Significance
In vivo fluorescence microscopy with resolution well beyond the diffraction limit entails complexities that challenge the attainment of sufficient image brightness and contrast. These challenges have so far hampered investigations of the nanoscale distributions of synaptic proteins in the living mouse. Here, we describe a combination of stimulated emission depletion microscopy and endogenous protein labeling, providing high-quality in vivo data of the key scaffolding protein PSD95 at the postsynaptic membrane, which frequently appeared in extended distributions rather than as isolated nanoclusters. Operating in the far-red to near-IR wavelength range, this combination promises reduced photostress compared with prior in vivo nanoscopy at much shorter wavelengths.
Keywords: in vivo fluorescence nanoscopy, superresolution imaging, synaptic architecture, PSD95, protein labeling
Abstract
Extending superresolution fluorescence microscopy to living animals has remained a challenging frontier ever since the first demonstration of STED (stimulated emission depletion) nanoscopy in the mouse visual cortex. The use of fluorescent proteins (FPs) in in vivo STED analyses has been limiting available fluorescence photon budgets and attainable image contrasts, in particular for far-red FPs. This has so far precluded the definition of subtle details in protein arrangements at sufficient signal-to-noise ratio. Furthermore, imaging with longer wavelengths holds promise for reducing photostress. Here, we demonstrate that a strategy based on enzymatic self-labeling of the HaloTag fusion protein by high-performance synthetic fluorophore labels provides a robust avenue to superior in vivo analysis with STED nanoscopy in the far-red spectral range. We illustrate our approach by mapping the nanoscale distributions of the abundant scaffolding protein PSD95 at the postsynaptic membrane of excitatory synapses in living mice. With silicon-rhodamine as the reporter fluorophore, we present imaging with high contrast and low background down to ∼70-nm lateral resolution in the visual cortex at ≤25-µm depth. This approach allowed us to identify and characterize the diversity of PSD95 scaffolds in vivo. Besides small round/ovoid shapes, a substantial fraction of scaffolds exhibited a much more complex spatial organization. This highly inhomogeneous, spatially extended PSD95 distribution within the disk-like postsynaptic density, featuring intricate perforations, has not been highlighted in cell- or tissue-culture experiments. Importantly, covisualization of the corresponding spine morphologies enabled us to contextualize the diverse PSD95 patterns within synapses of different orientations and sizes.
Unraveling the structure and molecular composition of synapses is paramount to elucidating the mechanisms of synaptic transmission. Since many neurological and psychiatric disorders are associated with alterations in synapse morphology, understanding synapses also has wide implications in the pathophysiology of the brain. Accurate information about the organization of key proteins on the nanoscale is essential to characterizing synaptic molecular architecture and to discovering the principles of its organization.
For decades, EM has essentially been the only method to study the ultrastructural morphology of synapses and their internal molecular organization (1). EM visualization methods, however, are limited in their molecular specificity and labeling coverage, as well as by the number of synapses typically investigated. The time demands of both EM data acquisition and analysis lead to a lower throughput compared with optical methods. Most importantly, EM cannot be applied to living cells, tissue, or animals. Fluorescence microscopy, in contrast, offers these features but has long lacked the required spatial resolution necessary to identify the distribution of molecular entities in the confined space of the synapse. Nevertheless, fluorescence imaging, and especially two-photon excitation microscopy, has been one of the most powerful and widely used imaging techniques in the neurosciences due to its outstanding ability for live-cell and in vivo applications with specific molecular recognition (2–4).
The advent of lens-based subdiffraction (superresolution) fluorescence microscopy methods such as those called stimulated emission depletion (STED) (5, 6) and reversible saturable/switchable optical linear fluorescence transitions (RESOLFT) (7, 8), and later also photoactivated localization microscopy/stochastic optical reconstruction microscopy (PALM/STORM) (9–11), jointly referred to as fluorescence nanoscopy (12), has dramatically changed the outlook on resolution. These approaches provide minimally invasive access to the composition and function of synapses (13, 14). Live-cell nanoscopy in particular has begun to enable explorations across the neurosciences (15–20), opening up research directions which could not be addressed by electron or conventional optical microscopy. These studies, however, investigated neuronal morphology and synaptic protein organization and function in model cell or tissue systems, and hence may not have fully reflected the physiological environment and complexity in a living animal. The ideal approach to characterizing the spatial distribution of a synaptic protein is therefore to map its appearance in vivo, excluding artifacts induced by culturing cells or tissues (21). However, in vivo nanoscopy comes with additional complexities of the experiment. Major challenges are the higher susceptibility to optical aberrations and to inherent motion of the animal due to heartbeat and breathing, which motivates the use of fast data acquisition. Additionally, light scattering reduces the signal collection efficiency with increasing imaging depth.
STED nanoscopy is so far the only superresolution fluorescence microscopy approach which has been demonstrated in living brain slices (16, 22) and in the brain of anesthetized live mice (23–26). These previous in vivo STED imaging studies examined dendritic features based on (over)expression of fluorescent proteins (FPs) either as volume labeling or to mark the actin network. A notable exception is the most recently reported application of in vivo STED imaging to a synaptic protein endogenously tagged with EGFP (27). The use of green or yellow FPs in STED nanoscopy, however, brings about increased hazards of tissue photodamage due to the higher photon energies of the required excitation and STED light in the green–yellow range. Indeed, previous in vivo STED imaging experiments using enhanced YFP (EYFP) exhibited instances of phototoxic swellings in dendritic morphologies (23, 24). Considering that common natural absorbers in tissue have their highest absorption at visible wavelengths (28, 29), working in the far-red to near-IR spectral range is favorable. In addition, imaging with light at red-shifted wavelengths benefits from reduced scattering, better optical penetration, and improved image contrast due to lower autofluorescence (30). A recent report of in vivo STED nanoscopy in mice with far-red-emitting FPs (mNeptune2 and tagRFP657) did not observe any obvious tissue photodamage (25). At the same time, red to far-red FPs show lower performance in terms of photostability and quantum yield compared with their green/yellow analogs (30). FPs generally have poorer photophysical properties compared with synthetic organic-fluorophore (dye) labels. To overcome the above limitations, a promising direction is the application of far-red to near-IR organic fluorescent dyes for in vivo STED nanoscopy.
Intracellular labeling with organic dyes in live-cell experiments requires membrane permeability, nontoxicity, and a high, nonperturbative affinity to the target structure within the cell. The latter requirement can only be fulfilled if the protein of interest or/and the fluorescent probe exhibit specific binding characteristics, as is the case for small molecular probes or enzymatic protein tags. Recently, several far-red and near-IR dyes with excellent STED compatibility have become available which label intracellular proteins with high specificity in living cells, for example by small molecular probes (31–35) or by means of the self-labeling techniques of SNAP-, Halo-, or Clip-Tag fusion proteins (36–40). For the latter class of labeling systems, the protein of interest is genetically fused with an engineered enzyme tag which covalently binds the corresponding fluorescent substrate (41–43). A major benefit of this kind of fusion tag is that the fluorescent dye ligand can be freely chosen, depending on the requirements of the imaging.
