Significance
Reactive oxygen species (ROS) increase with age and have been shown to negatively impact age-related diseases. However, the physiological roles they might play in development have not been extensively characterized. Here, we show that ROS have essential functions as oocytes complete meiosis, the specialized cell division that generates haploid eggs, and transition to embryonic development. Meiotic progression and early embryonic divisions are defective when ROS is misregulated. Furthermore, we document the effects of ROS on specific proteins. Our identification of proteins altered in redox state as the oocyte transitions to an embryo provides a valuable resource to guide future exploration of ROS functions in early development. The regulatory system described here has important implications for female fertility.
Keywords: Drosophila, thiol reactivity profiling, chemical proteomics, meiosis, metabolomics
Abstract
The metabolic and redox state changes during the transition from an arrested oocyte to a totipotent embryo remain uncharacterized. Here, we applied state-of-the-art, integrated methodologies to dissect these changes in Drosophila. We demonstrate that early embryos have a more oxidized state than mature oocytes. We identified specific alterations in reactive cysteines at a proteome-wide scale as a result of this metabolic and developmental transition. Consistent with a requirement for redox change, we demonstrate a role for the ovary-specific thioredoxin Deadhead (DHD). dhd-mutant oocytes are prematurely oxidized and exhibit meiotic defects. Epistatic analyses with redox regulators link dhd function to the distinctive redox-state balance set at the oocyte-to-embryo transition. Crucially, global thiol-redox profiling identified proteins whose cysteines became differentially modified in the absence of DHD. We validated these potential DHD substrates by recovering DHD-interaction partners using multiple approaches. One such target, NO66, is a conserved protein that genetically interacts with DHD, revealing parallel functions. As redox changes also have been observed in mammalian oocytes, we hypothesize a link between developmental control of this cell-cycle transition and regulation by metabolic cues. This link likely operates both by general redox state and by changes in the redox state of specific proteins. The redox proteome defined here is a valuable resource for future investigation of the mechanisms of redox-modulated control at the oocyte-to-embryo transition.
Although life is driven by reduction–oxidation (redox) reactions (1), remarkably little is known about how metabolic state interfaces with normal development. Reactive oxygen species (ROS) were first described as a byproduct of metabolism and a hallmark of disease and aging. Exciting new research, however, implicated ROS more directly in cell signaling and regulation (2). In a manner analogous to posttranslational modifications, ROS can alter the oxidation status of cysteine residues and thus affect protein stability, activity, and localization or protein–protein interactions. In this respect, how ROS link to the spatiotemporal regulation of development via downstream targets has not been explored thoroughly (3, 4).
The redox state in the cell exists in a dynamic balance between ROS production and removal. The source of ROS is primarily oxidative phosphorylation in mitochondria. The antioxidants catalase and superoxide dismutase (SOD) neutralize ROS in a direct enzymatic reaction. Further major ROS scavenger systems are glutathione (GSH), which exists as GSH in its reduced state and as glutathione disulfide (GSSG) in its oxidized state, and thioredoxins (5, 6). GSH and thioredoxin utilize reducing power provided by oxidative metabolism, tied to redox couples such as NADPH/NADP+, to counteract oxidation via ROS. Under the redox-optimized ROS balance hypothesis, energy metabolism, antioxidants, and redox couples are in equilibrium to allow physiological ROS signaling under varying conditions (7, 8). Ultimately, the redox balance sets the oxidation level of downstream targets via an interplay of scavenger systems with downstream substrates such as protein and lipids (3, 9, 10).
A key developmental event is the oocyte-to-embryo transition, when at fertilization the highly specialized oocyte becomes a totipotent embryo. The oocyte-to-embryo transition, occurring in the absence of transcription, relies on posttranscriptional and posttranslational control. In this respect, could redox modulation of protein function bring a highly dynamic, additional level of regulation? Indeed, Dumollard et al. (11) showed that metabolic activity and thus the redox potential varies during mouse oogenesis and fertilization and is critical for early development. In addition, several aspects of oogenesis and early embryogenesis were blocked if proper redox balance was altered (12–14). Although these studies suggest a direct role for ROS, no comprehensive analysis has been carried out, and the downstream targets have not been investigated rigorously.
Intriguingly, Drosophila expresses an oocyte-specific thioredoxin, Deadhead (DHD). DHD is required for early embryogenesis and timely protamine-to-histone exchange in the male pronucleus in fertilized eggs (15–18). DHD redox activity is essential for its function (17). A further exciting observation was that a ubiquitous thioredoxin, Trx-2, did not recognize protamines as substrates. This indicates that DHD has at least one specific target in early development (15, 19). Because the protamine exchange defect did not fully account for the developmental block in the mutant, more roles and thus additional specific substrates are postulated for the functions DHD likely controls through redox regulation in the oocyte-to-embryo transition in Drosophila.
In this study, we describe an integrated approach to understand the role of redox in oogenesis and early embryogenesis in Drosophila. We show that proper redox balance is essential for meiosis completion, fertilization, and early embryogenesis. We make use of recent technological breakthroughs enabling quantitative examination of the redox Cys-proteome (20–23). Our data corroborate the emerging view that redox systems selectively maintain a nonequilibrium redox dynamic in distinct sets of the redox Cys-proteome (3). We propose that the developmentally regulated thioredoxin DHD controls a specific set of substrates by interpreting global changes at the level of energy metabolism. Our findings provide a paradigm for understanding redox-sensitive targets and pathways in other organisms or systems where cell-state transitions are accompanied by global metabolic remodeling.
Results
The Oocyte-to-Embryo Transition Is Marked by a Change in Redox State.
To characterize metabolic changes at the Drosophila oocyte-to-embryo transition, we optimized a targeted metabolomics method and compared ovaries enriched for mature oocytes (stage 14), unfertilized activated 0- to 1-h eggs, and 0- to 1-h embryos. Drosophila unfertilized eggs undergo the events at egg activation, including completion of meiosis, mRNA translation, and proteomic changes, but do not initiate embryonic divisions (24, 25). Around 100 high-confidence polar metabolites spanning major metabolic pathways were identified (SI Appendix, Supporting Materials and Methods). Statistical examination by principal component analysis showed that the ovary samples were significantly different from fertilized or unfertilized eggs (SI Appendix, Fig. S1A). In contrast, the fertilized and unfertilized egg metabolomes clustered together. Results from two different extraction protocols yielded consistent results (Dataset S1).
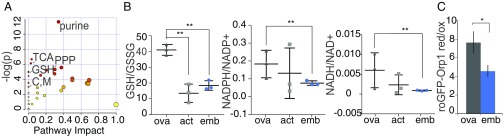
Pairwise comparisons between ovaries and 0- to 1-h embryos identified several developmentally regulated metabolic pathways (Dataset S2). We noticed that prominent among those were redox-related pathways (Fig. 1A and SI Appendix, Fig. S1B). To examine the redox state further, we focused on the redox couples GSH/GSSG, NADPH/NADP+, and NADH/NAD+, as measured directly by LC-MS (Fig. 1B). Decreases in these ratios revealed that 0- to 1-h embryos had a more oxidized environment than stage-14 enriched ovaries. Interestingly, unfertilized activated 0- to 1-h eggs displayed GSH/GSSG ratios that were comparable to those in embryos. NADPH/NADP+ and NADH/NAD+ also show a similar trend in activated eggs and embryos relative to the ovaries. This strongly suggests that the oocyte-to-embryo transition is sufficient to trigger a change in the oxidation state.
