Significance
Introns are noncoding DNA sequences interspersed among the coding sequences of genes. Shortly after transcription, the intronic sequences are spliced out of the primary RNA transcript as lariat RNAs (circular molecules with a short tail). Most of these lariats are destroyed within minutes in the cell nucleus. We report here that many such intronic RNAs are, in fact, exported to the cytoplasm, where they remain as stable circular molecules. These cytoplasmic introns are derived from hundreds of different genes of widely different functions. We find them in cells of human, mouse, chicken, frog, and zebrafish. The widespread occurrence of so many stable lariat RNAs in the cytoplasm suggests that they play some as-yet unexpected role in cell metabolism.
Keywords: RNA, intron, cytoplasm, vertebrate
Abstract
Most intronic RNAs are degraded within seconds or minutes after their excision from newly formed transcripts. However, stable intronic sequence RNAs (sisRNAs) have been described from oocytes of the frog Xenopus, from Drosophila embryos, and from human cell lines. In Xenopus oocytes, sisRNAs are abundant in both the nucleus and cytoplasm, they occur in the form of lariats, and they are stable for days. In this study we demonstrate that cytoplasmic sisRNAs are also found in human, mouse, chicken, and zebrafish cells. They exist as circular (lariat) molecules, mostly 100–500 nucleotides in length, and are derived from many housekeeping genes. They tend to have an unusual cytosine branchpoint (with the exception of those from the frog). Stable lariats are exported from the nucleus to the cytoplasm by the NXF1/NXT1 system, demonstrating that their presence in the cytoplasm is not due to passive diffusion. Lariats in the cytoplasm are not associated with transcripts of the genes from which they are derived. The biological significance of cytoplasmic sisRNAs remains obscure.
Intronic RNAs represent a large fraction of the unstable transcriptome of higher eukaryotes. These RNAs are spliced from pre-mRNA as branched circular RNAs (lariats). Within minutes of splicing, a dedicated debranching enzyme (DBR1) recognizes the branchpoint and linearizes the lariat, promoting its rapid degradation (1). This pathway is also necessary for processing noncoding RNAs such as small nucleolar RNAs (2). An earlier study from our laboratory revealed the existence of unusually abundant intronic RNAs in the nuclei of Xenopus oocytes (3). Many of these intronic RNAs persist for as long as 2 d and were given the name stable intronic sequence RNA, or sisRNA. In a later study we found thousands of different sisRNAs in the cytoplasm of the Xenopus oocyte (4). We were certain of the cytoplasmic localization, because the gigantic nucleus can be removed manually from the oocyte before the cytoplasmic RNAs are isolated and sequenced. The cytoplasmic sisRNAs are stored as circular molecules (lariats without tails).
Until now, however, it has remained unknown whether cytoplasmic circular sisRNAs are unique to frog oocytes or are more widely distributed. A major technical problem in searching for cytoplasmic sisRNAs is the difficulty of obtaining cytoplasmic fractions completely free of nuclear contamination. For this reason, we began our search for cytoplasmic sisRNAs in other species by examining mammalian red blood cells (RBCs). Mammalian RBCs lose their nuclei during their maturation from precursor erythroblasts and so provide a convenient source of pure cytoplasm. We found numerous circular intronic sequences in RBCs, thereby demonstrating the existence of sisRNA in mammalian somatic cells. We also found circular sisRNAs in cultured cells and in tissues from the mouse, human, chicken, and zebrafish. We discuss functions that cytoplasmic circular sisRNAs may perform independently of their host genes.
Results
Circular SisRNAs in RBCs.
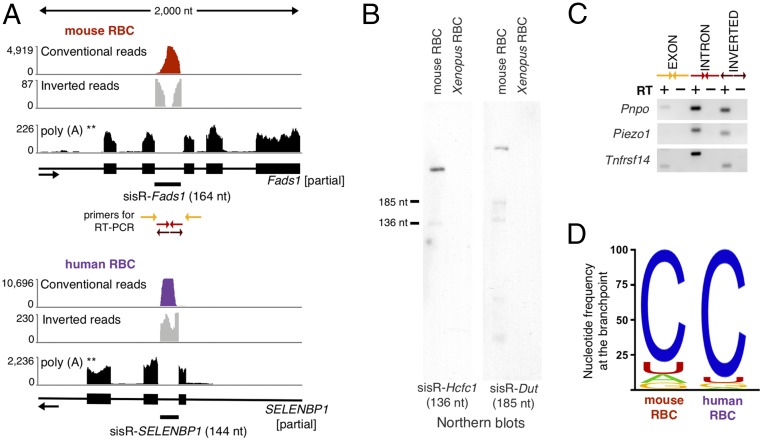
Mammalian RBCs extrude their nuclei before entering the circulation and hence are a source of pure cytoplasm. Although extensive RNA degradation occurs during RBC maturation (5), some RNAs persist (6), including circular RNA or circRNA (7, 8). Intronic RNA has not yet been analyzed in these cells. We purified RBCs from mouse blood by fractionation on a Percoll gradient (9) and subjected the small amount of recovered RNA to high-throughput sequencing. The 77 million single-end 100-bp reads that we obtained were analyzed using a method developed by Taggart et al. (10). Of particular interest, this method allows one to recover inverted reads that would otherwise be thrown out by standard pipelines. We searched for and found 524 mouse introns that were mapped by conventional reads and also by inverted reads that contained sequences from both ends of the intron (Fig. 1A and Dataset S1). Because we were looking for circular sisRNA, we disregarded any intron with conventional reads but no inverted reads. We confirmed the circularity of the intronic RNA by RT-PCR and Northern blotting of selected RNAs (Fig. 1 B and C). Two classes of circular sisRNA were detected. The most abundant circular sisRNAs presumably arise from failure of the excised intronic RNA to be linearized by the debranching enzyme DBR1. An unusual feature of these stable lariats was the presence of a cytosine at the branchpoint rather than the more common adenine (Fig. 1D). Approximately 80% of the stable lariats contained cytosine at the branchpoint compared with the 10% expected rate (10). On the other hand, a fraction of the circular sisRNAs did not resemble conventional spliced lariats. Specifically, the junctions of these circles occurred at the 3′ splice site of the intron, at the G or A nucleotide, and not at the branchpoint (SI Appendix, Fig. S1 A and B). These circular intronic RNAs have been previously observed in human cell culture. Presumably, they result from a postdebranching circularization event (10). In RBCs, the circularized full-length intronic RNAs were less abundant than stable lariats (SI Appendix, Fig. S1C). Finally, we observed that for the vast majority of genes encoding a sisRNA, most reads were derived from introns, with very few exonic reads. This result suggests that linear mRNA molecules are lost at some point during maturation of mammalian RBCs, whereas circular RNAs are retained.