Exploiting this benefit, we now demonstrate a versatile approach for imaging the nanoscale distribution of a synaptic protein in vivo, specifically in dendritic spines of the neocortex of anesthetized live mice. To this end, we used a transgenic knock-in mouse line engineered to express endogenous postsynaptic density 95 (PSD95) protein in fusion with the HaloTag enzyme. The HaloTag sequence allowed us to covalently label PSD95 in vivo with the ligand silicon-rhodamine-Halo (SiR-Halo). We stereotactically injected SiR-Halo into layer I of the visual cortex during the surgical preparation and performed in vivo superresolution imaging on a home-built upright STED nanoscope with excitation of far-red fluorescence at 640 nm and near-IR 775-nm deexcitation. The utilization of the near-IR SiR-Halo ligand resulted in highly specific labeling of PSD95 and imaging with superior signal-to-noise ratio and increased photostability compared with recently described FP-based in vivo experiments. We found that endogenous PSD95 formed diverse nanoscale arrangements with large variations in shape and size among individual synapses, as revealed by confocal covisualization of the dendritic morphology relative to the superresolved PSD95 scaffolds. In many synaptic sites, PSD95 resided in compact, round or ovoid assemblies with dimensions often considerably smaller than the diffraction limit. Frequently, we observed the PSD95 scaffolds as complex, oval or ring-like arrangements with irregular borders and internal perforations, which were not visible in the confocal mode and have not been reported in such detail in experiments with cultured neurons or brain slices. Our study underscores the importance of in vivo superresolution microscopy and introduces a powerful method for future investigations of the molecular organization and function of the brain in living animals.
Results
In Vivo Labeling of Endogenous PSD95.
To address the signal and photostability limitations of FPs in in vivo superresolution STED imaging, we explored an approach that exploits the molecular specificity of a genetically encoded self-labeling enzyme tag fused to the protein of interest, combining it with the superior photophysical performance of organic fluorophores. We utilized a transgenic PSD95 mouse, PSD95-HaloTag, as a knock-in with the HaloTag enzyme, which has the highest reported binding rate among the widely used enzymatic self-labeling protein tags (SNAP, Halo, and Clip). PSD95 is the major constituent of the PSD in dendritic spine heads, where it is known to act as a scaffold for numerous other proteins (44, 45) and controls synaptic plasticity and learning (46). The HaloTag was fused to the C terminus of PSD95 using a previously described genome engineering approach (45). In this mouse line, PSD95-HaloTag is expressed under the control of the regulatory elements of the native PSD95 gene, with the mice showing normal protein expression, localization, and synaptic physiological function. Hereafter we simply refer to PSD95-HaloTag as PSD95. For the staining of endogenous PSD95 fusion proteins with organic HaloTag ligands, we established an in vivo labeling strategy based on intracortical injections (Fig. 1). We chose SiR as the most appropriate membrane-permeable near-IR dye, since it features efficient excitation at 640–650 nm (36) and has enabled excellent nanoscopy performance in the far-red spectrum in living cultured cells, including neurons, at the popular STED wavelength of 775 nm (20, 32, 37, 47).
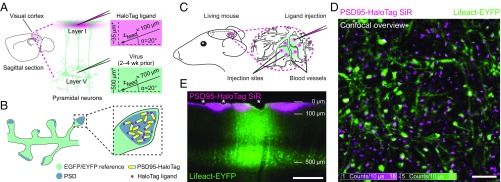
Fig. 1.
In vivo labeling of endogenous PSD95-HaloTag with organic fluorophores. (A–C) Schematic illustrations of the in vivo labeling of the postsynaptic protein PSD95 in combination with a morphological reference (see text). (A) Transgenic PSD95-HaloTag mice were transduced with recombinant adeno-associated viral particles encoding either Lifeact-EYFP or cytosolic EGFP, 2–4 wk before the in vivo labeling. The viral vectors were stereotactically injected into layer V of the visual cortex for FP expression in the pyramidal neurons (SI Appendix, Virus Transduction), whose dendritic arborizations expand upward to the cortical surface (green). SiR-Halo (750 nM, 100–250 nL) was injected at an angle of = 20° to the brain surface and a feed of zfeed = 100 µm, resulting in an injection site at a final depth of ∼35 µm below the cortical surface (magenta). *Depths below cortical surface are also affected by the curvature of the brain, see SI Appendix, Fig. S3A. (B) Schematic of molecular components within dendritic spine heads. Intracortical injections of SiR-Halo into layer I resulted in the specific binding of the SiR-Halo-ligand to the endogenously expressed PSD95-HaloTag fusion proteins. (C) Injections of SiR-Halo were set at multiple sites (magenta) in the region of EGFP/EYFP reference expression (green), in between superficial blood vessels. (D) Representative confocal overview image of SiR-Halo-labeled PSD95 fusion proteins (magenta), together with the actin reference (Lifeact-EYFP, green) in an anesthetized live mouse about 1 h after ligand injection. The labeling revealed PSD95 at synapses throughout the dendritic network. White in the overlay corresponds to colocalization of SiR-Halo–labeled PSD95 within spine heads of the morphological reference. In some experiments, the Lifeact-EYFP labeling was replaced by cytosolic EGFP expression (SI Appendix, Fig. S2). The overview image in D represents raw data. Imaging parameters are listed in SI Appendix, Table S1. (Scale bar: 5 µm.) (E) Epifluorescence image of a PFA-fixed coronal brain section (70-µm thickness) revealing the extent of the SiR-Halo label penetration in the visual cortex 1 h after injection in relation to the Lifeact-EYFP reference. Cell bodies of the Lifeact-EYFP–expressing pyramidal neurons are located predominantly at the depth of ∼500 µm below the cortical surface targeted by virus injection. The SiR-Halo labeling extended down to 100 µm with significant lateral broadening from the injection sites. Asterisks: superficial blood vessels. (Scale bar: 250 µm.)
The imaging approach involved the injection of neuron-specific recombinant adeno-associated viral particles (rAAV, mixed serotype 1 and 2), encoding either Lifeact-EYFP or cytosolic EGFP, into layer V of the intended imaging region 2–4 wk before the in vivo labeling and STED nanoscopy (SI Appendix, Virus Transduction). Lifeact-EYFP labeling of actin served as a dendritic morphology reference for the localization of the PSD95 fusion proteins in dendritic spines of the pyramidal neurons (Fig. 1 A and B). In some experiments cytosolic volume-filling EGFP expression was used instead of the Lifeact labeling. To label PSD95, we injected SiR as a HaloTag-ligand (SiR-Halo) stereotactically into layer I of the visual cortex at multiple sites in between the superficial blood vessels (SI Appendix, Surgical Preparation for In Vivo STED Imaging and In Vivo Labeling and Fig. 1 A and C).
About an hour after the SiR-Halo injection, we tested the labeling of PSD95 by in vivo recordings on our home-built upright STED nanoscope. This 775-nm STED system (SI Appendix, In Vivo STED Nanoscope and Fig. S1) was equipped with a high-N.A. 63× glycerol-immersion objective lens. Confocal-mode imaging of the molecular layer of the visual cortex demonstrated the high specificity of endogenous PSD95 labeling, with imaging at excellent signal-to-background for depths of 5–25 μm below the cortical surface (Fig. 1D). This can be ascribed to the fluorogenic character of SiR, which shows an increased fluorescence emission only when bound to the target protein, while it is almost nonfluorescent in its unbound state (36). As expected, PSD95 showed the characteristic punctate distribution throughout the cortical dendritic network, with Lifeact-EYFP (Fig. 1D) or cytosolic volume-filling EGFP (SI Appendix, Fig. S2) expressed in a sufficient number of dendrites to enable morphological recognition of the spines in many cases. The confocal puncta exhibited different dimensions ranging from small and round to larger ones with irregular shapes. Not all PSD95 puncta colocalized with an EYFP/EGFP-labeled spine head, which is expected as not all pyramidal neurons were transduced by the virus, whereas PSD95 was endogenously expressed at the excitatory synapses of all neuronal cells.