Fig. 1.
The redox state changes at the oocyte-to-embryo transition. (A) Pathway impact analysis from MetaboAnalyst comparing control stage-14 enriched ovaries and 0- to 1-h embryo collections using the formic acid extraction method. Several pathways that are statistically significantly different (P < 0.01 and false discovery rate (FDR) < 0.05) are indicated. C, M, cysteine and methionine metabolism; GSH, glutathione metabolism; PPP, pentose phosphate pathway; TCA, citrate cycle. (B) NADPH/NADP+, NADH/NAD+, or GSH/GSSG ratios measured by LC-MS analysis of formic acid-treated or Ellman’s reagent-derivatized extracts from ovary, 0- to 1-h unfertilized activated eggs, or 0- to 1-h embryo collections from OrR control flies. Error bars represent the SD from three independent biological replicates; **P < 0.05. NADH/NAD+ and NADPH/NADP+ measurements were done with formic acid-treated extracts, and GSH/GSSG was analyzed from Ellman’s reagent-derivatized extracts. (C) Live-cell imaging of the roGFP-Orp1 H2O2 sensor in oocytes (average of stages 9–14) or 0- to 1-h embryos from OrR flies. Values were normalized to fully oxidized samples as depicted in SI Appendix, Fig. S1 C and D. Error bars represent the SD from three independent biological replicates. *P < 0.1.
To validate these observations, we determined the redox state in vivo in oocytes and in early embryos by redox-sensitive GFP (roGFP) imaging. We utilized a previously characterized H2O2 sensor, cytoplasmic roGFP-Orp1 (26, 27). After modifying existing protocols to circumvent the problem of yolk autofluorescence (SI Appendix, Fig. S1 C and D), we compared the degree of oxidation with the sensor in oocytes and 0- to 1-h embryos. As evidenced by the reduction in the ratio of reduced/oxidized roGFP-Orp1, we detected higher levels of H2O2 in early embryos compared with oocytes (Fig. 1C). These measurements corroborate our metabolomics analysis and demonstrate that early embryos become oxidized as compared with mature oocytes. Our data thus reveal major metabolic remodeling at the oocyte-to-embryo transition accompanied by prominent redox state changes.
Thiol-Reactivity Profiling Demonstrates Global Cys-Proteome Remodeling at the Oocyte-to-Embryo Transition.
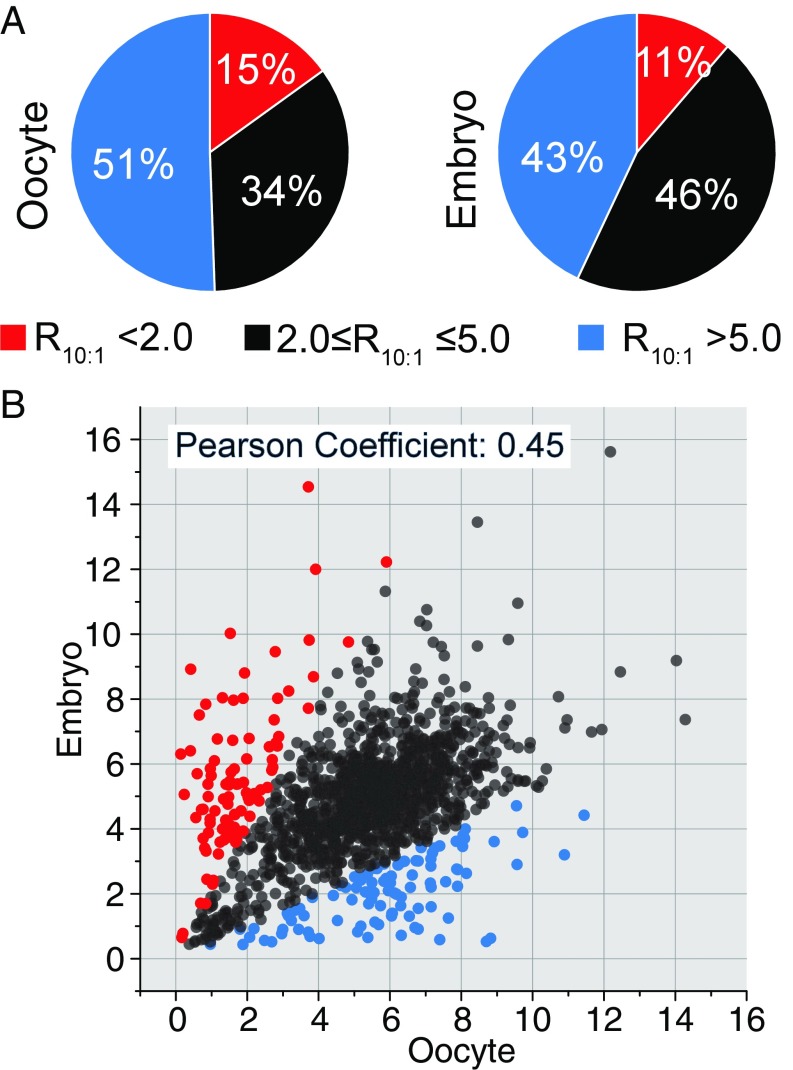
As the reactive thiol proteome (RTP) is the intrinsic target of redox state changes, we made use of recent technological breakthroughs to quantify globally and compare the site-specific reactivity of the RTP in late oocytes and early embryos (21–23). High-sensitivity mass spectrometry was used to identify labeled targets site-specifically after selective enrichment (20, 28–30). We employed a dose-dependent labeling of the proteome with low and high levels of a “clickable” thiol-reactive probe, alkynyl iodoacetamide (IPM) (SI Appendix, Fig. S2A). This allowed us to quantify hyperreactive cysteines, enriched with low-dose labeling, along with less reactive cysteine sites that require higher concentrations of the reactive probe for labeling. We defined hyperreactive cysteines as those having a ratio (R10:1) of heavy-to-light fractions (100 µM versus 10 µM, respectively) below 2. In total, we identified and quantified around 2,000 cysteine sites in wild-type stage-14 enriched ovaries and in 0- to 1-h embryos (Fig. 2A and SI Appendix, Fig. S2B). The hyperreactive cysteines account for 15% (315) and 11% (214) of the total quantified sites in stage-14 enriched ovaries and early embryos, respectively (Fig. 2A).
Fig. 2.
Cysteine-reactivity profiling of the oocyte-to-embryo transition. (A) Pie charts showing the distribution of cysteine reactivity of the Drosophila stage-14 enriched ovary and 0- to 1-h embryo proteome. (B) Correlation of proteomic cysteine reactivity quantified in stage-14 enriched ovaries and 0- to 1-h embryos. The determined R10:1 values of the cysteines present in both ovary and embryo samples are plotted against each other for comparison. Cysteines with an Rova10:1/Remb10:1 ratio >2 are in blue; cysteines with an Rova10:1/Remb10:1 ratio <0.5 are in red. Heavy/light (100 μM/10 μM) ratios were calculated from at least three biological replicates, with two technical runs for each.