Fig. 1.
Typical sisRNAs found in mouse and human RBCs. (A) The two Upper tracks show conventional reads (red and purple) and inverted reads (gray) of total RNA from mature RBCs. The Lower tracks correspond to public RNA sequencing data of polyadenylated transcripts from RBC precursor cells (erythroblasts) (11). (B) Northern blots against two mouse stable lariats. In each case, the major band ran slower than expected for a linear molecule, consistent with the circular nature of the intronic RNA. (C) RT-PCR using conventional exonic (yellow) and intronic primers (red). Mouse RBC intronic RNAs can be amplified with the conventional (red) as well as inverted (outward facing) primers (brown), consistent with the RNAs being circular. (D) Nucleotide frequency at the branchpoint for all 374 mouse and 99 human stable lariats.
Both stable lariats and circularized full-length intronic RNAs occur in human RBCs as well. As with mouse blood, we separated RBCs from white cells on a Percoll gradient. High-throughput analysis revealed 143 circular intronic RNAs similar to the two types of mouse sisRNAs (Fig. 1 and Dataset S1). Among the 137 human sisRNA host genes, 27 are common with mouse, and seven “orthologous” introns encode a sisRNA in both species.
Because there were very few exonic reads in the RNA from purified RBCs, one possibility was that the sisRNAs did not originate from the RBCs themselves. To test this unlikely scenario, we analyzed publicly available data from mouse and human erythroblasts, the nucleated cells in the bone marrow that give rise to RBCs (11). For each sisRNA detected in mouse and human RBCs, we found that the cognate mRNA was expressed in erythroblasts (Fig. 1A). Hence, during RBC maturation, it is likely that mRNAs are degraded, whereas lariat introns from the same transcripts persist in the cytoplasm.
SisRNAs Are Present in Cultured Cells (Mouse 3T3 Cells).
The demonstration of stable circular introns in the cytoplasm was made possible by two unique circumstances. First, it is possible to remove the nucleus manually from frog oocytes (12), and second, mammalian RBCs do not contain nuclei. In both cases, therefore, cytoplasm can be analyzed biochemically in the absence of contaminating nuclei. To demonstrate introns in the cytoplasm of other cell types is much more challenging, because there is no general way to prepare completely pure cytoplasmic fractions. Instead, we chose to study stable introns in cultured cells, on the assumption that they might be a reasonable proxy for cytoplasmic introns. We emphasize, however, that the designation sisRNA refers to the stability of the sequences, not their location.
We began our search for sisRNAs in other cell types by examining total RNA from mouse 3T3 cells. We treated the cells with α-amanitin to inhibit transcription and allow degradation of transient intronic RNA. Untreated cells served as controls. Total extracted RNA was depleted of rRNA and treated with RNase R or water before high-throughput sequencing. RNase R degrades linear RNA molecules but does not attack circular ones. This procedure enriches for stable circular RNA molecules but does not distinguish nuclear from cytoplasmic RNA. We will show later by in situ hybridization that selected sisRNAs are, in fact, in the cytoplasm.
In actively transcribing cells (no α-amanitin), we detected ∼6,000 RNase R-resistant intronic RNAs between 65 and ∼2,000 nt in length. These molecules were derived from about one third of the introns of ∼2,000 genes, with a bias for introns from 0.1–1.5 kb. The bias is presumably due to the method of RNA purification and library preparation. Specifically, longer intronic RNAs are more likely to be randomly nicked during RNA purification (and therefore degraded by RNase R before library preparation), whereas shorter RNAs are less likely to be represented in the library. Most of these intronic reads probably represent unstable lariats bound for degradation in the nucleus.
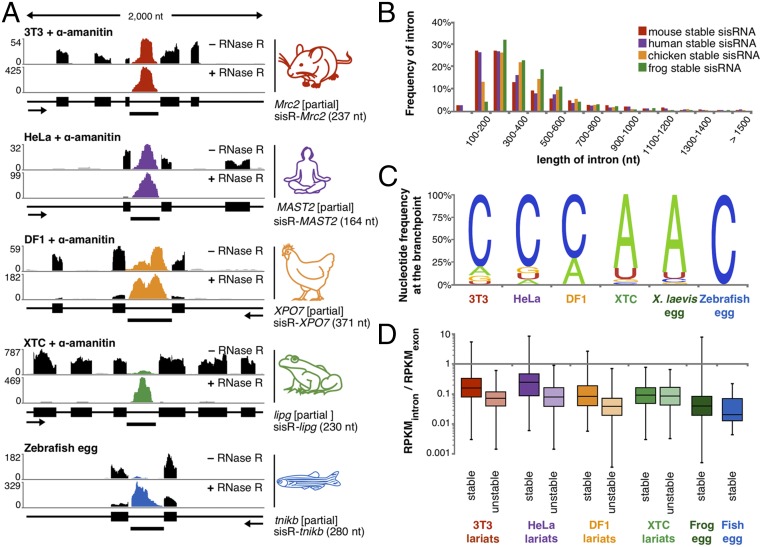
After inhibition of transcription with α-amanitin, 427 RNase R-resistant intronic RNAs were detected, evidence that mouse 3T3 cells produce stable intronic RNAs (Dataset S1). These sisRNAs were derived from 370 genes, mostly from one (∼85%) or two introns per gene. The nature of 210 sisRNAs was further investigated by analyzing the corresponding inverted reads. Among these, 10% represent circularized full-length intronic RNAs, three that were common to RBCs. The most abundant and diverse circular intronic RNAs were in the form of a lariat, including 59 that were common to RBCs (Fig. 2A). They showed a strong bias for short introns and cytosine at the branchpoint (Fig. 2 B and C). DBR1 has reduced activity toward C branchpoints (13), suggesting that a C branchpoint may be important for circular sisRNA formation.
Fig. 2.
Identification of lariat RNAs in five vertebrate species. (A) A typical prominent sisRNA detected in cultured cells of mouse (red), human (purple), chicken (orange), and frog (green) and zebrafish eggs (blue). RNase R was used to eliminate mRNA transcripts (black). Transcript annotation and orientation are shown below the coverage for each gene. (B) Percent of stable circular RNAs as a function of intron length (50-nt bins). Mouse (red), human (purple), chicken (orange), frog (green), and zebrafish (blue) sisRNAs derive from short introns. (C) Nucleotide frequency at the branchpoint of stable lariats. (D) Abundance of “stable” and “unstable” circular intronic RNA relative to their cognate mRNA in untreated cells.