With the labeling in place, starting at ∼1 h after dye injection, imaging was possible for hours, with our experiments typically completed within 4 h. Upon moving to a new region of interest (ROI), we encountered similar signal intensities as initially in prior ROIs, with no systematic signal reduction over the experiment. Local photobleaching at ROIs upon repeated imaging is discussed below. This persistence of signals is consistent with the finding in cultured neurons that endogenous PSD95 turnover is on the order of a few days (48), and that significant turnover leading to a partial replacement of the pool of labeled PSD95 is thus not expected to occur in the time frame of just 3–4 h postlabeling. Beyond considerations of protein turnover, the covalent attachment of SiR-Halo to a Halo-tagged protein is long-term and does not limit maximal experiment durations. Improved stability of the head fixation and cranial-window preparation by a new head-holder design, a more suitable composition of the dental cement used as an adhesive, and by an adapted wound-sealing protocol (SI Appendix, Surgical Preparation for In Vivo STED Imaging and Fig. S3) contributed to enabling prolonged imaging in our experiments. We kept the vital parameters of the mouse stable by adapting the depth of anesthesia and the artificial-ventilation parameters and by maintaining the body temperature (SI Appendix, Animal Procedures).
The spatial extent of the PSD95 labeling was verified by wide-field imaging of paraformaldehyde (PFA)-fixed brain sections. The SiR-Halo injections, at a depth of ∼35 μm below the cortical brain surface, produced concentrated fluorescent spots due to the penetration of the ligand from the cortical surface down to a depth of ∼100 μm, and a lateral extent of ∼100–300 μm (Fig. 1E). The direct intracortical injection of the SiR-Halo-ligand was thus highly effective to mark PSD95 in the visual cortex.
A Dedicated Platform for Far-Red in Vivo STED Nanoscopy.
Having established the labeling strategy, we aimed at resolving the native PSD95 nanoarchitectures using STED nanoscopy. To this end, we developed a compact in vivo STED nanoscope (SI Appendix, Fig. S1) for the far-red spectrum with a main design focus on mechanical stability, implementing short optical beam paths with groups of components decoupled from each other on separate breadboards in a modular fashion. Fibers delivered the laser light for excitation and deexcitation to the core module of the system (STED unit) and guided the fluorescence to the detection module. The STED unit was directly attached to the microscope stand, without the need for redirecting beams once they were coaxially aligned. An independent translation stage for mouse positioning, which was mechanically decoupled from the microscope body and other optomechanics, ensured that the impact of vibrations or thermal fluctuations on the associated optics was minimal. Our imaging system allowed us to adjust the lateral 2D resolution down to 40–50 nm (as determined with ∼20-nm fluorescent bead samples). It rendered SiR-Halo–labeled β-tubulin–HaloTag filaments in cultured living cells with a width of 66 nm FWHM for the maximally available STED average power of 177 mW (all powers stated in back aperture), suggesting an FWHM resolution of down to 50–60 nm (SI Appendix, Fig. S4).
In Vivo STED Nanoscopy of PSD95 Scaffolds with Excellent Structural Definition.
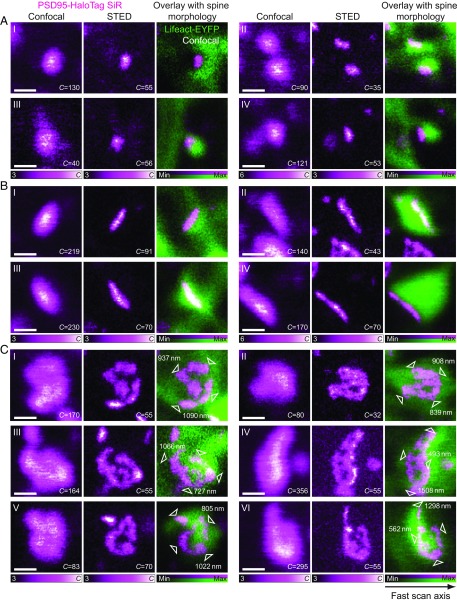
In vivo STED imaging was performed in ROIs identified from confocal overview images at 5- to 25-μm depth below the cortical surface, comparable to previous STED imaging in the mouse neocortex (24, 25). Whereas the confocal images showed PSD95 scaffolds as blurred puncta or elongated shapes of various dimensions and brightness, spanning from the diffraction limit up to ∼1.5 µm across, STED nanoscopy resolved the nanoscale architecture of individual PSD95 arrangements in unprecedented detail. We observed that PSD95 formed diverse nanoscale arrangements with large variations in size and shape among individual synapses (Figs. 2 and 3). These variations are putatively in part due to the differing orientations and axial positions with respect to the focal plane. The appearance of a PSD95 scaffold in a 2D recording can be drastically affected by its relative orientation to the imaging plane. In the 2D STED images, the PSD95 geometries typically appeared as small and round/ovoid, thin and elongated, or as larger structures with complex perforations (Fig. 3).
Fig. 2.
STED imaging of PSD95 scaffolds in vivo with excellent structural definition. (A) STED image of endogenous PSD95-HaloTag fusion proteins stained with SiR-Halo ligand (magenta), merged with the confocal image of the dendritic reference staining (Lifeact-EYFP, green). PSD95 appeared in various dimensions. (Scale bar: 2 µm.) (B) Magnified views (I–III) of the regions marked in the overview (A) are shown in confocal and STED mode, and as an overlay of the STED image of PSD95 with the confocal image of the dendritic reference staining. Superresolved PSD95 organizations appeared in different sizes and shapes. These include small round/ovoid (I), thin elongated (II), and complex-shaped (III) assemblies, which remain unresolved in the confocal counterpart images. Depending on the spine-head orientation with respect to the focal plane, intermediate variations of these basic appearances were imaged. The arrow in the bottom left indicates the fast scan axis for all images shown. Images were smoothed for display with a one-pixel-wide Gaussian filter. Imaging parameters are listed in SI Appendix, Table S1. (Scale bars: 500 nm.) (C) Normalized intensity profiles (five-pixel width) of the raw STED data from the indicated arrowheads of magnified views in B. Lorentzian (STED) and Gaussian (confocal) fits indicate the widths of individual PSD95 features as FWHM (I and II), and separations in the complex PSD95 pattern (III) are given, demonstrating an approximately of STED over confocal imaging (Inormalized, normalized signal intensity; r, distance).
Fig. 3.
Impressions of PSD95 scaffold nanoarchitectures in vivo. (A–C) Examples of the different appearances of PSD95 assemblies (SiR-Halo ligand, magenta) in 2D imaging in (Left) confocal mode, (Middle) STED mode, and (Right) the overlay of the STED image of PSD95 with the confocal image of the dendritic reference labeling (Lifeact-EYFP, green). (A) Examples of small round/ovoid PSD95 scaffolds, primarily located in small dendritic spine heads as revealed by the reference label. (B) Examples of thin elongated PSD95 scaffold side views in large spine heads. (C) Examples of complex PSD95 nanoarchitectures, with overall dimensions (white arrowheads) determined in the raw STED images from intensity profiles. Images represent magnified views extracted from full-field-of-view recordings. The fast scan axis for all images shown is indicated by an arrow in the bottom right. All images were smoothed for display with a one-pixel-wide Gaussian filter. Imaging parameters are listed in SI Appendix, Table S1. (All scale bars: 500 nm.)
The in vivo STED imaging was carried out in regions largely devoid of prominent vascularization, to reduce motion artifacts induced by pulsing blood vessels. Furthermore, rigid fixation of the mouse head and the cranial window (SI Appendix, Fig. S3B) largely prevented artifacts due to respiration-induced motions. Only rarely, abrupt lateral displacements were observed during the scan in imaging regions where the pulsing of blood vessels was sufficiently strong (SI Appendix, Fig. S3C). Such instances were recognizable by inspection of thin features oriented along the slow scan axis, and we moved to a different field of view (FOV) whenever this was observed.