We next sought to perform a global and side-by-side comparison of the reactivity of the same cysteines in the two developmental stages. As shown in Fig. 2B, although a fraction of cysteines (black) exhibits similar reactivity in both stage-14 enriched ovaries and 0- to 1-h embryos, 98 (blue) and 93 (red) cysteines exhibited higher reactivity in early embryos or oocytes, respectively. Gene Ontology (GO) category analysis identified the category mitotic nuclear division as significantly enriched (P = 0.0023) in the set of proteins that was more reactive in embryos than in oocytes and the category single organism reproductive process (P = 0.042) for the set of proteins that was more reactive in oocytes than in embryos (Dataset S3). It is of note that, unlike the RTP in mammals and worms (28, 29), the Drosophila cysteines with low R10:1 values (<2.0) were not enriched in residues with annotated function in either oocytes or embryos (SI Appendix, Fig. S2B), potentially due to poor annotation of residue and protein functions in the Drosophila UniProt database. The functions of most hyperreactive cysteine sites identified in this study are yet to be determined; however, our findings are consistent with a broad remodeling of the RTP during the oocyte-to-embryo transition paralleling global changes in metabolism and redox state.
DHD Has Additional Functions at the Oocyte-to-Embryo Transition.
Oxidative stress responses in Drosophila must rely on the thioredoxin system to recycle oxidized GSH because no GSH reductase activity has been found (31, 32). The redox changes we observed made it important to evaluate the role of the developmentally restricted, ovary-specific thioredoxin DHD (see the model in SI Appendix, Fig. S4H). DHD (SI Appendix, Supporting Results) has been shown recently to be required for the protamine-to-histone exchange and the dynamics of sperm decondensation at the onset of embryogenesis (15, 16). The defect, however, did not account for the complete developmental block observed in dhd mutants (17, 18). Using a combination of three independent null alleles (Materials and Methods) that allowed us to control for background effects, we investigated sperm-independent functions of DHD.
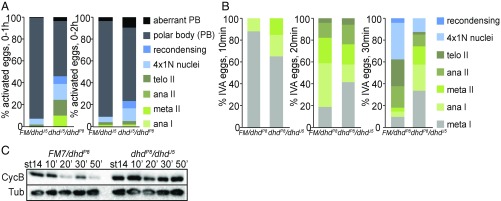
First, we scored completion of meiosis in a 0- to 1-h activated unfertilized egg collection from transheterozygous dhd-mutant mothers. A major fraction of the eggs delayed at various stages of meiosis: 46% of dhdJ5/dhdP8 eggs compared with 8% of the sibling controls (Fig. 3A). In 0- to 2-h egg collections, fewer meiotic stages were observed, but there was an accumulation of polar body defects (see also SI Appendix, Supporting Results). A similar trend was observed when parents that carried the null alleles dhdP8 or dhdJ5 and that were used to generate the transheterozygous mothers were interchanged, yielding the dhdP8/dhdJ5 transheterozygous combination (Materials and Methods and SI Appendix, Fig. S3A). Second, we performed time-course experiments with in vitro-activated eggs from transheterozygous dhdP8/dhdJ5 mutants and sibling controls (Fig. 3 B and C). We consistently scored a delay in meiotic progression. A similar reproducible trend was observed with the dhdJ5/dhdP8 transheterozygous combination (SI Appendix, Fig. S3B). The meiotic delay also was clearly mirrored in the analysis of degradation dynamics of Cyclin B, which decreased by 20–30 min in control eggs. However, reproducibly, no significant decrease was observed even after 50 min in dhd mutants (Fig. 3C and SI Appendix, Fig. S3C). This delay could not be solely due to a failure to activate mature oocytes from dhd-mutant mothers, as all analyzed eggs were bleach resistant and thus had a cross-linked vitelline envelope, an early activation event (33). Finally, we noted chromosome-organization defects in dhd mutants progressing through meiosis (SI Appendix, Fig. S3D). Indeed, we identified cohesin and condensin subunits in the RTP (Dataset S3). As SOD1 and SOD2 have recently been implicated in the maintenance of sister-chromatid cohesion in midprophase I and metaphase I Drosophila oocytes (34), we evaluated whether the metaphase I arrest would be similarly affected in dhd mutants. Examination of DNA morphology in mature stage-14 oocytes showed a significantly higher breakdown of the metaphase I alignment in transheterozygous Df(1)dhd81/dhdP8 but not Df(1)dhd81/dhdJ5 mutants compared with controls (SI Appendix, Fig. S3E). In conclusion, our data clearly demonstrate that DHD has additional roles during the completion of meiosis (see also SI Appendix, Supporting Results).
Fig. 3.
Phenotypic characterization of dhd mutants at the oocyte-to-embryo transition. (A) Quantification of stages in unfertilized activated eggs from 0- to 1-h or 0- to 2-h collections from dhdJ5/dhdP8 mothers and corresponding sibling controls mated to twineHB5 males that do not produce sperm. The eggs were stained with DAPI to score developmental stage. The meiotic stages are indicated as shown. The 4×1N nuclei are eggs that have completed meiosis. Ana, anaphase; FM, FM7 balancer; meta, metaphase; recondensing, meiotic products condensing their chromosomes after a transient interphase following the completion of meiosis; telo, telophase. This is a stage in which the polar body (PB) is formed. This experiment was performed once. (B) Quantification of meiotic progression 10, 20, or 30 min after in vitro activation (IVA) of mature, MI-arrested stage-14 oocytes from dhdP8/dhdJ5 virgins and corresponding sibling controls. This is a representative experiment from three biological replicates. FM, FM7 balancer. (C) Western blot analysis of Cyclin B (CycB) levels from the in vitro-activated stage 14 oocytes from dhdP8/dhdJ5 virgins and corresponding sibling controls for the indicated time points. α-Tubulin (Tub) was used as a loading control. This is a representative experiment from three biological replicates.
DHD Functions Are Linked to General Redox Control.
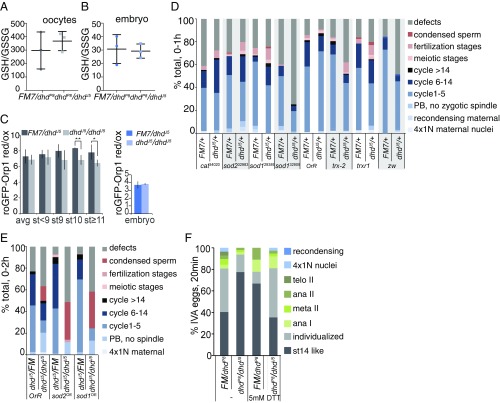
Next we asked if DHD function relates to the general redox state. We measured redox pairs by metabolomics in sibling control and mutant dhdP8/dhdJ5 stage-14 enriched ovaries and 0- to 1-h embryos. The GSH/GSSG ratio measurement remained unchanged in both embryos and ovaries from the dhdP8/dhdJ5 mutant (Fig. 4 A and B). However, we measured a significant difference in the NADH/NAD+ but not in the NADPH/NADP+ ratio in stage-14 enriched ovaries from the dhdP8/dhdJ5 mutant (SI Appendix, Fig. S4A). We then utilized the in vivo H2O2 sensor roGFP-Orp1, which indicated higher levels of H2O2 in dhdJ5-mutant late oocytes (Fig. 4C). H2O2 levels in mutant 0- to 1-h embryos remained unchanged, an observation consistent with our LC-MS demonstration that embryos are more oxidized than oocytes. Notably, oocytes were not oxidized to embryonic levels in the absence of DHD. We also did not observe major differences in the metabolomes of stage-14 enriched ovaries or 0- to 1-h embryos from dhd mutants (SI Appendix, Supporting Results). Taken together, our measurements indicate that DHD is partially required for the redox state in oocytes, where its protein levels are high, but not in early embryos, where its protein levels decreased (SI Appendix, Supporting Results). This observation is also consistent with our phenotypic characterization above.