To further investigate the significance of the branchpoint, we compared two classes of lariats detected in untreated cells: stable lariats (those resistant to α-amanitin) and unstable lariats (those sensitive to α-amanitin) (SI Appendix, Fig. S3A). In untreated cells, unstable lariats are less abundant relative to their cognate mRNA than are stable lariats (Fig. 2D). Also, we find that these two groups have distinct branchpoint signatures. Most stable lariats have a C branchpoint at a frequency similar to the C branchpoint detected in α-amanitin–treated cells, as expected. However, most unstable lariats have an A branchpoint (SI Appendix, Fig. S3B). Furthermore, we found that for equally abundant lariats, a C branchpoint predicts stability in the 3T3 cell line. On the other hand, the distance between the branchpoint and the 3′ splice site is comparable between the two groups (SI Appendix, Fig. S3C). Similarly, whereas stable lariats have a somewhat higher GC content, the average predicted minimum free energy is indistinguishable between the two groups (SI Appendix, Fig. S3 D and E).
In summary, we found that 3T3 cells produce stable circular intronic RNAs. These sisRNAs are characterized by (i) their short length, (ii) their C branchpoint, and (iii) their molecular ratio compared with their cognate mRNA.
SisRNAs in Different Vertebrate Species.
To determine whether sisRNAs are found in other vertebrate species, we extracted RNA from control and α-amanitin–treated HeLa cells, DF1 cells (a chicken cell line), and XTC cells (a Xenopus laevis cell line). For all samples, RNase R was used to enrich for circular RNA. We detected sisRNAs in all three species (Fig. 2A).
In HeLa cells, 318 sisRNAs were derived from 270 genes; 46 of these genes also hosted an sisRNA in mouse 3T3 cells (Dataset S1), and 14 sisRNA-encoding introns were orthologous. Similarities between human and mouse sisRNAs included the length of the sisRNAs (Fig. 2B) and the molecular ratio between the sisRNA and its cognate mRNA (Fig. 2D). Among these, most were stabilized as lariats with a bias for a C branchpoint (Fig. 2C), and a fifth (14) were common to human RBCs. In a previous study, Zhang et al. (15) analyzed 20 abundant lariats and determined that the branchpoint in untreated HeLa cells could be either an A or a C nucleotide with about equal frequency. We confirmed 16 out of the 20 and mapped the branchpoint of 13. Among these 13 lariats, 8 lariats persisted after transcription inhibition, all of which had a C branchpoint. Conversely, we found an A branchpoint in all five lariats that were detected in untreated cells only (SI Appendix, Fig. S2). These data suggest a common pathway for stability of human and mouse lariats.
Similar observations were made on chicken DF1 cells. We detected 277 circular sisRNAs derived from short introns of 214 genes (Dataset S1). Twenty-one of these sisRNAs had corresponding inverted reads, and most were stabilized as lariats with a cytosine branchpoint (71%), unlike unstable DF1 lariats with predominantly A branchpoints (65%). Among the chicken sisRNA host genes, 10 were the same as in HeLa cells and 15 were the same as in mouse 3T3 cells (including two orthologous introns). Overall six genes were host to at least one sisRNA in all three species. The six common genes have very different functions, including transcription regulation, motors, ribosome biogenesis, fatty acid synthesis, and protein localization.
In X. laevis XTC cells, we found a somewhat different pattern—about 4,000 circular sisRNAs derived from 2,000 genes (Dataset S1). In the most extreme case, one gene hosted 17 sisRNAs. We are sure of intronic RNA stability, because many unstable transcripts are no longer detectable after transcription is inhibited for 12 h (SI Appendix, Fig. S4). A total of 570 stable intronic RNAs had inverted reads, 98% of which were lariats. These lariats were barely detectable in RNase R minus samples, and most molecules had an A branchpoint and an AU-rich body, unlike those in mouse, human, and chicken. The sisRNAs from XTC cells resembled those from frog oocytes: thousands of circular sisRNAs, among which nearly 90% of mapped branchpoints were adenine (Fig. 2 C and D). Among the 4,000 circular sisRNAs, 20% were found in the cytoplasm of the X. laevis oocyte.
To study stable lariat RNAs from a fish, we extracted RNA from freshly expressed eggs of the zebrafish (Danio rerio). These eggs contain a large pool of stable RNAs used during early embryonic development. We treated egg RNA with RNase R to enrich for circular molecules but found only 13 examples among more than 10,000 spliced transcripts. Fig. 2A shows reads for the tnikb gene and the stable lariat derived from its intron. This sisRNA, which has a C branchpoint and an AU-rich body, is the most abundant zebrafish sisRNA in our sample. Unfortunately, we could not map the junction of other sisRNAs because of their low abundance. An important limitation of our zebrafish data is the dependence on genome annotation for sisRNA discovery. We anticipate that more sisRNAs will be detected when a more complete genome assembly becomes available.
In summary, we found hundreds of stable lariat intronic RNAs in six vertebrate species—two mammals, two amphibians, one bird, and one fish. These stable lariats derive from short introns and are characterized by a C branchpoint (except in Xenopus). They come from coding and noncoding genes related to nearly all known cellular functions.
In Situ Hybridization Detects Intronic RNAs in the Cytoplasm.
