Significance
Synonymous mutations in genes do not change protein sequence, but they may affect gene expression and cellular function. Here we describe an unexpected toxic effect of synonymous mutations in Escherichia coli, with potentially large implications for bacterial physiology and evolution. Unlike previously studied effects of synonymous mutations, the effect that we discovered is independent of translation, but it depends on the production of toxic mRNA molecules. We hypothesize that the mechanism we identified influences the evolution of endogenous genes in bacteria by imposing selective constraints on synonymous mutations that arise in the genome. Of interest for biotechnology and synthetic biology, we identify bacterial strains and growth conditions that alleviate RNA toxicity, thus allowing efficient overexpression of heterologous proteins.
Keywords: codon usage, RNA, synthetic biology, experimental evolution
Abstract
Many organisms are subject to selective pressure that gives rise to unequal usage of synonymous codons, known as codon bias. To experimentally dissect the mechanisms of selection on synonymous sites, we expressed several hundred synonymous variants of the GFP gene in Escherichia coli, and used quantitative growth and viability assays to estimate bacterial fitness. Unexpectedly, we found many synonymous variants whose expression was toxic to E. coli. Unlike previously studied effects of synonymous mutations, the effect that we discovered is independent of translation, but it depends on the production of toxic mRNA molecules. We identified RNA sequence determinants of toxicity and evolved suppressor strains that can tolerate the expression of toxic GFP variants. Genome sequencing of these suppressor strains revealed a cluster of promoter mutations that prevented toxicity by reducing mRNA levels. We conclude that translation-independent RNA toxicity is a previously unrecognized obstacle in bacterial gene expression.
Although synonymous mutations do not change the encoded protein sequence, they cause a broad range of molecular phenotypes, including changes of transcription (1), translation initiation (2, 3), translation elongation (4), translation accuracy (5, 6), RNA stability (7), and splicing (8). As a result, synonymous mutations are under subtle but nonnegligible selective pressure, which manifests itself in the unequal usage of synonymous codons across genes and genomes (9–11). Several recent experiments directly measured the effects of synonymous mutations on fitness in bacteria (2, 12–17). It has been commonly assumed that fitness depends primarily on the efficiency, accuracy, and yield of translation. Here we show that in the context of heterologous gene expression in Escherichia coli, large effects of synonymous mutations on fitness are translation-independent and are mediated by RNA toxicity.
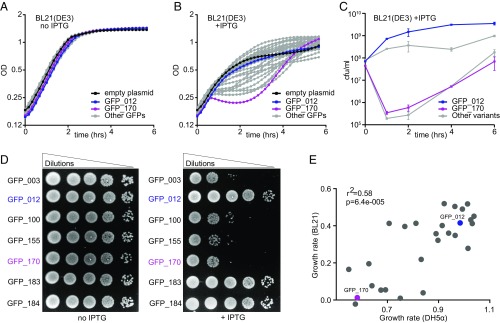
To study the effects of synonymous mutations on bacterial fitness, we used an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible, bacteriophage T7 polymerase-driven plasmid to express a collection of synonymous variants of the GFP gene (2) in E. coli BL21-Gold(DE3) (henceforth referred to as BL21) cells (Materials and Methods). Without IPTG induction, there were no discernible differences in growth between strains (Fig. 1A). When induced with IPTG, the growth rate of GFP-producing strains was reduced, consistent with the metabolic burden conferred by heterologous gene expression. The growth phenotype varied remarkably between strains expressing different synonymous variants of GFP (Fig. 1B and SI Appendix, Fig. S1). “Slow” variants caused a long lag phase postinduction, indicating that at this stage the cells either stopped growing or died, while “fast” variants showed growth rates closer to noninduced cells. Several hours after induction, the slow variants appeared to resume growth (Fig. 1B): we found that this was related to the emergence of suppressor strains that could tolerate the expression of these variants (SI Appendix, Fig. S1) (see below).
Fig. 1.
GFP variants are toxic in E. coli. (A and B) Growth curves of BL21 E. coli cells, noninduced (A) or induced with 1 mM IPTG at t = 0 h (B). Cells carrying GFP_012 (nontoxic variant, blue), GFP_170 (toxic variant, magenta), pGK8 (empty vector control, black), and 29 other variants (gray) are shown. Each curve represents an average of nine replicates (three biological × three technical). (C) Numbers of colony forming units per milliliter (cfu/mL) at specified time points after induction with 1 mM IPTG. Data points represent averages of four replicates, ±SEM. (D) Semiquantitative estimation of BL21 cell viability by spot assay. (E) Estimated growth rates of cells expressing GFP variants in DH5α and BL21 strains (averages of at least six replicates).