The SiR labeling facilitated STED recordings of PSD95 with robust signals of up to 100–120 counts per pixel in the brightest parts of the structure (typically exceeding 50–60 counts per pixel throughout), and a low background noise level of usually one to three counts. In a given FOV, the high photon budget and photostability of SiR under STED conditions allowed the recording of at least six sequential image scans with sufficient brightness and contrast to appreciate internal PSD95 scaffold morphologies (SI Appendix, Fig. S5). At the utilized average powers for deexcitation by STED of 53 or 66 mW in the in vivo experiments, a resolution of 90 nm was estimated based on the thinnest (∼80–95 nm) features in PSD95 scaffolds represented at robust signal intensities (compare examples in SI Appendix, Fig. S6). Further-enhanced resolution in vivo, at the level of 70 nm, was demonstrated for a higher STED power of 169 mW (SI Appendix, Fig. S6). Nevertheless, we typically limited the STED power to PSTED of 53 or 66 mW, except for occasional imaging at this higher STED power to explore current limits to the achievable resolution with our labeling and imaging approach. This allowed repeated imaging at still approximately fourfold improved resolution over confocal microscopy and with robust signal. Our resolution estimate of 90 nm is consistent with the experimentally determined STED-power dependence in our test experiments with cultured cells (SI Appendix, Fig. S4), indicating that the resolution performance of our setup can be fully harnessed in vivo.
Contrary to the SiR-Halo experiments, in vivo STED imaging of the far-red FPs mGarnet or mNeptune2 fused to PSD95 (virus-mediated expression) did not achieve sufficient resolution and image brightness to be able to resolve structural features to the same extent (SI Appendix, Fig. S7). Although the resolution enhancement between confocal and STED images of the far-red FPs was apparent, the overall performance did not reach that of SiR-based imaging.
A semiautomated analysis based on 2D STED images (n = 1,148 PSD95 scaffolds; Fig. 4 and SI Appendix, Fig. S8 and Semiautomated Analysis of PSD95 Scaffolds) revealed that the size of the arrangements spanned dimensions (minor × major axis, referred to as the short and the long axis) from 105 nm × 126 nm in the case of small puncta to 606 nm × 1,438 nm for the largest structures (Fig. 4A). The PSD95 arrangements featured mean dimensions of 237 nm × 458 nm, and a mean area of 0.12 µm2 (scaffold area: the area enclosed by the outermost perimeter of the scaffold) (Fig. 4B). In the majority of analyzed assemblies (57%; Fig. 4A), we observed small round or ovoid PSD95 arrangements (Figs. 2 B, I and 3A) and chose to designate scaffolds “small round/ovoid” if their long and short axes and were bounded by the condition (in nanometers) (Fig. 4A). For these scaffolds of long-axis diameters up to 500 nm, short-axis diameters were in the range of 103–335 nm (mean: 189 nm; SI Appendix, Fig. S9 A and B), and thus often considerably smaller than the diffraction limit.
Fig. 4.
Dimensions, areas, and estimated filling fractions of PSD95 scaffold nanoarchitectures extracted from a semiautomated image analysis (n = 1,148). (A) A 2D histogram of long-axis vs. short-axis diameters; 651 scaffolds (57%), for which the sum of their squared long and short axes was ≤500 nm2, were designated ‟small round/ovoid.” The remaining 497 scaffolds (43% of the total number) were larger and wider and often appeared as perforated complex-shaped structures. For separate representations of scaffolds in the small round/ovoid and larger categories, see SI Appendix, Fig. S9 A–H. (B) Frequency plot of scaffold areas; 17 of 1,148 scaffolds with area >0.5 µm2 are not included in the displayed histogram. (C) Frequency plot of estimated filling fraction (fraction of scaffold area filled by PSD95). Gray: all scaffolds; black: larger scaffolds as a subset of all scaffolds. (Inset) Enlarged view of larger-scaffold data.
In addition to the small round/ovoid scaffolds, the confocal images often showed large elliptical PSD95 assemblies, whereas with STED imaging these shapes were revealed as thin elongated (Fig. 2 B, II and Fig. 3B) and wider extended structures with complex perforations (Fig. 2 B, III and Fig. 3C), as well as intermediate shapes. For this more heterogeneous category of scaffolds (43%), the mean dimensions were 299 nm and 657 nm on the short and long axis, respectively (Fig. 4A and SI Appendix, Fig. S9 E and F). The complex-shaped, oval or ring-like structures with irregular borders and perforations were highly diverse, and without an apparent common structural geometry (Fig. 3C and SI Appendix, Fig. S10). We observed mostly continuous nanoarchitectures, with largely uniform signal intensity within the occupied domains. Rarely, hotspots of signal (i.e., protein density) were found within the extended continuous irregular distribution.
Noting that the overall areas bounded by the outer perimeters of scaffolds were not always entirely filled with PSD95 protein (black gaps, compare examples provided in Figs. 2 B and C (in parts III) and 3C and SI Appendix, Fig. S10), our semiautomated analysis also allowed us to extract a filling-fraction parameter. This filling fraction (Fig. 4C) quantifies the relative area (as an approximation to the volume) within the scaffold occupied by PSD95 (SI Appendix, Semiautomated Analysis of PSD95 Scaffolds). The scaffolds in the small round/ovoid category featured almost complete (close to 100%) filling throughout. For the larger scaffolds, the filling fractions were typically in a range from 60 to 100%, with a nonnegligible fraction of scaffolds with filling fractions of <80% and even as low as 50%.
In scaffolds taken from each of the most prominent geometric appearances within four large FOVs, the smallest measured widths (FWHM, intensity line profiles; SI Appendix, Fig. S6) were almost identical, with 88 ± 6 nm (mean ± 1 SD, = 58; PSTED = 53 or 66 mW) for the small round/oval-shaped, 92 ± 6 nm ( = 24; PSTED = 53 or 66 mW) for the thin elongated ones, and 91 ± 9 nm ( = 8; PSTED = 53 or 66 mW) for larger scaffolds of a more complex architecture. These sizes are approximately equal to the 90-nm in vivo resolution realized in this part of the imaging, suggesting the underlying protein distribution along the respective direction may thus be even more narrowly confined.
Relating the PSD95 Nanomorphology to the Spine Shape.