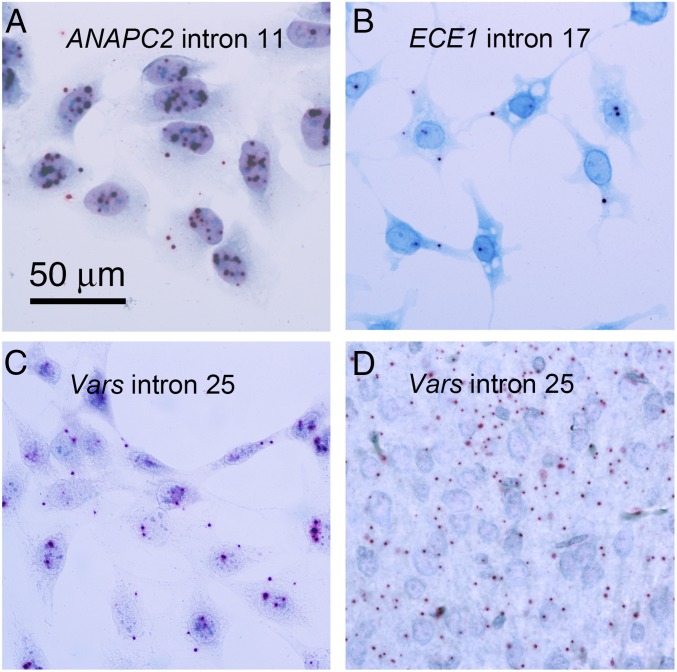
Definitive evidence that lariats are stored in the cytoplasm of vertebrate cells (other than RBCs) comes from single molecule in situ hybridization. We chose probes for three highly abundant sisRNAs, one each from human, mouse, and chicken (SI Appendix, Fig. S5). The human and mouse sisRNAs were known to be abundant in RBCs as well as in cultured HeLa and 3T3 cells. The chicken probe was selected as the most abundant sisRNA in cultured DF1 cells. Because all of these introns are less than 250 nucleotides in length (SI Appendix, Fig. S5), we used probes consisting of three to four short oligonucleotides, a multistep signal amplification series, and detection by Fast Red (BaseScope probes from Advanced Cell Diagnostics). Positive (exonic PpiB gene) and negative (bacterial dapB gene) controls were included. In each case, we detected weak but clear positive signals in the cytoplasm corresponding to the intronic probe (Fig. 3). Because the tissues had not been treated with α-amanitin to inhibit ongoing transcription, we also saw positive signals in the nucleus that probably correspond to nascent transcripts at the sites of transcription and to unstable spliced introns.
Fig. 3.
Single-molecule in situ hybridization of introns. (A) Human ANAPC2 intron 11 in cultured human cells (HeLa). (B) Chicken ECE1 intron 17 in cultured chicken cells (DF1). (C) Mouse Vars intron 25 in cultured mouse cells (3T3). (D) Mouse Vars intron 25 in section of mouse hind brain. Tissues were fixed in 4% paraformaldehyde and hybridized according to the protocol supplied by the probe manufacturer (Advanced Cell Diagnostics).
Although our molecular analysis of lariat RNAs began with examination of RBCs, we did not carry out confirming in situ hybridization experiments on RBCs. In a recently published study on 7SL RNA in amphibian and mammalian blood cells (16), we encountered unexpected and still unexplained resistance of these cells to standard in situ hybridization protocols. The simplest explanation is that conventional in situ hybridization probes do not penetrate the intact RBC membrane. But until these technical issues are overcome, we are unable to provide confirming in situ hybridization data for lariat RNAs in RBCs.
SisRNAs in Mouse Liver and Brain.
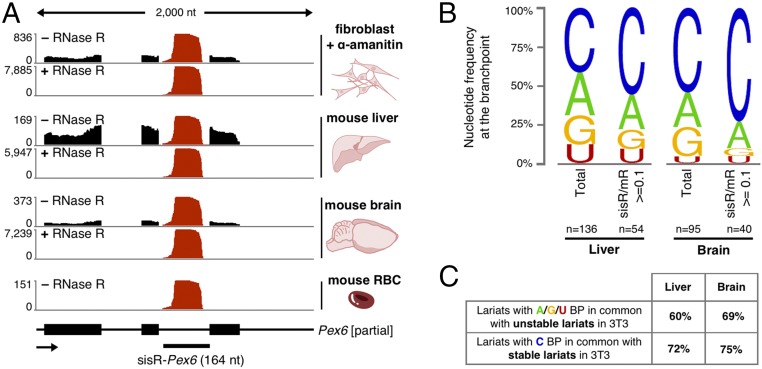
It was convenient to begin our study of sisRNAs with cell lines, which provide pure populations of cells in which transcription is easily inhibited. To examine whether stable lariats also occur in somatic tissues, we collected total RNA from mouse liver and brain. As before, we used RNase R to enrich for circular RNAs before sequencing the samples. We found ∼3,500 different circular intronic RNAs in the liver and ∼1,000 in the brain. Fig. 4A shows the most abundant circular RNA in these tissues, derived from an intron of Pex6.
Fig. 4.
SisRNAs in various mouse tissues. (A) A very abundant sisRNA, sisR-Pex6 (red), is detected in fibroblasts, liver, brain, and RBCs. (B) Branchpoint nucleotide frequency of all lariats detected in liver and brain. “n” represents the number of mapped branchpoints. (C, Upper) Lariats with A, G, and U branchpoints in common with lariats detected in control 3T3 cells and (Lower) lariats with C branchpoint in common with lariats detected in α-amanitin–treated 3T3 cells.
Unlike RNA from α-amanitin–treated cultured cells, which is enriched for metabolically stable molecules, total RNA derived from whole tissues is a mixture of short lived and more stable molecules. We analyzed intronic RNAs derived from short introns (<1,500 nt) and determined the branchpoint usage. In both the liver and brain, roughly half of the mapped branchpoints were cytosine, suggesting the existence of stable lariats in these tissues (Fig. 4B). Next, we examined intronic RNAs that were expressed at 10% or more of their mRNA level (as seen for most sisRNAs detected in cell culture). We found 946 and 242 such intronic RNAs in the liver and brain, respectively. These pools were enriched for C-branched lariats, consistent with their being stable lariats (Fig. 4B). Taken together, the data suggest that we detect both stable and unstable lariats, but importantly some of the lariats have the characteristics of those detected in α-amanitin–treated cells: They are short, they have a C branchpoint, and they are expressed at 10% or more of their cognate mRNA level. These data provide a suggestion that sisRNAs exist in liver and brain.
To explore these presumed sisRNAs further, we divided the lariats from liver and brain into two categories: those with a C branchpoint and those with A, G, or T. Most of the C-branched lariats (72% from the liver and 75% from the brain) were also detected in α-amanitin–treated 3T3 cells, strongly suggesting the presence of sisRNA in liver and brain. Conversely, many of the presumably unstable lariats (those with an A, G, or T branchpoint) were detected in untreated cells only (Fig. 4C). To confirm the presence of sisRNA in the brain, we carried out in situ hybridization on conventional formaldehyde-fixed tissue sections using a probe against intron 25 of the mouse Vars gene. Positive signals were evident in both the cerebrum and cerebellum (Fig. 3D).
Cytoplasmic SisRNAs Are Not Associated with Their Cognate mRNAs.