We quantified cell viability postinduction by assessing the colony-forming ability of cells (Fig. 1C). Fast variants showed the expected increase in cell numbers postinduction, but slow variants caused a 1,000-fold decrease in viable cell numbers. Similarly, spotting of noninduced cells onto LB plates with IPTG showed that the slow variants formed markedly fewer colonies than fast variants (Fig. 1D). Microscopic analysis of slow variants showed decrease in cell number, growth arrest, and in some cases massive cell death following IPTG induction. In the case of fast variants we observed normal increase in cell numbers and negligible cell death after induction (SI Appendix, Fig. S2). These results indicate that certain synonymous variants of GFP cause significant growth defects when overexpressed in E. coli cells, and we will henceforth refer to these variants as “toxic.”
To test if toxicity was specific to T7 promoter-driven overexpression, we analyzed growth phenotypes following the expression of a subset of GFP variants using a bacterial polymerase (trp/lac) promoter system (SI Appendix). Although the growth phenotypes measured with bacterial promoter constructs were not as dramatic as with T7-based constructs, presumably because of lower GFP expression levels, growth rates with both types of promoters were correlated with each other (Fig. 1E). Interestingly, toxicity increased at high temperature, and decreased at low temperature (SI Appendix, Fig. S1). Taken together, these results indicate that the toxic GFP variants cause growth defects in two different E. coli strains, with two types of promoters, possibly through a common mechanism.
To understand whether toxicity depends on the process of translation, we selected several toxic and nontoxic variants of GFP and mutated their Shine–Dalgarno sequences from GAAGGA to TTCTCT to prevent ribosome binding and block translation initiation. As expected, mutation of Shine–Dalgarno sequences completely inhibited the production of functional GFP protein from all tested constructs (Fig. 2A). To our surprise, GFP variants without Shine–Dalgarno sequences remained toxic, and their effects on growth were indistinguishable from variants with a functional Shine–Dalgarno sequence (Fig. 2B). Western blot analysis confirmed that mutation of the Shine–Dalgarno sequences ablates GFP expression (SI Appendix, Fig. S3). We considered the possibility that a cryptic Shine–Dalgarno element within the coding region allowed translation of a truncated fragment of GFP, which would be consistent with loss of GFP fluorescence and translation-dependent toxicity. However, analysis of the coding regions with the RBS Calculator (18) revealed no strong Shine–Dalgarno consensus sequences. These results raise the possibility that toxicity might arise at the RNA level, rather than at translation or protein level.
Fig. 2.
Toxicity of GFP variants is independent of translation. (A and B) Fluorescence (A) and growth rate (B) of BL21 cells expressing GFP variants with functional and nonfunctional ribosome-binding sites (RBS). (C and D) Fluorescence (C) and growth rate (D) of cells expressing full-length GFP variants, truncated variants, and variants containing internal stop codons or transcription terminators. Inset in C shows location of toxic sequence element in GFP_170, which was calculated based on an analysis of growth rates of 36 shuffled constructs. The y axis shows the statistical significance of the association of particular positions with slow growth. Variants derived from nontoxic GFP_012 are shown in blue and variants derived from toxic GFP_170 are shown in magenta. Full-length constructs, truncated constructs, and constructs with internal stop codons have similar growth rates, suggesting that the element of toxicity resides within the truncated fragment and that the mechanism of toxicity is independent of translation. FL, full-length construct; TT, T7 transcription terminator. All data are averages of nine replicates, ±SEM.
To identify sequence elements required for toxicity, we selected one of the toxic variants (GFP_170) and a nontoxic variant (GFP_012), and performed DNA shuffling (19) to generate constructs that consisted of random fragments of GFP_170 and GFP_012. All of the shuffled and nonshuffled constructs we generated encoded the same GFP protein sequence. Analysis of growth rate phenotypes of these shuffled constructs revealed a fragment near the 3′ end of the GFP_170 coding sequence (nucleotides 514–645) that was sufficient to elicit the toxic phenotype (Fig. 2C and SI Appendix, Fig. S4). Some mutations outside of the toxic region partially improved fitness, which might be explained by interactions of the RNA secondary structure between the toxic region and the mutated regions. The GFP_170 mRNA is predicted to have a very low translation initiation rate due to strong RNA secondary structure near the mRNA 5′ end (2). Nevertheless, replacement of the strongly structured 5′ region with an unstructured fragment did not affect toxicity (SI Appendix, Fig. S4).