Fig. 4.
Redox state control of the oocyte-to-embryo transition. (A and B) GSH/GSSG measured by LC-MS analysis of Ellman’s reagent-derivatized extracts from ovary (A) or 0- to 1-h embryo collections (B) from dhdP8/dhdJ5 mothers and corresponding sibling controls. Error bars are the SD from three independent biological replicates. (C) Live-cell imaging of roGFP-Orp1 in oocytes or 0- to 1-h embryos from dhdJ5 homozygous mutants or sibling control flies. Values were normalized to fully oxidized samples. *P < 0.1, **P < 0.05. (D) Embryonic progression of indicated RNAi-driven knockdowns from FM7/dhdJ5; Mat(alpha) GAL4 mothers that also carried the indicated UAS-TRIP transgenes. The numbers for each RNAi line are the Bloomington stock numbers. DAPI- and antitubulin–stained 0- to 1-h embryo collections were categorized based on their exact stage in early development, the presence of defects, or meiotic and fertilization stages. Fertilization stages included pronuclear migration and opposition (PB, polar body). This is a representative experiment from two biological replicates. Defects included large multipolar spindles, fused nuclei appearing to missegregate, free asters, and other microtubule or segregation defects (see examples in SI Appendix, Fig. S4F). (E) Embryonic progression of indicated GAL4-UAS–driven overexpression (OE) from FM7/dhdJ5 or dhdJ5/dhdJ5; Mat(alpha) GAL4 mothers with the indicated UAS transgenes. DAPI- and anti-tubulin–stained 0- to 1-h embryo collections were categorized based on exact stage in early development, presence of defects, or meiotic and fertilization stages. Fertilization stages included pronuclear migration and juxtaposition. FM, FM7 balancer; PB, polar body. This experiment was done once. (F) Quantification of meiotic progression at 20 min after in vitro activation (IVA) of mature, MI-arrested stage 14 oocytes from dhdP8/dhdJ5 young females and corresponding sibling controls in the presence of 0 or 5 mM DTT. As oocytes become activated, DNA morphology changes from the MI-arrest configuration (st14-like category) to chromosome becoming more condensed and individualized (individualized category). This is a representative experiment from three biological replicates.
We further reasoned that if the dhd-mutant phenotype resulted from a misbalance in the general redox state, we should be able to modify one or all of its phenotypes by changing redox independently. We first asked if embryonic progression would be modulated in progeny from FM7/dhdJ5; Mat(alpha)-GAL4 mothers that also had UAS-RNAi lines against genes encoding redox proteins. In addition to mitotic defects, we scored 0- to 1-h embryos for the presence of meiotic or fertilization stages. Depletions of trx-2, the ubiquitous thioredoxin, and, to a lesser extent, of trxr-1 or the pentose phosphate pathway gene zw (zwischenferment, a glucose-6-phosphate dehydrogenase) did not cause significant defects on their own but showed substantial synthetic interaction with dhdJ5 (Fig. 4D and SI Appendix, Fig. S4D). In addition, several RNAi lines showed meiotic and fertilization defects and an increase in defects if one gene copy of dhd was removed (Fig. 4D). The effect was strongest with sod1 (RNAi line 32909). Several examples of defective mitosis are shown in SI Appendix, Fig. S4E, with the most common being spindle defects and big multipolar spindles with an aberrantly large number of chromosomes. Furthermore, the most penetrant sod1 RNAi line led to a phenotype similar to that of a dhd complete null—a block in early embryogenesis, the presence of spindles with a haploid number of chromosomes, and an increase in polar body defects, indicative of defective meiosis completion (SI Appendix, Fig. S4E).
Next, we tested if mild overexpression of sod1 or sod2 would affect progression through embryogenesis in a dhd mutant. We crossed FM7/dhdJ5; Mat(alpha)-GAL4 females with males that were either wild-type or dhdJ5 and carried UAS-driven sod1 or sod2 transgenes. In 0- to 2-h embryos produced by mothers derived from this cross, we observed an increase in the number of embryos with condensed sperm if the mothers were dhdJ5/dhdJ5; Mat(alpha)-GAL4/UAS-sod1 or sod2. This suggested an exacerbated dhd phenotype upon mild overexpression of either sod1 or sod2 (Fig. 4E and SI Appendix, Fig. S4F). Thus, in this context, SODs could be increasing the concentration of H2O2, further collapsing the redox balance in the dhd mutant.
Finally, we focused on meiotic progression. We tested the effect of the reducing agent DTT on dhdP8/dhdJ5 or sibling control oocytes activated for 20 min in vitro. We first noted that the addition of 5 mM DTT markedly delayed meiotic progression and/or activation in control oocytes (Fig. 4F). Interestingly, the effect of DTT was suppressed in mutant dhdP8/dhdJ5 oocytes, which are more oxidized and thus are able to compensate for the change in reductive state caused by DTT. Thus, an increase in oxidation level is required to proceed from stage 14 to the onset of anaphase in vitro. We did not observe the same effect with the dhdJ5/dhdP8 allele combination, presumably due to stock-specific differences impinging on redox state (SI Appendix, Fig. S4G). These observations illustrate the need for a unique redox balance for proper progression through meiosis and embryogenesis and indicate that DHD functions as part of the global redox network (SI Appendix, Fig. S4H).
DHD Affects a Specific Set of Reactive Cysteine-Containing Proteins.
Together our data indicate proteome-wide redox regulation with indispensable functions at the oocyte-to-embryo transition and, in particular, a central role for DHD. Therefore the global RTP changes we report must be redox-driven, at least in part. To narrow down which proteins or cysteines depend on DHD, we turned to thiol-redox proteomics (SI Appendix, Fig. S5A). The RTP in transheterozygous mutant dhd stage-14 enriched ovaries or 0- to 1-h early embryos and their sibling controls were first labeled with 100-μM IPM probe, which has been demonstrated by our and other groups to label the native RTP completely (28, 30). The analyses evaluated Rmutant/control (Rmut/crl) values for up to ∼6,000 cysteines in each tissue from three biological replicates. Low Rmut/crl values are indicative of cysteines with decreased reactivity in dhd mutants, suggesting potential DHD-regulated targets. The majority of identified cysteines showed Rmut/crl values higher than 0.67 (Fig. 5A and Dataset S4).
Fig. 5.