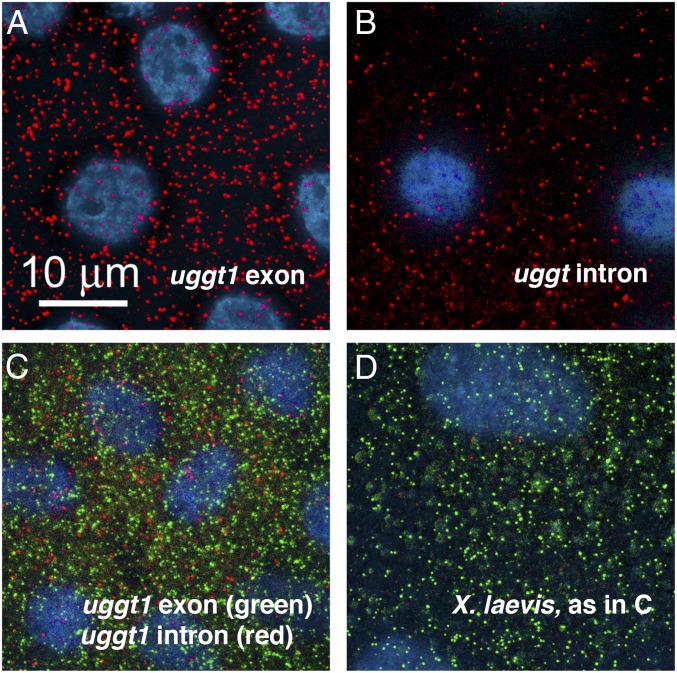
A possible function for cytoplasmic sisRNAs would be to regulate translation or localization of their cognate mRNAs. To explore this possibility, we tested whether sisRNAs might be physically associated with their cognate mRNAs in the cytoplasm. We carried out in situ hybridization for the products of two genes in the cytoplasm of Xenopus oocytes: uggt1 (UDP-glucose glycoprotein glucosyltransferase 1) and pphln1 (periphilin 1). These two genes each produce a very long sisRNA, making it possible to design probes consisting of multiple fluorescent oligonucleotides for maximal signal intensity. Probes were labeled with Quasar 570 (mRNA) or Quasar 670 (intron) and hybridized simultaneously. Very small transparent oocytes were chosen so that the probes could be visualized in whole mounts. In situ hybridization of probes against uggt1 mRNA and its longest intron (Fig. 5) show essentially no colocalization. Similar results were obtained for pphln1 mRNA and its longest intron. Although the mRNA and cognate sisRNA from each of these two genes might colocalize transiently in the cytoplasm, they do not form a permanent association detectable by in situ hybridization.
Fig. 5.
Single-molecule in situ hybridization of exon and intron probes from the X. tropicalis uggt-1 gene. Oocytes (200–400 µm diameter) were hybridized and examined as whole mounts. Images are deconvolved stacks extending from the follicle cells (blue nuclei, DAPI stain) surrounding the oocyte into the peripheral cytoplasm of the oocyte. All of the signal is in the oocyte itself. (A) X. tropicalis oocyte hybridized against the uggt-1 exon probe (red). (B) X. tropicalis oocyte hybridized against the uggt-1 intron probe (red). (C) X. tropicalis oocyte hybridized against both the exon probe (green) and the intron probe (red). The two probes give nonoverlapping signals. (D) X. laevis oocyte hybridized with both probes, as in C. Only the green exonic probe gives a positive signal in this species. Oocytes were fixed in 4% paraformaldehyde and hybridized according to the protocol supplied by the probe manufacturer (Stellaris probes from LGC Biosearch Technologies).
SisRNAs Are Exported from the Nucleus by NXF1/NXT1.
Because the oocyte nuclear envelope remains intact until the very end of oogenesis, it seemed probable that sisRNAs reach the cytoplasm by an active transport mechanism. To test this possibility, we injected purified sisRNA or pre-mRNA into the oocyte nucleus. However, these RNAs were rapidly degraded, suggesting that sisRNAs are normally stabilized at or near the time of their transcription. Hence, to study the export mechanism, we expressed sisRNAs from DNA constructs that we injected into the nucleus. After 48 h, we measured the amount of expressed sisRNAs that remained in the nucleus and the amount exported to the cytoplasm. Such experiments are possible because the oocyte stores large amounts of the proteins required for RNA export (14). Also, because export is a saturable process, one can perform export competition assays by injecting an RNA of interest along with large amounts of competitor RNA. After a few minutes, the oocytes can be dissected and cytoplasmic accumulation can be determined (17, 18).
In our system, we expressed ectopic sisRNAs independently of their endogenous cognate mRNA. X. tropicalis introns were inserted into a partial X. laevis ncl gene between exons 14 and 15, a combination that shows high splicing efficiency. The partial ncl gene was placed downstream of an mCherry gene (SI Appendix, Fig. S6A). Once injected into the nucleus, the DNA construct was transcribed, giving rise to both X. tropicalis sisRNA and mCherry mRNA (SI Appendix, Fig. S6C). By selecting for red oocytes, we could monitor the efficiency of injection, transcription, and translation (SI Appendix, Fig. S6B). Because intronic sequences are highly divergent between X. tropicalis and X. laevis, we could easily distinguish ectopic from endogenous sisRNAs.
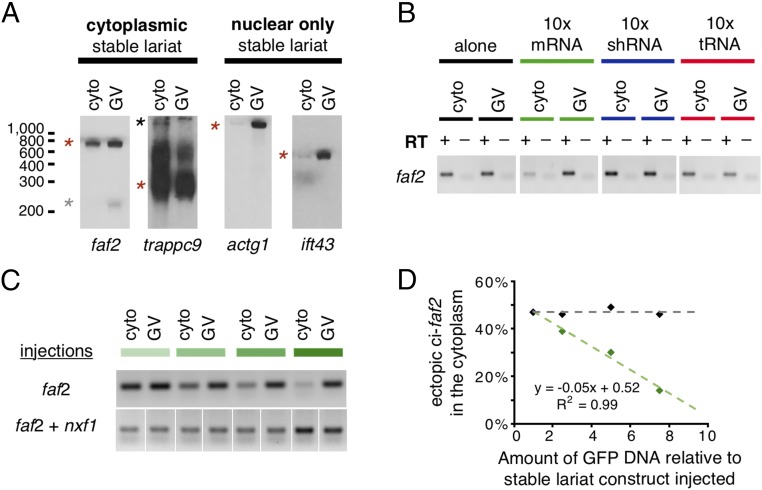
Two days after injection, we examined the amount of sisRNA that had accumulated in the cytoplasm (Fig. 6A). For two abundant X. tropicalis cytoplasmic sisRNAs, sisR-faf2 and sisR-trappc9, 50% of the newly transcribed lariats were detected in the cytoplasm. On the other hand, for sisR-actg1 and sisR-ift43, sisRNAs found primarily in the nucleus, only 5% of the ectopic transcripts accumulated in the cytoplasm. To determine which export system is used by sisRNA, we coinjected three constructs whose transcripts should compete for different export pathways: an mRNA (GFP), a Xenopus tRNA, and a short hairpin RNA (against a mouse transcript not conserved in Xenopus) (Fig. 6B). The export of sisRNA was inhibited only by overexpression of GFP mRNA, suggesting that mRNA and sisRNA share a common export pathway. Importantly, the fact that sisRNA export could be inhibited in this way strongly implies that sisRNA export is not due to simple leakage from the nucleus.