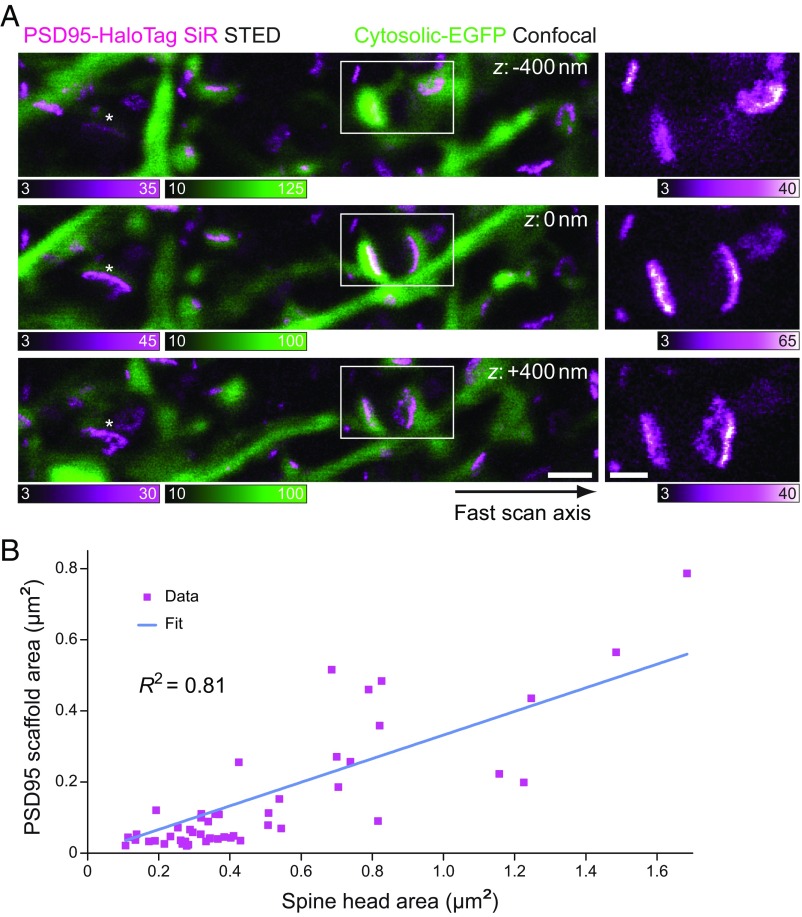
The appearance of PSD95 arrangements in STED recordings with lateral resolution enhancement can be highly dependent on the orientation and axial position of the synapse with respect to the focal plane. For small round/oval-shaped PSD95 distributions at or near the achieved resolution, the impact of orientation on appearance in the STED imaging is only marginal. We speculated that thin elongated PSD95 patterns were mainly a result of synapses observed as side views, whereas complex-shaped morphologies could represent top views of the PSD. We thus examined the 2D appearance of PSD95, putting it in context of the Lifeact-EYFP/cytosolic EGFP imaging channel to ascertain spine-head orientation. Where this determination was possible, our data showed thin elongated PSD95 scaffold structures arranged along the postsynaptic membranes of dendritic spine heads in side views ( = 9 for the examples in Figs. 2 B, II, 3B, and 5), as identified by the reference labeling showing the dendritic spine head connected to the neck and the shaft. Complex-shaped morphologies were observed in top or near-top views of synapses ( = 10 for the examples in Figs. 2 B, III, 3C, and 5), imaged as elliptical disks of the EYFP/EGFP reference. The lack of resolution enhancement along the direction complicated the unambiguous interpretation of observed 2D images. The disk-shaped PSD has been reported to have <30–50 nm extent in the direction perpendicular to its plane (49). The wide range of orientations in which the spine was encountered in our imaging led to different imaged projections of the PSD95 distribution located at the PSD. This explains the observation that examples of axial -stack STED imaging with a step size of 400 nm showed a thin elongated scaffold appearance in one focal plane and revealed a complex and perforated nanoscale arrangement in the adjacent plane (Fig. 5A). Such repeated imaging was not possible when utilizing far-red FPs instead of SiR (SI Appendix, Fig. S7) since, for PSTED almost equal to those of the SiR imaging, photobleaching prevented -stack recordings with multiple frames at high resolution for these labels.
Fig. 5.
PSD95 nanomorphologies related to spine shapes and sizes. (A) The 3D context of in vivo nanoscale PSD95 scaffold morphologies. (Left) Overlays of STED images of PSD95 (magenta) with the corresponding confocal images of cytosolic EGFP (green) from a -stack image series. Individual images were recorded with a 400-nm axial step size (acquisition order: : 0 nm, −400 nm, +400 nm). (Right) Magnified views of marked regions (white boxes) in the corresponding STED images on the left. The apparent morphologies of individual imaged PSD95 scaffolds are dependent on the orientation of the synapse with respect to the focal plane, such that the appearance of a PSD95 scaffold structure can change with the focal plane from a thin elongated appearance to a complex morphology. The asterisks mark an additional scaffold whose appearance changed with the focal plane. The fast scan axis for all images shown is indicated by an arrow. Counts stated are counts per 30 µs. All images were smoothed for display with a one-pixel-wide Gaussian filter. Imaging parameters are listed in SI Appendix, Table S1. [Scale bars: 1 μm (overlay), 500 nm (magnified views).] (B) Correlation of PSD95 scaffold area (imaged in STED mode) and corresponding spine-head area (imaged in confocal mode), both estimated from determinations of the respective short- and long-axis diameters. The blue line represents a linear fit of the data (N = 49 selected spines from a range of sizes; Pearson’s R2 = 0.81). PSD95 scaffolds without a matching Lifeact-EYFP spine head were not included.
Seeking to evaluate the effects of spine orientation, which we assume to be random, on the observed dimensions (Fig. 4A), we performed a simple geometrical simulation. Histograms of the measured data along the elliptic angle θ (SI Appendix, Fig. S9 I and J) reveal that the ellipticity (ratio of long-axis and short-axis diameters) of the PSD95 scaffold nanoarchitectures is distributed around preferred intermediate values of elliptic angle for either subset of scaffolds analyzed ( or in nanometers, respectively). Ellipticity is higher (i.e., tendency for lower θ) for the large scaffolds than for the small scaffolds. Although even perfectly round structures can appear elliptic when seen partially from the side, our data suggest that the PSD95 scaffold geometry is neither only round structures nor structures where the ellipticity can have any value with equal probability (both simulated under fully random orientation). While the extraction of exact values would require more extensive modeling, it is apparent from this analysis that the large, often complex nanoarchitectures are considerably more elliptical than the smaller scaffolds.
In general, dendritic spine size cannot be deduced from the size of a given PSD95 scaffold. Since previous studies have reported on a correlation of the spatial extent of the PSD with the size of the spine head (50, 51), we analyzed our data to explore any such relation. For this, the Lifeact-EYFP reference labeling was further used to verify the spine morphology in selected examples ( = 49) where the signal intensity in the EYFP channel allowed a reliable size determination. From fitting intensity line profiles and determining the FWHM, it became evident that the small round/ovoid PSD95 scaffolds (fulfilling , discussed above) were found in spine heads which tended to be small [minor axis: 488 ± 124 nm, major axis: 701 ± 145 nm, estimated area: 0.28 ± 0.11 µm2 ( = 22), based on confocal data]. In contrast, the larger assemblies were found in large spines [minor axis: 763 ± 259 nm, major axis: 1,048 ± 322 nm, estimated area: 0.67 ± 0.40 µm2 ( = 27)] (see examples in Figs. 2, 3, and 5). A strong positive correlation between spine head area and PSD95 scaffold area in images was confirmed (Pearson’s R2 = 0.81, = 49; Fig. 5B).
Discussion and Outlook
In vivo nanoscale fluorescence imaging has been applied relatively rarely and still represents a frontier, not only due to the complexity of the experiments but also due to a lack of adequate labeling tools. Whereas previous in vivo STED studies employed FPs, we here introduced an approach to overcome the signal and photostability limitations of FPs by exploiting the self-labeling HaloTag in combination with the near-IR organic dye SiR, a high-performance synthetic fluorophore.
With a typical focal-plane resolution of90 nm (or down to70 nm) in living mice, in vivo STED imaging allowed us to identify the nanoscale organization of PSD95 scaffolds in the molecular layer of the visual cortex, with appearances in various shapes and sizes among individual synapses (Figs. 2 and 3). Overall, the size of the PSD95 arrangements (Fig. 4A) spanned from dimensions of ∼100 nm × 130 nm in the case of small puncta to ∼600 nm × 1,400 nm for the largest structures and allowed us to visualize complex-perforated arrangements in great detail. In this aspect of a high diversity of PSD95 morphologies—from small to large—our observations are consistent with previous superresolution studies on the organization of PSD95 (27, 52).