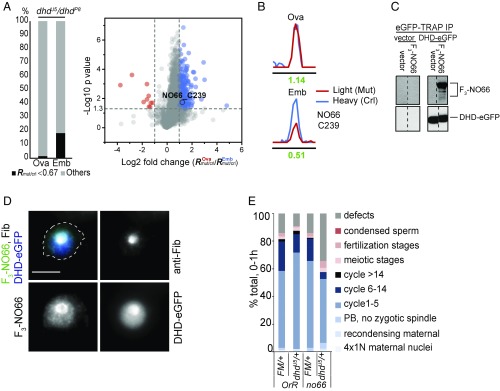
DHD interacts with a set of downstream targets. (A) Quantitative thiol-reactivity profiling from stage-14 enriched ovaries or 0- to 1-h embryos from dhdJ5/dhdP8-mutant females and corresponding sibling controls. (Left) The bar chart shows the distribution of cysteines with a DHD-dependent decrease in reactivity (Rmut/crl <0.67) from ovary and embryo extracts. (Right) The volcano plot shows the comparison of the Rmut/crl values determined in the two tissues. Log2 of fold change (ova/emb) for each cysteine identified is plotted against significance values in −log10. All data points are averages from at least three biological replicates. The cysteines with an Rmut/crl changed more than twofold in ovaries compared with embryos are highlighted in blue and red dots, respectively. If the fold change of Rova/Remb of a cysteine is higher than two (i.e., blue) it suggests that the cysteine has a greater DHD-dependent decrease in reactivity in embryos than in ovaries. The NO66 (C239) protein is indicated. (B) Extracted ion chromatograms showing changes in the IPM-tagged peptide from NO66 C239 in ovaries (Ova) and embryos (Emb) from the dhd mutant. The profiles for light- and heavy-labeled peptides are shown in red (dhd mutant) and blue (control). Light (dhd mutant) to heavy (control) ratios were calculated from at least three biological replicates and are displayed below the individual chromatograms. (C) Western blot analysis with anti-GFP– or FLAG-specific antibodies of immunoprecipitation (IP) experiments from S2 cells with stably integrated empty vector (Left) or GFP-tagged DHD (Right) and transiently transfected with FLAG3-tagged NO66 (F3) or vector control. (D) Immunofluorescence of S2 cells with anti-FLAG and anti-fibrillarin (Fib) antibodies and anti-GFP nanobodies. The S2 cells stably integrated with an inducible DHD-eGFP construct were transiently transfected with FLAG3-NO66. DNA was stained with DAPI. A single stained cell is shown. Both NO66 and DHD-GFP colocalize with fibrillarin at the nucleolus. (E) Embryonic progression of indicated RNAi-driven knock downs from FM7/dhdJ5 or FM7/+; Mat(alpha) GAL4 mothers carrying the indicated UAS-TRIP transgenes. DAPI- and anti-Tubulin–stained 0- to 1-h embryo collections were categorized based on exact stage in early development, presence of defects, or meiotic and fertilization stages. Fertilization stages included pronuclear migration and opposition. FM, FM7 balancer; PB, polar body.
We found a specific set of cysteines whose apparent reactivity decreased more than 1.5-fold (Rmut/crl <0.67) in dhd mutants (dhdJ5/dhdP8 transheterozygotes) compared with control samples from both stage-14 enriched ovaries and 0- to 1-h embryos. By comparing the Rmut/crl values determined in late oocytes and early embryos, we next identified 237 cysteines that exhibited a greater DHD-dependent decrease of reactivity in embryos from dhdJ5/dhdP8-mutant females than determined in oocytes (Fig. 5A, Right). However, in samples from dhdP8/dhdJ5 transheterozygous mutant females, only 17 cysteines demonstrated decreased reactivity in dhdP8/dhdJ5-mutant embryos versus oocytes (SI Appendix, Fig. S5B). These differences suggested that the origin of the dhdJ5 allele or the dhdP8 allele in the cross to generate transheterozygous mutant mothers mattered. Interestingly, the observed effects on the RTP tracked with observed phenotypic differences, as we consistently detected more penetrant phenotypic effects in embryos and unfertilized eggs when G0 mothers carried the dhdP8 allele (Fig. 3A and SI Appendix, Figs. S3A and S6D) and in oocytes when G0 mothers carried the dhdJ5 allele (compare Fig. 3B and SI Appendix, Fig. S3B).
To gain a better understanding of the cellular distribution and biological functions of potential DHD targets, we performed a bioinformatics analysis based on GO classification. The GO analysis revealed that the potential targets of DHD revealed by analysis of dhdJ5/dhdP8 mutants were enriched in various intracellular compartments, such as microtubule-associated complex (P = 6.3 × 10−7) and nucleus (P = 0.0056) (Dataset S5). For oxidized proteins, among the top GO-enriched biological process categories in dhdJ5/dhdP8 0- to 1-h embryos were cell cycle process and cytoskeleton organization (P = 5.4e-11; P = 3.8e-7) (Dataset S5). In summary, our substrate analyses reveal a highly specific list of DHD-dependent, potentially developmentally regulated targets. The general redox network likely controls a broader set of substrates (Fig. 2), and DHD plays a key role possibly to provide developmental specificity.
DHD-Specific Redox Targets Are Required in Early Embryogenesis.
To investigate direct substrates of DHD, we performed pulldowns from either stage-14 enriched ovaries or 0- to 2-h embryo extracts incubated with recombinant mutant DHD in which the conserved resolving cysteine residue (Cys-34) was mutated to serine. This is predicted to trap thioredoxin targets covalently so that bound proteins can be eluted with a high concentration of reducing agent (35). A similarly generated mutant for Trx-2 allowed us to distinguish potential shared versus DHD-specific targets by quantitative mass spectrometry (Dataset S6). Relative enrichment was quantified for roughly 130 proteins. Among the top enriched interactors of Trx-2 in both late oocyte and early embryo samples were the redox proteins peroxiredoxin 3 and jafrac1 (thioredoxin peroxidase 1). DHD, however, associated with more and diverse proteins in both oocytes and embryos. A major fraction of DHD interactors was ribosome or ribosome-associated. Prominent among these, shared between Trx-2 and DHD, was CG2982 (nucleolar protein 66, NO66).
We carried out additional validation of DHD targets using an eGFP-dhd transgenic line (SI Appendix, Supporting Results). We performed eGFP-Trap immunoprecipitations from extracts generated from either whole ovaries enriched for stage 14 oocytes or 0- to 1-h embryo collections. We identified 57 and 73 proteins, respectively, that were significantly associated with DHD in oocytes or embryos by significance analysis (Significance Analysis of INTeractome; SAINT), respectively (SI Appendix, Fig. S5 C and D and Dataset S6). Underscoring the robustness of our assays, we identified thioredoxin reductase-1 as well as the previously described interactor nucleoplasmin like (nucleoplasmin only in the embryo experiment) (16). As with the DHD (C34S) pulldown, a pronounced characteristic was that a significant fraction of interactions seemed under developmental control, as they were present in either late oocytes or early embryos (SI Appendix, Fig. S5C). Thirty-two interactors were common between oocyte and embryo, whereas 25 were oocyte-specific, and 41 were embryo-specific.
We identified which of the immunoprecipitation interactors contain reactive cysteines with a DHD-dependent decrease of reactivity (SI Appendix, Fig. S5E and Dataset S6). Several ribosomal proteins were among these. We also noted that DHD interactors in embryos and oocytes were enriched for ribonucleoprotein or translation-related GO categories (translation, P = 5.32 × 10−11 in embryos and P = 9.45 × 10−6 in oocytes; ribonucleoprotein complex assembly, P = 4.93 × 10−8 in embryos and P = 5.2 × 10−6 in oocytes) (Dataset S6). One related interactor, present in all three datasets, was NO66. In particular, Cys239 of NO66 exhibited a DHD-dependent decrease of reactivity in embryos (Fig. 5B and SI Appendix, Fig. S5F). Moreover, NO66 could be immunoprecipitated with DHD-eGFP in S2 cells and colocalized with DHD-eGFP in the nucleolus (Fig. 5 C and D).
