivalis was de-energized by prior treatment with sodium azide, exhibited a uniform distribution (Fig. 5B). The nonmotile bacteria were actively transported via the swarms as cargo (Fig. 5C). Polymicrobial aggregates of nonmotile bacteria that were transported by C. gingivalis changed their structures while moving with a swarm (Fig. 5D and Movie S14). Tracking of individual bacterial species revealed that some nonmotile bacteria moved distances of up to 30 µm in a swarm. Some nonmotile bacteria, such as F. nucleatum, used this mode of transport more efficiently than others (Fig. 6 A–E). Since equal numbers of cells of each species were included in the mixture added to the agar pad (Methods), the transport efficiency, the relative number of tracks for each species that appear in Fig. 5C, can be found from the areas of the distributions in Fig. 6. Additional information can be obtained from the lengths of distribution tails. Differences in the composition of the polysaccharides or cell-surface proteins of the nonmotile bacteria could have an impact on their ability to be transported as cargo. Based on our results, we propose a model where a microbial community containing gliding bacteria can use surface motility and cargo transport to find a specific niche. Once a niche is established, the bacteria can arrange in well-defined spatial structures (Fig. 7). Evidently, cargo transport plays a role in forming the spatial features of polymicrobial biofilms that contain Capnocytophaga sp. Such biofilms are widespread in the human gingival regions. It is known that cooperation in the form of mutualism among bacterial species occurs via the sharing of public goods (33). We demonstrate that public transport enables bacterial cooperation and shapes communities of bacteria found in the human body.
Fig. 5.
Long-range cargo transport shapes a microbial community. (A) A microbial community containing fluorescently labeled bacteria. Eight bacteria (seven nonmotile and one motile) of the human microbiome attained a shape in which the nonmotile bacteria were organized in islands by the swarming bacteria. This is a still image from Movie S13. (B) A control in which the experiment of A was repeated with de-energized cells of C. gingivalis. (Scale bar: 5 µm.) Experiments done at 37 °C on an agar pad. (C) Twenty-minute trajectories of the displacement of nonmotile bacteria showing that the swarming microbe transports nonmotile bacteria over long distances. (D) Time-lapse images taken at intervals of 1 min of a small region from Movie S13 showing that dynamically forming polymicrobial aggregates of nonmotile bacteria move long distances in a swarm.
Fig. 6.
(A–G) Frequency distributions of the distances that seven species of nonmotile bacteria were transported by a swarm of C. gingivalis. Different species are indicated in the color code defined in Fig. 4. The frequency distributions are for cells, the tracks of which appear in Fig. 5C.
Fig. 7.
A model depicting how a gliding bacterium from the human microbiome can carry polymicrobial cargo, form mobile aggregates, and find a spatial niche where a polymicrobial biofilm can develop.
Methods
Strains and Growth.
Cells of C. gingivalis ATCC 33624 were used for gliding motility assays. The strain was originally isolated from the periodontal lesion of a human subject. C. gingivalis cells were motile when grown on TSY agar, which contains trypticase soy broth (TSB) 30 g/L, yeast extract 3 g/L, and 1.5% Bacto Agar (Difco). Growth was carried out in a CO2-rich anaerobic environment at high relative humidity. This was achieved by placing the inoculated agar plates in an AnaeroPack System jar (Mitsubishi Gas Chemical) containing three candles and an ignited sheet of Kimtech paper at 37 °C for 48 h. Nonmotile bacteria were originally isolated from the human oral cavity, and they are cataloged in the Human Oral Microbiome Database (26). F. nucleatum, Actinomyces sp. oral taxon-169, P. micra, S. sanguinis, P. oris, and Veilonella parvula were grown on a medium containing TSB 20 g/L, brain heart infusion broth 26 g/L, yeast extract 10 g/L, hemin 5 mg/L, 5% sheep blood, and agar 22 g/L at 37 °C in an anaerobic chamber. P. endodontalis was grown on a medium containing TSB 37 g/L, hemin 5 mg/L, NaHCO3 1 g/L, yeast extract 1 g/L, 5% sheep blood, and 1.5% agar at 37 °C in an anaerobic chamber.
Single-Cell Motility Assay.
C. gingivalis cells were suspended in TSY broth that contained TSB 30 g/L and yeast extract 3 g/L. The cell suspension was injected into a tunnel slide (the space between a slide and a coverslip separated by one layer of double-stick Scotch tape) and was allowed to stand for 5 min. Then cells in the tunnel slide were washed with 0.25% methyl-cellulose (Methocel 90 HG Sigma-Aldrich catalog no. 64680; viscosity of a 2% solution in water at 20 °C 4,000 cP) containing TSY broth. After another 5 min, the gliding cells were imaged using a Nikon Plan 40× BM N.A. 0.65 objective (Nikon) and a ThorLabs DCC1545M-GL camera (ThorLabs). Images were analyzed using a custom MATLAB (The MathWorks) code. To image and analyze tethered and rotating cells, a suspension of C. gingivalis cells was injected into a tunnel slide and washed with TSY broth. Some of the cells tethered to glass and displayed rotation. Images of such cells were captured and analyzed using the method described above.