As a main challenge to the interpretation of observed structures, the relative orientation to the imaging plane drastically affected the appearance of a PSD95 scaffold. Although the identification of a single scaffold’s orientation was ambiguous, two classes of scaffolds were distinguished by introducing a bound on their size. We found that ∼57% of the PSD95 assemblies were relatively small and round/ovoid (Figs. 3A and 4A and SI Appendix, Figs. S6A and S9). The other, larger structures (43%) appeared either thin and elongated or extended and complex with irregular borders and perforations (Figs. 3 B and C and 4A and SI Appendix, Figs. S6 B and C, S9, and S10), or as intermediate shapes. The PSD95 scaffolds which appeared thin elongated are plausible side views of the (predominantly complex) assemblies, orientated perpendicular to the imaging plane. The morphological reference imaging allowed us to confirm this in several examples. For all three main classes of appearance, the width of smallest features was ∼90 nm (or ∼70 nm for the higher STED power; SI Appendix, Fig. S6). This width was indicative of the resolution achieved at the respective STED power. Importantly, the distinctive complex morphologies were resolved in such detail only in the STED imaging mode, whereas confocal imaging did not resolve the internal structure.
Contrary to recent reports (18, 19, 52–56), our data at 70- to 90-nm resolution showed no indications of sparse, isolated PSD95 clustering. Notably, the complex-shaped scaffolds appeared to be continuous (albeit convoluted) structures, not fragmented or clustered. PSD95 “nanoclusters” or “nanodomains” have been reported in several superresolution studies of living or fixed dissociated rat hippocampal or cortical neurons (18, 19, 53, 54, 56) and in fixed brain sections of the mouse hippocampus (52). All these studies based conclusions on the number of nanoclusters per synaptic site, identified as elevated density of localizations in PALM/STORM nanoscopy data, or as particularly bright image regions in STED images. In consensus, these studies reported spines containing typically one to two PSD95 clusters, much fewer with three or more clusters and rarely observed more extensive patterns (containing up to five individual clusters). In these works the structures were analyzed within the cluster paradigm, rather than considering the existence of distributed, continuous arrangements. The studies diverge in their conclusions on the spatial extent of clusters, with determinations ranging from 80 to 150 nm. Furthermore, the cluster terminology has also been used to include rather large domains of up to 400–600 nm in diameter (mean: ∼300 nm) (53). As an important aspect, the studies concluded that clusters were separated by substantial distances of 100–300 nm, that is, farther apart than the resolution of the various fluorescence nanoscopy experiments of between 25 and 80 nm. The resolution down to ∼70 nm in our PSD95 analysis in the living mouse would have readily resolved clusters at the reported spacings. The discrete clustering in all these reports, which included studies in living cell-culture systems (18, 19, 53, 56), with separations almost as large as the diffraction limit, is not supported by our observations in vivo. The different resolutions between the aforementioned reports and our present in vivo study cannot account for the qualitatively different observations, given that even a report claiming 25-nm resolution in live cells quantified nanoclusters to be ∼80 nm in size individually (18).
Instead, possible explanations for the discrepancy include the effects of culturing neurons in a flat monolayer without the presence of other cell types such as astrocytes. A further difference is that our in vivo imaging explored dendritic spines in the molecular layer (layer I) of neurons from deeper cortical layers, rather than neurons from hippocampal regions. Furthermore, different rodent species (mouse vs. rat in the other reports except ref. 52) were used. On a technical level, the majority of past reports imaged transiently (over)expressed PSD95, whereas our present study examined the mouse brain with endogenous protein expression, that is, without possibly uncontrolled effects of the PSD95 overexpression on interacting proteins, as pointed out for AMPA receptors in one study (19). Choices of image processing and presentation, in particular thresholding, may additionally affect the PSD95 data and may lead to a clustered appearance by unduly emphasizing the hotspots which we also observed in a small number of scaffolds.
In the PALM/STORM reports, clusters of PSD95 were surrounded by a small but nonzero baseline density of localizations throughout the PSD. These localizations were not further discussed in refs. 18, 19, and 54, but they may be interpreted as indicative of PSD95’s being present at more moderate levels also in the proximity of clusters. In contrast, our STED data suggest that PSD95 protein is distributed as continuous “bands” of varying length, as often seen within the complex, convoluted structures. STED nanoscopy revealed PSD95 scaffolds as heavily segregated into spatial domains either containing or entirely lacking the protein. In the PSD95-containing regions, its concentration appeared rather homogenous. Note that STED imaging renders protein concentration differences in the specimen linearly, from very low to highest encountered density. This is a strength of our approach, and the localization of PSD95 in well-separated clusters, as observed in refs. 18, 19, and 52–56, can be excluded for our in vivo conditions based on our results.
To summarize, while small round/ovoid scaffolds were commonly observed, our in vivo data also suggest the existence of many large PSD95 scaffolds with complex internal organization, rather than small numbers of isolated nanoclusters. The present resolution did not allow us to exclude the possibility that larger numbers of clusters are densely spaced within the highly convoluted scaffold, but certainly not just one to three nanoclusters. Therefore, it will be necessary to further scrutinize the smallest structural units of PSD95 in vivo with yet higher resolution, a challenge that will likely be successfully met by STED nanoscopy or related concepts.
From our data we can further discern the relation of the PSD95 nanoscale morphology to the spine head shape. By utilizing information from a second imaging channel showing the spines’ morphologies, our results indicate that small round/ovoid PSD95 assemblies are located in relatively small dendritic spines (∼600-nm mean spine-head diameter), whereas the complex-shaped PSD95 scaffolds and their (partial) side views tend to occur in larger spine heads. This is in agreement with previous EM data, which have shown that small spine heads harbor small round postsynaptic densities, while large (mushroom) spines contain complex-shaped and perforated PSDs, and that their appearance depends on the angle of view (57, 58).
Our results provide one of the first nanoscale views of protein distributions in living mice, and the quality of the image data demonstrates the strength of our chosen labeling and imaging approaches. Previous in vivo superresolution studies used the most common and conventional labeling method based on the expression of FPs. Compared with our data achieved with far-red FPs, we observed a substantial contrast and resolution improvement of the superresolved SiR-Halo–labeled PSD95 structures (compare Figs. 2 and 3 to SI Appendix, Fig. S7). For the far-red FPs, photobleaching limited the imaging to single frames (SI Appendix, Fig. S7), without the ability to record -stacks or perform time-lapse analyses.
This pronounced difference can be largely attributed to the higher photostability of the organic dye compared with FPs in STED nanoscopy. SiR enabled -stack acquisition (Fig. 5) and repeated imaging at STED resolution (SI Appendix, Fig. S5). These capabilities allowed us to better ascertain the 3D character of the PSD95 assemblies, which appeared static on the few-minutes timescale. Although the SiR-Halo ligand was directly injected into the brain and was most likely also present in an unbound fraction during STED imaging, the background in the images was exceptionally low. This can be ascribed to the high degree of fluorogenicity of SiR, which shows an increase in fluorescence only upon covalent binding to the biomolecular target structure (59), in our case to the HaloTag protein. A comparison of our imaging results to the recent in vivo STED data of PSD95 labeled with EGFP (27) shows that our approach revealed the PSD95 organizations in finer detail and with improved image contrast, benefitting the visualization of the complex scaffolds. The data in ref. 27 are broadly consistent with our findings, suggesting that the specifics of our labeling approach cannot explain the diverging results from prior cell- and tissue-culture studies of PSD95. The images reported in ref. 27 exhibit ∼10–15 counts per pixel in the maxima of rendered structures (for the examples where count data are provided), whereas our SiR-based approach featured signals typically exceeding ∼50–60 counts per pixel in the PSD95-containing regions. The brightest parts of the structures exhibited signals of up to 100–120 counts per pixel. The differing imaging parameters in both studies were chosen, out of practical considerations, with multiple image acquisitions in mind for performing time-lapse or -stack recordings. In either study, the main focus was not set on obtaining merely one best image in a single-shot approach. Even so, the presented datasets let us conclude that the endogenous near-IR synthetic fluorophore labeling demonstrated herein enables superior and robust nanoscale imaging performance.