Fig. 6.
(A) Northern blots against two cytoplasmic and two nuclear X. tropicalis sisRNAs expressed ectopically in X. laevis. Red stars mark circular transcripts. The gray star marks the linearized sisR-faf2, and the black star marks the unprocessed intronic RNA trappc9. (B) RT-PCR against sisR-faf2 on cytoplasmic and nuclear fractions. Oocytes were injected with a sisR-faf2 intron construct alone (black) or with GFP DNA (mRNA, green), an shRNA construct (blue), and a tRNA construct (red). All 16 lanes were run on the same gel. After imaging, the lanes were cut (digitally) and rearranged to give the final panel. (C) Upper lanes: increasing amounts of GFP DNA were injected into the nucleus along with the sisR-faf2 intron construct. The amount of cytoplasmic sisR-faf2 decreases as more GFP DNA is injected. Lower lanes: coinjection of nxf1 DNA along with the sisR-faf2 intron construct rescues the cytoplasmic accumulation of sisR-faf2. The samples were run on nonadjacent lanes on one gel. (D) Semiquantitative analysis of the RT-PCR shown in C.
Most mRNAs are exported via NXF1/NXT1 (19). To test whether NXF1 is sufficient for sisRNA export, we coinjected a sisR-faf2 DNA construct along with increasing concentrations of GFP DNA constructs with or without nxf1 DNA. As expected, increasing amounts of GFP mRNA led to a decrease in sisRNA export (Fig. 6 C and D, green data points). On the other hand, when NXF1 was overexpressed, the export of sisRNA was restored, presumably because there was now an excess of the correct export system (Fig. 6 C and D, black data points). From these experiments, we conclude that sisRNAs are selectively exported to the cytoplasm using the NXF1/NXT1 machinery.
Discussion
During or shortly after transcription, introns are generally spliced out of new transcripts in the form of lariats (circles with a tail). They are then attacked by the nuclear enzyme DBR1 (1), which hydrolyzes the 2′–5′ phosphodiester bond (branchpoint) to give rise to a linear molecule that is degraded within minutes in the nucleus. A variety of exceptions to this canonical pathway have been described. If the excised intron is not debranched, it can remain as a circular molecule in the nucleus. Zhang et al. (15) described numerous circular intronic long noncoding RNAs (ciRNAs) in cultured human cells and suggested that these RNAs have a general role as regulators of their parent coding genes.
Previously, we described stable circular introns from oocytes and early cleavage stages of the frog Xenopus. Stable introns from thousands of genes were found in the cytoplasm of both immature and mature oocytes. They persist during the maturation stages of the oocyte and are transmitted to the early embryo. Hence, they were given the name sisRNA. Molecular analysis showed that these sisRNAs in the cytoplasm are circular in form.
The studies reported here were undertaken to determine whether cytoplasmic sisRNAs are limited to oocytes and early embryos or exist in other organism and other cell types. We began by examining stable transcripts in mouse and human RBCs. Mature RBCs lack nuclei and hence provide an ideal source of pure cytoplasm. Circular sisRNAs were readily detected, ∼500 in the mouse and 150 in human RBCs. To extend our search, we examined cultured cells of four species—human, mouse, chicken, and frog. The cells were treated for 12 h with α-amanitin, an inhibitor of transcription, to allow time for unstable introns to be degraded. In each case, we found numerous transcripts that were resistant to the inhibitor. The cytoplasmic localization of a few transcripts was demonstrated by single-molecule in situ hybridization. From these studies, we feel confident that cytoplasmic sisRNAs are widespread in normal cells of a variety of vertebrate species.
Cytoplasmic sisRNA from human, mouse, and chicken cells share several general characteristics. They are usually derived from a single short intron per gene with a strong bias for a C branchpoint instead of the canonical A. It is known that the debranching enzyme DBR1 is relatively inefficient in linearizing C-branched lariats. We speculate that a delay in linearization of short, C-branched introns may contribute to the formation of a stable complex that is more resistant to debranching. However, about 10% of circular sisRNAs in mammals are A-branched, and not all C-branched lariats are stabilized. Therefore, other features, such as folding of the lariat, must also be involved in the failure to linearize.
The final step in stabilizing the sisRNA is its export to the cytoplasm, where there is no DBR1 enzyme. Export does not occur by simple diffusion; instead, sisRNAs are actively transported to the cytoplasm by the NXF1/NXT1 export pathway. In summary, C-branched sisRNAs may avoid debranching by a combination of resistance to the DBR1 enzyme and dedicated export to the cytoplasm, where there is no enzyme.
Xenopus sisRNAs, which have a preponderance of A branchpoints in both cultured cells and oocytes, are 10 times less abundant on average than mammalian sisRNAs, perhaps reflecting their sensitivity to DBR1. Nevertheless, there are 4,000 different sisRNAs in Xenopus XTC cells, compared with only 500 in 3T3 cells and 400 in HeLa cells. Xenopus sisRNAs, like those of other species, escape debranching in part by export to the cytoplasm by way of the NXF1/NXT1 pathway.
Two other chemically distinct sisRNA species have been observed in the cytoplasm. The first occurs in Drosophila, where Pek et al. (20) detected a linear sisRNA, sisR-1, in the cytoplasm of the early embryo. SisR-1 is processed from the fourth intron of the rga gene, presumably after splicing and linearization of the full-length intronic RNA. The sisR-1 export pathway remains unknown. The second example involves an unexpected source of stable introns derived from tRNA transcripts. Intronic RNAs are spliced out of the pretransfer RNA by an endonuclease and released as linear molecules. Instead of being targeted for degradation by Xrn1, some molecules are circularized and stabilized in the cytoplasm (21).
Postsplicing circularization has also been observed for pol II intronic transcripts, including the stable ones reported here. Intronic RNAs are presumably linearized and can then be either recircularized without further trimming (5′ss to 3′ss circle) (10) or trimmed before recircularization (22). The stability or localization of these circular molecules has not yet been examined.