Generation of Microbubbles and Tracking Fluid Flow.
The method for producing surfactant-encapsulated air bubbles is given in ref. 34. About 70 μL water-insoluble surfactant Span 83 (Sorbitan sesquioleate, S3386; Sigma-Aldrich) was mixed with 100 mL sterile deionized water in a glass bottle with two inlets. Air was injected into the mixture for 1 h, turning the solution into a milky white suspension. To prepare an agar pad, about 700 µL of melted 1% TSY agar was applied to a glass slide (#48300–026, VWR, 25 × 75 mm, 1.0 mm thick). A 1-μL sample of the Span 83 suspension was placed on top of the agar pad, and the slide was placed in a high-relative-humidity chamber at 37 °C for about 40 min. After the water in the sample was absorbed by the agar, the Span 83 droplet spread out explosively and soon contracted into an array of micrometer-sized air bubbles with surfactant walls. Then, 1 μL of C. gingivalis cells suspended in sterile deionized water were added near the edge of the area containing the surfactant. After incubation for about 20 min, C. gingivalis started to swarm and reached the area with the bubbles. The microbubbles formed by the surfactant were used as tracers to image the patterns of motion of the top layer of a swarm. Microbubbles were imaged using the Nikon microscope and ThorLabs camera system described above, and their motion was tracked using ImageJ and a custom MATLAB script.
Cargo Transport and Polymicrobial Interactions.
To study cargo transport by single cells, aliquots of cultures of C. gingivalis at OD 0.6 were mixed with similar aliquots of cultures of each nonmotile species. The mixture of cell pairs was incubated for 10 min after which they were added to a tunnel slide, incubated for 5 min, washed with 0.25% methyl-cellulose (defined above) containing TSY broth, and imaged using the Nikon microscope and ThorLabs camera system described above. Motion of cargo cells on the transporter cells was tracked using ImageJ, and the motion of the transporter cell was tracked using a custom MATLAB script. To obtain the trajectory of a moving cargo cell with respect to the transporter cell, the motion of the transporter cell was subtracted from the motion of the cargo cell using a custom MATLAB script. To test if the adhesin SprB is present on the surface of a C. gingivalis cell, 5 µL of affinity-purified antibody raised against F. johnsonaie SprB was added to 100 µL of C. gingivalis cells suspended in PBS. After incubation for 10 min at 25 °C, the preparation and control with 5 µL PBS were injected into tunnel slides and were imaged using the Nikon microscope and ThorLabs camera system described above.
To image collective motion in a polymicrobial community, the cell surface of live bacterial cells was stained with an Alexa Fluor NHS ester (succinimidyl ester) dye (35). The cell suspensions were at OD 0.6. The following bacteria-dye combinations were used: P. endodontalis—Alexa 514; P. oris—Alexa 555; P. micra—Alexa 568; Actinomyces sp. oral taxon-169—Alexa 594; F. nucleatum—Alexa 633; S. sanguinis—Alexa 647; and V. parvula—Alexa 660. C. gingivalis was not labeled. Labeling was performed by mixing 1 µL of 4 µg/µL dye with 10 µL of the respective bacterial cell suspension, incubated in the dark and shaken gently for 1 h and washed twice with sterile deionized water by centrifugation to remove cell-free dye. Equal volumes of cultures at the same OD (of C. gingivalis and the seven fluorescently labeled bacteria) were mixed, and 1 µL of the mixture was added to an agar pad and incubated for 20 min at high relative humidity. To maintain an anaerobic environment, the agar pad was sealed with a cover glass and paraffin wax. After the incubation, swarming was observed. Cells were imaged with a Zeiss LSM 880 Airyscan Confocal microscope in a chamber set at 37 °C. Bacteria labeled with the dyes described above were used for calibration, and spectral imaging was used to separate the seven fluorescent signals. The images were analyzed using ImageJ, and superimposition of the fluorescent signal onto a differential interference contrast image provided the location of C. gingivalis. The motion of individual bacteria was tracked using ImageJ and was analyzed using a custom MATLAB script. The above experiment was repeated using cells of C. gingivalis that had been de-energized by treatment with 1 mM sodium azide for 10 min followed by two washes with deionized water.
Imaging of Fluorescent Cells in a Swarm.
One microliter of C. gingivalis cells at OD 0.6 was added to an agar pad and incubated at 37 °C for 20 min in a chamber at high relative humidity. The edge of a swarm had smaller motile groups of cells (slugs) that were imaged using the Nikon microscope and ThorLabs camera system described above. Motion of the slugs was tracked using a custom MATLAB script. An aliquot of C. gingivalis cells suspended in sterile deionized water was labeled with Alexa Fluor 633 as described above. The labeled cells were mixed with unlabeled cells so that only 1% of the cells in the final mixture were labeled. This mixture was placed on an agar pad, and fluorescently labeled cells were imaged using a Zeiss LSM 880 Airyscan Confocal microscope. Motion of the cells was tracked using a custom MATLAB script.