To test if NO66 functions together with DHD at the oocyte-to-embryo transition, we crossed a UAS-RNAi line against no66 to FM7/dhdJ5; Mat(alpha)-GAL4 and analyzed early embryonic progression in 0- to 1-h embryo collections from females with or without the no66 UAS-RNAi or dhdJ5 mutation. Interestingly, although RNAi against no66 alone did not cause significant defects in the embryo, no66 showed delays and defects in mitotic progression in the absence of one copy of dhd (Fig. 5E and SI Appendix, Fig. S5G). This epistatic interaction is consistent with NO66 and DHD acting in parallel to control the same process.
Discussion
Although redox state changes have been described to occur in oogenesis and embryogenesis in diverse systems, no comprehensive analysis of the regulators and targets of redox has been carried out. In this work, we demonstrate that global redox changes occur at the oocyte-to-embryo transition in Drosophila. Redox pair measurements by LC-MS or in vivo H2O2 sensor imaging showed that embryos are more oxidized. The developmentally regulated thioredoxin DHD contributes to these changes so that the GSH/GSSG and NADH/NAD+ ratios and H2O2 levels in oocytes partially depend on its proper function. DHD works in conjunction with other redox proteins to ensure the fidelity of meiosis completion, fertilization, and early embryogenesis. DHD likely exerts its function via specific substrates, and we found that DHD interacts with other proteins in addition to the previously described protamines. Importantly, we linked global changes in redox state to corresponding changes in the redox-thiol proteome and documented a set of specific DHD-dependent redox targets. We found that one such substrate, NO66, contributes to DHD function during early embryogenesis. Our study leads to the hypothesis that metabolic changes occurring in the context of a developmental transition can be utilized to modify the function of specific downstream proteins and protein networks via dedicated redox systems (see model in SI Appendix, Fig. S5H).
Our work extends previous studies describing metabolic changes in Drosophila during oogenesis and embryogenesis (36, 37). The metabolome changes as the Drosophila fertilized oocyte is activated to proceed through embryonic development. Within a very short time frame, 0–1 h after egg-laying, there are metabolic changes that affect, among others, the TCA cycle, amino acid metabolism, the pentose phosphate pathway, and purine metabolism. Although the mechanism by which metabolic changes and redox interact in early embryogenesis in Drosophila is yet to be determined, it is possible that this could occur via modulation of mitochondrial functions. Recently, mitochondria were shown to exist in a quiescent state in Drosophila oocytes and to reactivate in embryos (36). It will be interesting to investigate in the future how mitochondrial function, calcium signaling, and ROS generation and signaling are coupled at the oocyte-to-embryo transition.
The oocyte-specific thioredoxin DHD is a key developmentally regulated node in the redox control of the oocyte-to-embryo transition in Drosophila. DHD protein levels are regulated, increasing markedly during oocyte maturation and sharply decreasing at egg activation (SI Appendix) (25, 38). Interestingly, sperm-specific thioredoxins have been described in mammals as well as in Drosophila (39, 40). Developmental control of thioredoxins could thus be a broader strategy to modulate cell-state transitions in other systems.
A key question that our study poses is whether DHD exerts its essential roles at the oocyte-to-embryo transition via modulation of global redox state and/or via specific downstream targets. We found that DHD is partially required for redox state in late oocytes. The effect on the GSH/GSSG ratio could be via thioredoxin’s direct contribution in Drosophila to the GSH cycle, as no GSH reductase has been found (31, 32). Interestingly, our genetic analysis of interaction between dhd and trx-2 revealed an overlap of function, such that the proteins could cooperate in setting the proper GSH/GSSG ratio in late oocytes and early embryos. The genetic interactions between dhd and other genes involved in redox control argue for a global role for general redox in the oocyte-to-embryo transition. This conclusion is supported further by the observation that modulation of the redox state balance by the addition of DTT affected progression through meiosis in the in vitro system.
In addition to redox control at a global level, we showed both by physical interactions and reactive-thiol mass spectroscopy profiling that specific proteins are DHD targets. Along with TrxR1, a well-known DHD partner, NO66 was one of the strongest interactors. NO66 was identified by our thiol-redox proteomics analysis as having a cysteine residue (C239) whose reactivity diminished, indicating thiol-oxidation, in dhdJ5/dhdP8-mutant embryos. NO66 is a JmjC domain-containing histone demethylase, shown recently to act as a suppressor of variegation and to localize to the nucleolus in flies (41). A link to redox control is that JmjC domain-containing proteins have been described as requiring ascorbate, a reducing agent, for optimal activity in vitro (42, 43). The colocalization between DHD and NO66 in the nucleolus in S2 cells is consistent with a functional relationship. Importantly, the genetic interaction between no66 and dhd further supported this notion. DHD interacts with and/or modulates ribosomal and RNA-binding proteins and thus could control aspects of nucleolar or ribosomal function. Interestingly, the human NO66 has been described as having ribosome oxygenase activity (44). In the future, it will be important to mutate redox-sensitive cysteine residues on potential DHD targets and examine functions in vivo.
Intriguingly, the mitochondrial NADH dehydrogenase ND-75 is among the top changed reactive cysteine-containing proteins in dhd mutants. Although the same residue (C193) is not present in our wild-type reactive Cys-proteome, the conserved cysteine Cys712 changes reactivity between oocytes and embryos. As ND-75, along with other complex I subunits, leaks into the cytoplasm in late-stage oocytes (36), an exciting possibility is that DHD-dependent global redox changes occur via modulation of mitochondrial metabolism. Furthermore, in our interaction partner analysis, we noted the presence of SOD1, a thioredoxin partner previously described in plants (45). These observations suggest that the effect of DHD on global H2O2 as well as on NADH/NAD+ levels could be via modulation of the function of downstream targets. Thus, global and specific redox roles are likely interwoven. Overall, we propose a model whereby DHD affects specific targets in a developmental context, thus “translating” the metabolic and redox changes to control important regulators of the oocyte-to-embryo transition (SI Appendix, Fig. S5H).
Thiol-redox proteomic studies have been widely successful in understanding thiol-redox dynamics in various systems but have not been broadly applied in a developmental context (21–23). In this regard, model organisms offer the advantage of applying integrated approaches. For example, Knoefler et al. (46) monitored thiol-redox changes as a function of life span in Caenorhabditis elegans. Menger et al. (47) showed the dramatic effect of fasting on the Drosophila reactive-thiol proteome. In this study, we demonstrate the use of thiol-redox proteomics to investigate the oocyte-to-embryo transition in Drosophila. In addition to identifying specific targets of DHD, 500 cysteines were found to become differentially reactive at egg activation. This dataset will be a valuable resource for the community. Our results provide evidence that a defined set of regulators along with general redox state control the oocyte-to-embryo transition. We conclude that remodeling of the reactive thiol proteome should be considered a fundamental part of this critical developmental window.
Materials and Methods
Fly Stocks and Transgenic Lines.