We considered the hypothesis that sisRNAs are derived from and somehow regulate a specific class of mRNAs in the cytoplasm. Using the admittedly imprecise gene ontology categories, we did not find that sisRNAs are derived from any specific class of genes. It seems unlikely, therefore, that sisRNAs regulate mRNAs in a specific metabolic pathway or pathways.
One simple hypothesis is that each cytoplasmic sisRNA interacts directly with and regulates the mRNA derived from the same gene. To look for such physical association between a sisRNA and its cognate mRNA, we carried out single-molecule FISH on two mRNAs and the sisRNAs from the same genes (uggt1 and pphln1). We found no tendency for the sisRNA to be associated physically with its cognate mRNA. These observations rule out a permanent association between the mRNA and its cognate sisRNA but leave open the possibility of more transient interactions.
Clues about the function of cytoplasmic sisRNA may be extrapolated from studies of intronic lariat RNAs that accumulate in cells with a minimal level of the DBR1 enzyme. These induced or artificial sisRNAs are associated with RNA-binding proteins (RBPs)—dicer, TDP-43, and small nuclear ribonucleoproteins (snRNPs)—and they contribute to an abnormal depletion of RBPs (23–25). Therefore, it is possible that cytoplasmic sisRNAs play a role in the regulation of RBPs.
Whatever may be the function(s) of cytoplasmic sisRNA in normal somatic tissues, they cannot be involved in transcription or translation in RBCs, which lack these functions entirely. Cytoplasmic sisRNAs in RBCs must have some as-yet undetermined function, or they may simply represent stable molecules that will be discarded when the RBCs themselves are later degraded.
In summary, our studies document the existence of stable intronic RNAs in the cytoplasm of somatic tissues from five vertebrate species (human, mouse, chicken, fish, and frog). These intronic RNAs are circular (lariat) in form and are characterized by a predominance of an unusual C branchpoint (except in the frog). A specific function for these lariat intronic RNAs remains to be determined.
Materials and Methods
Cell Culture.
Human HeLa and mouse 3T3 cells were cultured at 37 °C in DMEM (Dulbecco’s modified Eagle’s medium; Gibco) with 10% FBS, l-alanyl-l-glutamine (Glutamax; Gibco), and antibiotics. Chicken DF1 cells were cultured at 39 °C in the same DMEM. X. laevis XTC cells were cultured in 66% Leibovitz’s l-15 medium (Gibco) with 10% FBS, Glutamax, and antibiotics at room temperature.
Tissue Collection.
Mouse tissues were provided by Safia Malki from the Bortvin laboratory, Department of Embryology, Carnegie Institution for Science, Baltimore, MD. Blood was collected from recently euthanized CD-1 or C57BL/6 animals by cardiac puncture with an EDTA-coated 1 mL syringe and a 22-gauge needle. Brain and liver were dissected from the same animals and immediately processed for RNA extraction. Human RBCs were purchased from the Interstate Blood Bank (through Zen-Bio, Inc.). Before shipment, the sample tested negative for hepatitis B, hepatitis C, HIV, and syphilis. The samples were stored between 2 °C and 8 °C in anticoagulant citrate dextrose solution. Eggs from wild-type zebrafish were provided by the Halpern laboratory, Department of Embryology, Carnegie Institution for Science, Baltimore, MD.
RBC Purification.
Blood samples were centrifuged at 300 g for 10 min at room temperature. The supernatant was removed, and the pelleted cells were resuspended in 1× PBS. The suspension was layered on a 50–80% Percoll gradient and centrifuged at 1,000 g for 30 min at room temperature. RBCs settled near the bottom of the tube and were collected free of white blood cells. Before usage, RBCs were washed three times in 1× PBS.
RNA Purification.
RNA was extracted with TRIzol reagent (Ambion) and purified with the Direct-zol kit (Zymo Research). DNase treatment was performed on the Direct-zol column. RNA was quantitated with a Nanodrop instrument (Thermo Fisher Scientific) and its quality assessed on a Bioanalyzer 2100 electrophoresis instrument (Agilent).
RNase R.
RNA was denatured 5 min at 72 °C in 8 µL of RNase R 1× buffer. Then, 1 µL of RNase R (Epicentre) was added, and the reaction was incubated overnight at 37 °C. RNA degradation was prevented with RNasin (Promega).
Library Preparation, Sequencing, and Sequence Analysis.
Libraries were prepared using TrueSeq stranded total RNA sample preparation (Illumina). Sequencing was performed on an Illumina HiSEq. 2000 sequencer with either 100 bp or 150 bp single-end reads. Reads were aligned with TopHat (v2.0.7) to the mouse genome (v10), the human genome (v19), the chicken genome (v5), or the X. laevis genome (v9.0). Intronic reads were quantified using Bedtools (v2.15.0), and lariat reads were obtained using lariat_find.pl (10).
RT-PCR Analysis.
To confirm the bioinformatic prediction, we performed RT-PCR analysis according to a previously published protocol (4). In short, cDNA was made using Episcript (Epicenter) according to the manufacturer’s protocol and amplified with Taq polymerase (Qiagen) and various primers. In vitro transcribed RNA was used as a control to demonstrate the efficiency of the primers used.
Northern Blots.
Up to 5 μg of RNA was separated on an 8% polyacrylamide–8 M urea gel and transferred onto a nylon membrane (Zeta Probe GT; Bio-Rad). RNA was probed with dsDNA labeled with digoxigenin (DIG)-dUTP in hybridization buffer and detected using an anti-DIG antibody conjugated with alkaline phosphatase and CDP-Star chemiluminescent substrate (Roche).
Single-Molecule in Situ Hybridization.
The probes consisted of “ZZ” oligonucleotide pairs from Advanced Cell Diagnostics, which are complementary to the following gene regions:
Mus musculus Vars intron 25, bases TGAGGATCTTGTCGGTGGCCATCGGTGACAGTGCCT GTGCCCACACAGTCTGTGGCTCCCTCCACTCAGTGTCAAGACTAAAGCTGCTGTCCCCTGAGATCCTCCTTTTAGTCAGGGACAGACAGGTGGAAGTTGATGATTGATGGGGAAGTCTCTGCAGCTTGGGGCC
Gallus gallus ECE1-intron17, bases GCCCATCCATGCCTTTCCTTCCATCCACGATGGTTTTGGGAGAGCATGACAGCAAAGCCGAGGTGTCCAGAGATGCCCCAGAGAATGAGATCAGGGAGGTGCTGGTTAAAGAGCGAGCCAGGCTTGGCGAGAGTGGGGTCCTGCAGCTGCGGGCATCA
Homo sapiens ANAPC2-intron11, bases AGCTGGCCTGCCCCGGCCACACGGCCCAGCTGTGGGTGAGGGGCTGTTCCCCAGTGAGGGTGTGGTTTGGGGCAGCTGCGCCAGCAGGCGGCACCCTTCCCT and TGACGGGAGGCAGGGGAGGTGCAGCCCGTGCCCA
The ZZ oligonucleotide pairs are attached to a proprietary amplification target, which in turn is detected by a proprietary set of oligonucleotides. The latter are finally detected by Fast Red dye. Each red “dot” in the final preparation presumably represents a single RNA molecule.