Another notable advantage of our approach is the option of attaching a synthetic fluorescent ligand of choice. This allows adapting the approach to any spectral window and affords great flexibility with regard to, for example, pulse-chase analyses. We opted for SiR as a red label, since in the far-red to near-IR spectral range autofluorescence, absorbance, and scattering are reduced in tissue, aspects that are prominent drawbacks for blue-shifted wavelengths. While former in vivo STED studies of EYFP-labeled structures reported phototoxic effects, such as swellings in dendrites (23, 24), a recent study with the far-red FP mNeptune2 (560-nm excitation and 732-nm STED wavelengths) discussed no signs of photodamage (25). We confirmed this observation. The wavelengths utilized in the present work (640-nm excitation and 775-nm STED) are shifted even further to the near-IR. Based on the evaluation of confocal images of the Lifeact-EYFP reference labeling, we observed no ligand-related toxicity or effects from the intracortical injections on the dendritic morphology, or phototoxicity or tissue damage from the repeated overview, confocal, and STED imaging. Whereas we used SiR-Halo for our experiments, the HaloTag technology would also allow the utilization of other fluorescent ligands. Ongoing developments of new dyes for the far-red to the near-IR spectral range are of special interest for deep-tissue and in vivo optical imaging and might yield ligands with even better photophysical properties, including enhanced brightness and photostability, while also being cell-permeable. A step toward the near-IR spectrum was made by the design of the membrane-permeable silicon-rhodamine derivatives 680SiR ( = 679 nm, = 697 nm; ref. 34) and SiR700 ( = 690 nm, = 715 nm; ref. 33), which were shown to work well in cultured cells with an 810-nm and 775-nm near-IR STED laser, respectively. Whereas our approach using intracortical injections of the fluorescent dye was limited by its tissue penetration, resulting in concentrated labeled spots (Fig. 1E), this could be remedied if the membrane-permeant ligand were to be blood–brain-barrier-permeable. Then, the ligand could be i.v. injected to obtain a homogenous staining of the entire brain, as recently reported for the Janelia Fluor 585-HaloTag ligand (ref. 60; STED performance not demonstrated so far). With improvements in the photostability of organic dyes, the use of higher STED intensities to increase spatial resolution will become feasible. Although our data provide a representative view of the PSD95 nanostructure in vivo, a higher resolution could further help to resolve the smallest structural units of PSD95.
Dyes with increased stability will also prove beneficial for 3D STED recordings, because considerably more image frames would need to be recorded and the cumulative exposure on the fluorophores would be higher. Finally, protein tags open up new opportunities to design experiments in different ways compared with FPs. Next to the freedom of choice for the tag ligand, the protein tag itself can be freely chosen (e.g., SNAP-, Halo-, or CLIP-tag proteins), and the tag systems can even be used for multicolor experiments implementing two protein tags simultaneously.
In conclusion, we developed a highly effective and versatile labeling and imaging approach, which enabled one of the first superresolution studies of protein distributions in the native environment of the living mouse. The flexibility and wide spectrum of available substrates make the use of self-labeling protein tags by synthetic fluorophores highly attractive for in vivo nanoscale imaging. Our work reveals unprecedented details of PSD protein PSD95 scaffolds in vivo, which were concealed in prior cell- or tissue-culture studies. The results therefore highlight the importance of in vivo superresolution microscopy, as well as the advantages of far-red and near-IR organic dyes for future nanoscopy studies in the living animal.
Materials and Methods
Details of the mouse line, plasmid and viral vector production, animal procedures, virus transduction, surgical preparation, in vivo labeling and histology, and in vivo STED image acquisition and associated analysis are described in SI Appendix, SI Materials and Methods. In brief, we performed STED nanoscopy of nanoscale distributions of the PSD protein PSD95 in fusion with the HaloTag and enzymatically self-labeled by SiR fluorophores in the visual cortex of anesthetized live mice. All imaging was carried out on a home-built upright STED microscope (SI Appendix, Fig. S1) with a 63× glycerol-immersion objective lens, scanning a pair of laser spots for 640-nm fluorescence excitation and 775-nm STED deexcitation of SiR (as a focal-plane doughnut), and 473-nm excitation for EGFP/EYFP (confocal). The far-red FPs, also explored for PSD95 labeling and STED imaging, were excited by a 594-nm laser. Image acquisition of 5 × 5 µm2 to 20 × 20 µm2 FOVs was typically performed with 20-nm pixel size and 30-µs dwell time, resulting in acquisition times of ∼2–30 s for the large STED images and ∼1.3–5.6 s for image portions containing the individual PSD95 assemblies (depending on the acquisition mode and width of the FOV). Further details on image acquisition parameters are in SI Appendix, Table S1.
All animal procedures were conducted in accordance with the Animal Welfare Law of the Federal Republic of Germany and the regulation about animals used in experiments and were approved and authorized by the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit.
Supplementary Material
Acknowledgments
We thank Tanja Gilat for technical assistance, Dr. Carola Gregor for engineering the PSD95-mNeptune2 and the EGFP plasmids, Dr. Alexey N. Butkevich for providing silicon-rhodamine as a HaloTag ligand, and the animal facility of the Max Planck Institute for Biophysical Chemistry for taking care of the mice. We thank Jaydev Jethwa for a critical reading of the manuscript and Dr. Gražvydas Lukinavičius for helpful discussions. We thank the laboratories of Prof. Karl Deisseroth (Stanford University), Prof. G. Ulrich Nienhaus (Karlsruhe Institute for Technology), Prof. Yasushi Okada (RIKEN), and Prof. Tobias Bonhoeffer (Max Planck Institute of Neurobiology) for providing plasmids as indicated and Prof. Frank Kirchhoff (University of Saarland) for providing the Thy1.2-EYFP mouse line, which served as a comparison with in vivo imaging with far-red FPs (SI Appendix, Fig. S7A). We thank Prof. Nils Brose (Max Planck Institute of Experimental Medicine) for making us aware of S.G.N.G.’s mouse lines. This work was supported by the Deutsche Forschungsgemeinschaft Cluster of Excellence 171 “Center for Nanoscale Microscopy and Molecular Physiology of the Brain” and SFB 1286 and European Research Council Advanced Grant 695568 (to S.G.N.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807104115/-/DCSupplemental.