Oregon R was used as the wild-type control. dhdJ5, dhdP8, and Df(1)dhd81 are null alleles and have been described previously (18). Table 1 outlines the crosses between the various dhd alleles used to generate dhd transheterozygous flies. twineHB5 (48) males were used to generate unfertilized activated eggs. roGFP-Orp1 flies were described previously (27). Flies were maintained at 22 or 25 °C on standard Drosophila cornmeal molasses food. The strains carrying the maternal Mat(alpha)-GAL4 driver carried Wolbachia.
Table 1.
Crosses of dhd alleles and F1 genotypes analyzed
| G0 female | G0 male | F1 control females | F1 transheterozygous females |
| w dhdJ5/FM7 | w dhdP8/Y | w dhdP8/FM7 | w dhdP8/w dhdJ5 |
| w dhdP8/FM7 | w dhdJ5/Y | w dhdJ5/FM7 | w dhdJ5/w dhdP8 |
| w dhdJ5/FM7 | Df(dhd81)/Y | Df(dhd81)/FM7 | Df(dhd81)/w dhdJ5 |
| w dhdP8/FM7 | Df(dhd81)/Y | Df(dhd81)/FM7 | Df(dhd81)/w dhdP8 |
Only the sex chromosome genotypes are shown.
Immunoprecipitation to Identify DHD Interactors.
The 0- to 1-h embryos were collected from mothers with either eGFP-dhd transgenes (SI Appendix, Supporting Results and Supporting Materials and Methods) or OrR controls. Ovaries were isolated from females of the same genotypes. These ovaries were enriched for stage-14 oocytes by using virgin females fattened on yeast for 2 d and starved for an additional 2 d. The embryos or ovaries were frozen in liquid nitrogen. Extracts were prepared as described by Hara et al. (49), and the protein concentration was adjusted to 3 µg/µL. eGFP-DHD was immunoprecipitated from 3 mg of the protein extracts with 25 µL GFP-TRAP_MA beads (ChromoTek) for 30 min. After three washes with Nonidet P-40 lysis buffer [25 mM Tris (pH 7.4), 0.5 mM EDTA, 0.5% Nonidet P-40], proteins were eluted by a 10-s incubation in 15 µL 0.2 M glycine, pH 2.5. The eluate was neutralized with 1 µL of 1-M Tris⋅HCl (pH 8), and proteins were precipitated by 10% TCA. The pellet was washed with ice-cold acetone and frozen in liquid nitrogen.
Cysteine Proteome Labeling.
For quantitative comparisons of changes in cysteine reactivity between dhd mutants and corresponding sibling controls, ∼3 mg total tissue (ovaries or embryos) was used per replicate. Stage-14 enriched ovaries or 0- to 1-h embryo collections were collected as for immunoprecipitation and stored at −80 °C. The tissue was homogenized in 500 µL Nonidet P-40 buffer containing 200 units/mL catalase (Sigma-Aldrich) and 100 µM IPM (EVU111; Kerafast) in a Bioruptor (Diagenode) at 4 °C for 15 cycles of 30 s on and 30 s off, incubated on ice for 20 min, and further incubated at RT for 1 h. The protocol then was followed as described previously (see also SI Appendix, Supporting Materials and Methods) (20, 30). Reactive cysteine profiling was performed in a similar way, with one-half of the samples treated with 10 µM IPM and one half with 100 µM for 1 h at RT.
roGFP Live-Cell Imaging.
A method was developed to avoid the background from yolk autofluorescence (SI Appendix, Supporting Materials and Methods). Embryos or oocytes were transferred to two wells on Lab-Tek slides in 500 µL of Grace’s unsupplemented medium and were imaged immediately. Then 500 µL of Grace’s medium supplemented with either 2 mM diamide (D3648; Sigma Aldrich), 0.6% H2O2 (H1009; Sigma Aldrich), or 10 mM DTT (D9779; Sigma Aldrich) was added as specified, and samples were imaged again. Imaging took around 30 min in total, which corresponded to treatment with the oxidizing or reducing reagent for 7 min. Images were taken with fixed exposure times at both 405/535 nm and 488/535 nm excitation/emission settings on a Nikon ECLIPSE Ti microscope with Plan Fluor 10× or Plan Apo 20× objectives using NIS-Elements software. Three regions of interest away from any yolk autofluorescence signal were chosen per oocyte or embryo, and intensities were measured using ImageJ (https://imagej.nih.gov/ij). Ratios were averaged in Excel.
Supplementary Material
Acknowledgments
We thank Mary-Lou Pardue (MIT) and Tobias Dick (German Cancer Research Center) for providing antibodies and stocks, respectively; Caroline Lewis, Amanda DiRosario, Eric Spooner, and Yeelin Bacchus for technical assistance; Michael Welte and Erinc Hallacli for sharing protocols and reagents; and Satyaki Rajavasireddy, Rebecca Povilus, Grzegorz Sienski, and Masatoshi Hara for comments on the manuscript. This work was supported by National Key Research and Development Program of China Grant 2016YFA0501303; National Natural Science Foundation of China Grants 31770885, 31500666, and 81573395; Beijing Nova Program Grant Z171100001117014 (to J.Y.); and NIH Grant GM118098 (to T.L.O.-W.). T.L.O.-W. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807918115/-/DCSupplemental.