Supplementary Material
Acknowledgments
We thank the Harvard Center for Biological Imaging for infrastructure and support and Karen A. Fahrner for comments on the manuscript. This research was supported by NIH National Institute of Dental and Craniofacial Research K99/R00 Pathway to Independence Award DE026826 (to A.S.) and NIH National Institute of Allergy and Infectious Diseases Grant R01 AI016478 (to H.C.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808966115/-/DCSupplemental.
References
- 1.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Son K, Brumley DR, Stocker R. Live from under the lens: Exploring microbial motility with dynamic imaging and microfluidics. Nat Rev Microbiol. 2015;13:761–775. doi: 10.1038/nrmicro3567. [DOI] [PubMed] [Google Scholar]
- 3.Tamar E, Koler M, Vaknin A. The role of motility and chemotaxis in the bacterial colonization of protected surfaces. Sci Rep. 2016;6:19616. doi: 10.1038/srep19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottemann KM, Miller JF. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 5.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leadbetter ER, Holt SC, Socransky SS. Capnocytophaga: New genus of gram-negative gliding bacteria. I. General characteristics, taxonomic considerations and significance. Arch Microbiol. 1979;122:9–16. doi: 10.1007/BF00408040. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson J, et al. Characterization of motility and piliation in pathogenic Neisseria. BMC Microbiol. 2015;15:92. doi: 10.1186/s12866-015-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerner-Smidt P, et al. Lautropia mirabilis gen. nov., sp. nov., a gram-negative motile coccus with unusual morphology isolated from the human mouth. Microbiology. 1994;140:1787–1797. doi: 10.1099/13500872-140-7-1787. [DOI] [PubMed] [Google Scholar]
- 9.Berg HC. Random Walks in Biology. Princeton Univ Press; Princeton, NJ: 1993. pp 10, 56. [Google Scholar]
- 10.Grossart HP, Dziallas C, Leunert F, Tang KW. Bacteria dispersal by hitchhiking on zooplankton. Proc Natl Acad Sci USA. 2010;107:11959–11964. doi: 10.1073/pnas.1000668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelshtein A, Roth D, Ben Jacob E, Ingham CJ. Bacterial swarms recruit cargo bacteria to pave the way in toxic environments. MBio. 2015;6:e00074-15. doi: 10.1128/mBio.00074-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samad T, et al. Swimming bacteria promote dispersal of non-motile staphylococcal species. ISME J. 2017;11:1933–1937. doi: 10.1038/ismej.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ioannou CC, Guttal V, Couzin ID. Predatory fish select for coordinated collective motion in virtual prey. Science. 2012;337:1212–1215. doi: 10.1126/science.1218919. [DOI] [PubMed] [Google Scholar]
- 14.Vicsek T, Zafeiris A. Collective motion. Phys Rep. 2012;517:71–140. [Google Scholar]
- 15.Berg HC. Swarming motility: It better be wet. Curr Biol. 2005;15:R599–R600. doi: 10.1016/j.cub.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 16.Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrivastava A, Roland T, Berg HC. The screw-like movement of a gliding bacterium is powered by spiral motion of cell-surface adhesins. Biophys J. 2016;111:1008–1013. doi: 10.1016/j.bpj.2016.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faure LM, et al. The mechanism of force transmission at bacterial focal adhesion complexes. Nature. 2016;539:530–535. doi: 10.1038/nature20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nan B. Bacterial gliding motility: Rolling out a consensus model. Curr Biol. 2017;27:R154–R156. doi: 10.1016/j.cub.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson EL, Heaver SL, Walters WA, Ley RE. Microbiome and metabolic disease: Revisiting the bacterial phylum Bacteroidetes. J Mol Med (Berl) 2017;95:1–8. doi: 10.1007/s00109-016-1492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 23.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubin K, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride MJ, Zhu Y. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J Bacteriol. 2013;195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA. 2014;111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao H, et al. Variations in oral microbiota associated with oral cancer. Sci Rep. 2017;7:11773. doi: 10.1038/s41598-017-11779-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teles FR, et al. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J Periodontal Res. 2012;47:95–104. doi: 10.1111/j.1600-0765.2011.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kita D, et al. Involvement of the type IX secretion system in Capnocytophaga ochracea gliding motility and biofilm formation. Appl Environ Microbiol. 2016;82:1756–1766. doi: 10.1128/AEM.03452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrivastava A, Lele PP, Berg HC. A rotary motor drives Flavobacterium gliding. Curr Biol. 2015;25:338–341. doi: 10.1016/j.cub.2014.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abby SS, et al. Identification of protein secretion systems in bacterial genomes. Sci Rep. 2016;6:23080. doi: 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol. 2014;24:50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Hosu BG, Berg HC. Microbubbles reveal chiral fluid flows in bacterial swarms. Proc Natl Acad Sci USA. 2011;108:4147–4151. doi: 10.1073/pnas.1016693108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.