References
- 1.Harris KM, Weinberg RJ. Ultrastructure of synapses in the mammalian brain. Cold Spring Harb Perspect Biol. 2012;4:a005587. doi: 10.1101/cshperspect.a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 3.Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Yang W, Yuste R. In vivo imaging of neural activity. Nat Methods. 2017;14:349–359. doi: 10.1038/nmeth.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 6.Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grotjohann T, et al. Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature. 2011;478:204–208. doi: 10.1038/nature10497. [DOI] [PubMed] [Google Scholar]
- 9.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 10.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahl SJ, Hell SW, Jakobs S. Fluorescence nanoscopy in cell biology. Nat Rev Mol Cell Biol. 2017;18:685–701. doi: 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- 13.Maglione M, Sigrist SJ. Seeing the forest tree by tree: Super-resolution light microscopy meets the neurosciences. Nat Neurosci. 2013;16:790–797. doi: 10.1038/nn.3403. [DOI] [PubMed] [Google Scholar]
- 14.Willig KI, Barrantes FJ. Recent applications of superresolution microscopy in neurobiology. Curr Opin Chem Biol. 2014;20:16–21. doi: 10.1016/j.cbpa.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Westphal V, et al. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 16.Nägerl UV, Willig KI, Hein B, Hell SW, Bonhoeffer T. Live-cell imaging of dendritic spines by STED microscopy. Proc Natl Acad Sci USA. 2008;105:18982–18987. doi: 10.1073/pnas.0810028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Testa I, et al. Nanoscopy of living brain slices with low light levels. Neuron. 2012;75:992–1000. doi: 10.1016/j.neuron.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 18.MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78:615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair D, et al. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J Neurosci. 2013;33:13204–13224. doi: 10.1523/JNEUROSCI.2381-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Este E, Kamin D, Göttfert F, El-Hady A, Hell SW. STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep. 2015;10:1246–1251. doi: 10.1016/j.celrep.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Kirov SA, Sorra KE, Harris KM. Slices have more synapses than perfusion-fixed hippocampus from both young and mature rats. J Neurosci. 1999;19:2876–2886. doi: 10.1523/JNEUROSCI.19-08-02876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urban NT, Willig KI, Hell SW, Nägerl UV. STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophys J. 2011;101:1277–1284. doi: 10.1016/j.bpj.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berning S, Willig KI, Steffens H, Dibaj P, Hell SW. Nanoscopy in a living mouse brain. Science. 2012;335:551. doi: 10.1126/science.1215369. [DOI] [PubMed] [Google Scholar]
- 24.Willig KI, et al. Nanoscopy of filamentous actin in cortical dendrites of a living mouse. Biophys J. 2014;106:L01–L03. doi: 10.1016/j.bpj.2013.11.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wegner W, et al. In vivo mouse and live cell STED microscopy of neuronal actin plasticity using far-red emitting fluorescent proteins. Sci Rep. 2017;7:11781. doi: 10.1038/s41598-017-11827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ter Veer MJT, Pfeiffer T, Nägerl UV. Two-photon STED microscopy for nanoscale imaging of neural morphology in vivo. In: Erfle H, editor. Super-Resolution Microscopy: Methods and Protocols. Springer; New York: 2017. pp. 45–64. [DOI] [PubMed] [Google Scholar]
- 27.Wegner W, Mott AC, Grant SGN, Steffens H, Willig KI. In vivo STED microscopy visualizes PSD95 sub-structures and morphological changes over several hours in the mouse visual cortex. Sci Rep. 2018;8:219. doi: 10.1038/s41598-017-18640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.König K. Multiphoton microscopy in life sciences. J Microsc. 2000;200:83–104. doi: 10.1046/j.1365-2818.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 29.Weissleder R. A clearer vision for in vivo imaging. Nat Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- 30.Nienhaus K, Nienhaus GU. Fluorescent proteins for live-cell imaging with super-resolution. Chem Soc Rev. 2014;43:1088–1106. doi: 10.1039/c3cs60171d. [DOI] [PubMed] [Google Scholar]
- 31.Erdmann RS, et al. Super-resolution imaging of the Golgi in live cells with a bioorthogonal ceramide probe. Angew Chem Int Ed Engl. 2014;53:10242–10246. doi: 10.1002/anie.201403349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukinavičius G, et al. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods. 2014;11:731–733. doi: 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 33.Lukinavičius G, et al. Fluorogenic probes for multicolor imaging in living cells. J Am Chem Soc. 2016;138:9365–9368. doi: 10.1021/jacs.6b04782. [DOI] [PubMed] [Google Scholar]
- 34.Butkevich AN, Lukinavičius G, D’Este E, Hell SW. Cell-permeant large stokes shift dyes for transfection-free multicolor nanoscopy. J Am Chem Soc. 2017;139:12378–12381. doi: 10.1021/jacs.7b06412. [DOI] [PubMed] [Google Scholar]
- 35.Lukinavičius G, et al. Fluorescent dyes and probes for super-resolution microscopy of microtubules and tracheoles in living cells and tissues. Chem Sci (Camb) 2018;9:3324–3334. doi: 10.1039/c7sc05334g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukinavičius G, et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat Chem. 2013;5:132–139. doi: 10.1038/nchem.1546. [DOI] [PubMed] [Google Scholar]
- 37.Bottanelli F, et al. Two-colour live-cell nanoscale imaging of intracellular targets. Nat Commun. 2016;7:10778. doi: 10.1038/ncomms10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butkevich AN, et al. Fluorescent rhodamines and fluorogenic carbopyronines for super-resolution STED microscopy in living cells. Angew Chem Int Ed Engl. 2016;55:3290–3294. doi: 10.1002/anie.201511018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butkevich AN, et al. Hydroxylated fluorescent dyes for live-cell labeling: Synthesis, spectra and super-resolution STED. Chemistry. 2017;23:12114–12119. doi: 10.1002/chem.201701216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butkevich AN, et al. Two-color 810 nm STED nanoscopy of living cells with endogenous SNAP-tagged fusion proteins. ACS Chem Biol. 2018;13:475–480. doi: 10.1021/acschembio.7b00616. [DOI] [PubMed] [Google Scholar]
- 41.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 42.Los GV, Wood K. The HaloTag: A novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 43.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SGN. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 45.Fernández E, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Migaud M, et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 47.D’Este E, et al. Subcortical cytoskeleton periodicity throughout the nervous system. Sci Rep. 2016;6:22741. doi: 10.1038/srep22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen LD, et al. Metabolic turnover of synaptic proteins: Kinetics, interdependencies and implications for synaptic maintenance. PLoS One. 2013;8:e63191. doi: 10.1371/journal.pone.0063191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hussain NK, Huganir RL. Structure and molecular organization of the postsynaptic density. In: Stuart G, Spruston N, Häusser M, editors. Dendrites. 3rd Ed Oxford Univ Press; New York: 2016. [Google Scholar]
- 50.Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: Serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9:2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gray NW, Weimer RM, Bureau I, Svoboda K. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Broadhead MJ, et al. PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci Rep. 2016;6:24626. doi: 10.1038/srep24626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukata Y, et al. Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J Cell Biol. 2013;202:145–161. doi: 10.1083/jcb.201302071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang A-H, et al. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature. 2016;536:210–214. doi: 10.1038/nature19058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dzyubenko E, Rozenberg A, Hermann DM, Faissner A. Colocalization of synapse marker proteins evaluated by STED-microscopy reveals patterns of neuronal synapse distribution in vitro. J Neurosci Methods. 2016;273:149–159. doi: 10.1016/j.jneumeth.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Hruska M, Henderson N, Le Marchand SJ, Jafri H, Dalva MB. Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat Neurosci. 2018;21:671–682. doi: 10.1038/s41593-018-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popov VI, et al. Remodelling of synaptic morphology but unchanged synaptic density during late phase long-term potentiation (LTP): A serial section electron micrograph study in the dentate gyrus in the anaesthetised rat. Neuroscience. 2004;128:251–262. doi: 10.1016/j.neuroscience.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 58.Arellano JI, Benavides-Piccione R, Defelipe J, Yuste R. Ultrastructure of dendritic spines: Correlation between synaptic and spine morphologies. Front Neurosci. 2007;1:131–143. doi: 10.3389/neuro.01.1.1.010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nadler A, Schultz C. The power of fluorogenic probes. Angew Chem Int Ed Engl. 2013;52:2408–2410. doi: 10.1002/anie.201209733. [DOI] [PubMed] [Google Scholar]
- 60.Grimm JB, et al. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods. 2017;14:987–994. doi: 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.