References
- 1.Cosmol JAJ. Redox homeostasis in the emergence of life. On the constant internal environment of nascent living cells. J Cosmology. 2010;10:3362–3373. [Google Scholar]
- 2.Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 3.Go Y-M, Chandler JD, Jones DP. The cysteine proteome. Free Radic Biol Med. 2015;84:227–245. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covarrubias L, Hernández-García D, Schnabel D, Salas-Vidal E, Castro-Obregón S. Function of reactive oxygen species during animal development: Passive or active? Dev Biol. 2008;320:1–11. doi: 10.1016/j.ydbio.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 5.Holmgren A, et al. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 6.Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aon MA, Cortassa S, O’Rourke B. Redox-optimized ROS balance: A unifying hypothesis. Biochim Biophys Acta. 2010;1797:865–877. doi: 10.1016/j.bbabio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nickel A, Kohlhaas M, Maack C. Mitochondrial reactive oxygen species production and elimination. J Mol Cell Cardiol. 2014;73:26–33. doi: 10.1016/j.yjmcc.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Netto LES, Antunes F. The roles of peroxiredoxin and thioredoxin in hydrogen peroxide sensing and in signal transduction. Mol Cells. 2016;39:65–71. doi: 10.14348/molcells.2016.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhee SG, Kil IS. Multiple functions and regulation of mammalian peroxiredoxins. Annu Rev Biochem. 2017;86:749–775. doi: 10.1146/annurev-biochem-060815-014431. [DOI] [PubMed] [Google Scholar]
- 11.Dumollard R, Ward Z, Carroll J, Duchen MR. Regulation of redox metabolism in the mouse oocyte and embryo. Development. 2007;134:455–465. doi: 10.1242/dev.02744. [DOI] [PubMed] [Google Scholar]
- 12.Urner F, Sakkas D. Involvement of the pentose phosphate pathway and redox regulation in fertilization in the mouse. Mol Reprod Dev. 2005;70:494–503. doi: 10.1002/mrd.20222. [DOI] [PubMed] [Google Scholar]
- 13.Chaube SK, Khatun S, Misra SK, Shrivastav TG. Calcium ionophore-induced egg activation and apoptosis are associated with the generation of intracellular hydrogen peroxide. Free Radic Res. 2008;42:212–220. doi: 10.1080/10715760701868352. [DOI] [PubMed] [Google Scholar]
- 14.Tripathi A, et al. Intracellular levels of hydrogen peroxide and nitric oxide in oocytes at various stages of meiotic cell cycle and apoptosis. Free Radic Res. 2009;43:287–294. doi: 10.1080/10715760802695985. [DOI] [PubMed] [Google Scholar]
- 15.Tirmarche S, Kimura S, Dubruille R, Horard B, Loppin B. Unlocking sperm chromatin at fertilization requires a dedicated egg thioredoxin in Drosophila. Nat Commun. 2016;7:13539. doi: 10.1038/ncomms13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emelyanov AV, Fyodorov DV. Thioredoxin-dependent disulfide bond reduction is required for protamine eviction from sperm chromatin. Genes Dev. 2016;30:2651–2656. doi: 10.1101/gad.290916.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellicena-Pallé A, Stitzinger SM, Salz HK. The function of the Drosophila thioredoxin homologue encoded by the deadhead gene is redox-dependent and blocks the initiation of development but not DNA synthesis. Mech Dev. 1997;62:61–65. doi: 10.1016/s0925-4773(96)00650-8. [DOI] [PubMed] [Google Scholar]
- 18.Salz HK, et al. The Drosophila maternal effect locus deadhead encodes a thioredoxin homolog required for female meiosis and early embryonic development. Genetics. 1994;136:1075–1086. doi: 10.1093/genetics/136.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avilés-Pagán EE, Orr-Weaver TL. Activating embryonic development in Drosophila. Semin Cell Dev Biol. February 21, 2018 doi: 10.1016/j.semcdb.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu L, et al. Systematic and quantitative assessment of hydrogen peroxide reactivity with cysteines across human proteomes. Mol Cell Proteomics. 2017;16:1815–1828. doi: 10.1074/mcp.RA117.000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Carroll KS, Liebler DC. The expanding landscape of the thiol redox proteome. Mol Cell Proteomics. 2016;15:1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojdyla K, Rogowska-Wrzesinska A. Differential alkylation-based redox proteomics–Lessons learnt. Redox Biol. 2015;6:240–252. doi: 10.1016/j.redox.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsen CE, Carroll KS. Cysteine-mediated redox signaling: Chemistry, biology, and tools for discovery. Chem Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doane WW. Completion of meiosis in uninseminated eggs of Drosophila melanogaster. Science. 1960;132:677–678. doi: 10.1126/science.132.3428.677. [DOI] [PubMed] [Google Scholar]
- 25.Kronja I, et al. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 2014;7:1495–1508. doi: 10.1016/j.celrep.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albrecht SC, Barata AG, Grosshans J, Teleman AA, Dick TP. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab. 2011;14:819–829. doi: 10.1016/j.cmet.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Albrecht SC, et al. Redesign of genetically encoded biosensors for monitoring mitochondrial redox status in a broad range of model eukaryotes. J Biomol Screen. 2014;19:379–386. doi: 10.1177/1087057113499634. [DOI] [PubMed] [Google Scholar]
- 28.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martell J, et al. Global cysteine-reactivity profiling during impaired Insulin/IGF-1 signaling in C. elegans identifies uncharacterized mediators of longevity. Cell Chem Biol. 2016;23:955–966. doi: 10.1016/j.chembiol.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, et al. Global, in situ, site-specific analysis of protein S-sulfenylation. Nat Protoc. 2015;10:1022–1037. doi: 10.1038/nprot.2015.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Z, Arscott LD, Ballou DP, Williams CH., Jr The relationship of the redox potentials of thioredoxin and thioredoxin reductase from Drosophila melanogaster to the enzymatic mechanism: Reduced thioredoxin is the reductant of glutathione in Drosophila. Biochemistry. 2007;46:7875–7885. doi: 10.1021/bi700442r. [DOI] [PubMed] [Google Scholar]
- 32.Kanzok SM, et al. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 33.Page AW, Orr-Weaver TL. Activation of the meiotic divisions in Drosophila oocytes. Dev Biol. 1997;183:195–207. doi: 10.1006/dbio.1997.8506. [DOI] [PubMed] [Google Scholar]
- 34.Perkins AT, Das TM, Panzera LC, Bickel SE. Oxidative stress in oocytes during midprophase induces premature loss of cohesion and chromosome segregation errors. Proc Natl Acad Sci USA. 2016;113:E6823–E6830. doi: 10.1073/pnas.1612047113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motohashi K, Romano PGN, Hisabori T. Identification of thioredoxin targeted proteins using thioredoxin single cysteine mutant-immobilized resin. Methods Mol Biol. 2009;479:117–131. doi: 10.1007/978-1-59745-289-2_8. [DOI] [PubMed] [Google Scholar]
- 36.Sieber MH, Thomsen MB, Spradling AC. Electron transport chain remodeling by GSK3 during oogenesis connects nutrient state to reproduction. Cell. 2016;164:420–432. doi: 10.1016/j.cell.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tennessen JM, et al. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 2014;4:839–850. doi: 10.1534/g3.114.010652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kronja I, et al. Quantitative proteomics reveals the dynamics of protein changes during Drosophila oocyte maturation and the oocyte-to-embryo transition. Proc Natl Acad Sci USA. 2014;111:16023–16028. doi: 10.1073/pnas.1418657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svensson MJ, Chen JD, Pirrotta V, Larsson J. The ThioredoxinT and deadhead gene pair encode testis- and ovary-specific thioredoxins in Drosophila melanogaster. Chromosoma. 2003;112:133–143. doi: 10.1007/s00412-003-0253-5. [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, Oko R, Miranda-Vizuete A. Developmental expression of spermatid-specific thioredoxin-1 protein: Transient association to the longitudinal columns of the fibrous sheath during sperm tail formation. Biol Reprod. 2002;67:1546–1554. doi: 10.1095/biolreprod.102.004838. [DOI] [PubMed] [Google Scholar]
- 41.Shalaby NA, et al. Systematic discovery of genetic modulation by Jumonji histone demethylases in Drosophila. Sci Rep. 2017;7:5240. doi: 10.1038/s41598-017-05004-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young JI, Züchner S, Wang G. Regulation of the epigenome by vitamin C. Annu Rev Nutr. 2015;35:545–564. doi: 10.1146/annurev-nutr-071714-034228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 44.Ge W, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol. 2012;8:960–962. doi: 10.1038/nchembio.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montrichard F, et al. Thioredoxin targets in plants: The first 30 years. J Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Knoefler D, et al. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol Cell. 2012;47:767–776. doi: 10.1016/j.molcel.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menger KE, et al. Fasting, but not aging, dramatically alters the redox status of cysteine residues on proteins in Drosophila melanogaster. Cell Rep. 2015;11:1856–1865. doi: 10.1016/j.celrep.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121:101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hara M, Petrova B, Orr-Weaver TL. Control of PNG kinase, a key regulator of mRNA translation, is coupled to meiosis completion at egg activation. eLife. 2017;6:e22219. doi: 10.7554/eLife.22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.