Xenopus Oocytes and Injections.
X. laevis females were anesthetized with 1% MS222 (tricaine methane sulfonate, pH 7), and pieces of ovary were surgically removed and cultured in OR2 medium (26) at 16°. Manually separated oocytes were microinjected at the animal pole with 100 pg of plasmid in a 2.4 nL volume using the Nanoject II microinjection apparatus (Drummond Scientific). Needles were pulled from capillary tubes (0.5-mm inner diameter, 1.2-mm outer diameter) with a horizontal micropette puller (model P-97; Sutter Instrument). Different amounts of competitor plasmid were coinjected: 100, 250, 500, 750, and up to 1,000 pg. Injection of more than 1,000 pg of plasmids was often lethal for oocytes. At 48 h after injection, oocytes were dissected in OR2 isolation solution (83 mM KCl, 17 mM NaCl, 6.0 mM Na2HPO4, 4.0 mM KH2PO4, 1 mM MgCl2, 1.0 mM DTT, pH 7.8). Nuclei and cytoplasms were collected on dry ice before adding TRIzol for RNA extraction.
Plasmids.
A construct that expresses sisRNAs was generated as follows: The mCherry-CAAX gene, kindly provided by the Halpern laboratory, Department of Embryology, Carnegie Institution for Science, Baltimore, MD, was subcloned into the pGEM-T plasmid. Multicloning sites were used to add a minimal CMV promoter and a partial X. laevis gene (exon 14–15 of ncl) in which the intron was replaced by intron 2 of the X. tropicalis faf2 gene.
A plasmid that encoded an HA-tagged (N terminus) human NXF1 gene was purchased from ABclonal. The pCS2 GFP plasmid was generated in the Brown laboratory, Department of Embryology, Carnegie Institution for Science, Baltimore, MD. An X. tropicalis tRNA gene was amplified by PCR and cloned in the pGEM-T plasmid. An shRNA construct was kindly provided by the Zhao Zhang laboratory, Department of Embryology, Carnegie Institution for Science, Baltimore, MD.
Data Deposition.
RNA sequencing data have been deposited in the NCBI Sequence Read Archive (SRA BioProject ID: PRJNA479418).
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award R01 GM33397 (to J.G.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.G.G. is an American Cancer Society Professor of Developmental Genetics.
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA sequencing data have been deposited in the NCBI Sequence Read Archive (SRA BioProject ID PRJNA479418).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808816115/-/DCSupplemental.
References
- 1.Ooi SL, et al. RNA lariat debranching enzyme. Methods Enzymol. 2001;342:233–248. doi: 10.1016/s0076-6879(01)42548-1. [DOI] [PubMed] [Google Scholar]
- 2.Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: Intron length effects and activity of a precursor snoRNA. RNA. 1998;4:1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner EJ, Nizami ZF, Talbot CC, Jr, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26:2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talhouarne GJ, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA. 2014;20:1476–1487. doi: 10.1261/rna.045781.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertles JF, Beck WS. Biochemical aspects of reticulocyte maturation. I. Fate of the ribonucleic acid. J Biol Chem. 1962;237:3770–3777. [Google Scholar]
- 6.Doss JF, et al. A comprehensive joint analysis of the long and short RNA transcriptomes of human erythrocytes. BMC Genomics. 2015;16:952. doi: 10.1186/s12864-015-2156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alhasan AA, et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2016;127:e1–e11. doi: 10.1182/blood-2015-06-649434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian Z, et al. Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine. 2018;27:18–26. doi: 10.1016/j.ebiom.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sedgwick AD, Morris T, Russell BA, Lees P. Single step purification procedure for the rapid separation of equine leucocytes. Vet Res Commun. 1986;10:445–452. doi: 10.1007/BF02214007. [DOI] [PubMed] [Google Scholar]
- 10.Taggart AJ, et al. Large-scale analysis of branchpoint usage across species and cell lines. Genome Res. 2017;27:639–649. doi: 10.1101/gr.202820.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An X, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123:3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gall JG, Wu Z. Examining the contents of isolated Xenopus germinal vesicles. Methods. 2010;51:45–51. doi: 10.1016/j.ymeth.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nam K, et al. Yeast lariat debranching enzyme. Substrate and sequence specificity. J Biol Chem. 1994;269:20613–20621. [PubMed] [Google Scholar]
- 14.Panté N. Use of intact Xenopus oocytes in nucleocytoplasmic transport studies. Methods Mol Biol. 2006;322:301–314. doi: 10.1007/978-1-59745-000-3_21. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Talhouarne GJS, Gall JG. 7SL RNA in vertebrate red blood cells. RNA. 2018;24:908–914. doi: 10.1261/rna.065474.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dargemont C, Kühn LC. Export of mRNA from microinjected nuclei of Xenopus laevis oocytes. J Cell Biol. 1992;118:1–9. doi: 10.1083/jcb.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delaleau M, Borden KL. Multiple export mechanisms for mRNAs. Cells. 2015;4:452–473. doi: 10.3390/cells4030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pek JW, Osman I, Tay ML, Zheng RT. Stable intronic sequence RNAs have possible regulatory roles in Drosophila melanogaster. J Cell Biol. 2015;211:243–251. doi: 10.1083/jcb.201507065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Z, et al. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panda AC, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45:e116. doi: 10.1093/nar/gkx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armakola M, et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet. 2012;44:1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, et al. Intron lariat RNA inhibits MicroRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLoS Genet. 2016;12:e1006422. doi: 10.1371/journal.pgen.1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han B, et al. Human DBR1 modulates the recycling of snRNPs to affect alternative RNA splicing and contributes to the suppression of cancer development. Oncogene. 2017;36:5382–5391. doi: 10.1038/onc.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace RA, Jared DW, Dumont JN, Sega MW. Protein incorporation by isolated amphibian oocytes. III. Optimum incubation conditions. J Exp Zool. 1973;184:321–333. doi: 10.1002/jez.1401840305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.