The above results led us to hypothesize that the toxicity associated with GFP expression was independent of translation, but depended on the presence of a specific fragment of RNA. To test this hypothesis, we performed growth rate measurements with a series of constructs. First, we isolated the 132-nt toxic region identified in the DNA shuffling experiment and expressed it on its own, with or without start and stop codons. The expression of the 132-nt fragment of GFP_170 was sufficient for toxicity, whereas the corresponding fragment of GFP_012 did not cause toxicity. The effect of the 132-nt fragments on growth did not depend on the presence of translation start and stop codons (Fig. 2 C and D), the fragments contained no cryptic translation initiation signals, and FLAG-tag fusions showed no detectable protein expression from the GFP_170 fragment in any of the three reading frames (SI Appendix, Fig. S3). Second, we introduced stop codons upstream of the toxic fragment in the GFP_170 coding sequence and in the corresponding positions of GFP_012. This placement of stop codons ensures that ribosomes terminate translation before reaching the putative toxic region of the RNA, while still allowing a full-length transcript to be produced. As expected, internal stop codons abrogated GFP protein production (Fig. 2C), but despite the presence of premature stop codons, GFP_170_Stop still caused toxicity to bacterial cells while GFP_012_Stop remained nontoxic (Fig. 2D). To remove possible out-of-frame translation, we inserted stop codons into GFP_170 in all three frames, before and after the toxic region, and toxicity remained the same in all cases (SI Appendix, Fig. S4). Third, we introduced an efficient synthetic T7 transcription terminator (20) upstream of the toxic region in GFP_170 and in the corresponding location in GFP_012. Notably, we found that both variants with internal transcription terminators became nontoxic, and GFP_170_TT grew slightly faster than GFP_012_TT (Fig. 2D). The GFP_170 fragment also caused toxicity when fused to FLAG-tags (in any of the three reading frames), and when fused to fluorescent protein mKate2 it caused toxicity and reduced expression of mKate2 by 50-fold (SI Appendix, Fig. S4). Overall, these data suggest that toxicity is caused by the RNA itself, rather than the process of translation or by the protein produced.
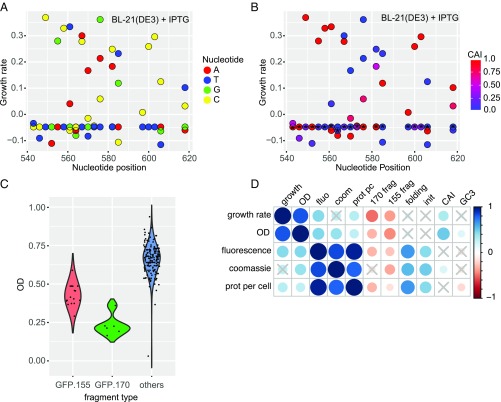
To investigate the sequence determinants of RNA-mediated toxicity, we measured the growth phenotypes of single synonymous mutations within the 132-nt region of GFP_170. Close to half of these mutations reduced or abolished the toxic phenotype, whereas the remaining mutations had no effect (Fig. 3A). There was no clear relationship between the position of mutations within the region and their effect on growth, nor was there any relationship between the type of nucleotide introduced and growth. RNA toxicity associated with triplet repeats has been described in Eukaryotes (21), but we found no triplet repeats in the toxic GFP mRNAs. Consistent with our observation that the toxic effect does not require translation, codon adaptation index (CAI) was not associated with toxicity (Fig. 3B). RNA folding energy, measured either in the immediate vicinity of each mutation or for the entire 132-nt mutagenized region, was not correlated with toxicity, and we were unable to identify any RNA structural elements associated with the toxic phenotype. We further probed the effects of sets of several mutations within the 132-nt toxic region. Of 98 sets of mutations we introduced within the region, 75 reduced or abolished toxicity, whereas 23 of 98 sets had no effect (SI Appendix, Fig. S5). In almost all cases, the phenotypes of sets could be deduced from the effects of individual mutations in a simple way: if any mutation in a set abolished toxicity, then the set also did. Four sets did not conform to this rule, indicating potential epistatic interactions between mutations. Mutations near the 3′ end of the 132-nt fragment had no effect on toxicity, identifying a minimal toxicity-determining region of about 100 nucleotides that either consists of a single functional element or contains multiple elements whose cooperative action causes toxicity.
Fig. 3.
Multiple sequence elements determine RNA-mediated toxicity. (A) Growth rates of single synonymous mutants of GFP_170, measured in BL21 strain (averages of nine replicates). Mutations located throughout the toxic region reduce or abolish toxicity. (B) Relationship between the CAI and the growth rate of GFP mutants. Asterisk-marked codons represent the original codon in GFP_170. (C) Growth estimate (OD) of BL21 cells expressing GFP variants containing fragments: GFP_155 nucleotides 490–720 (n = 16, red), GFP_170 nucleotides 514–645 (n = 6, green), and other variants (n = 163, blue). (D) Spearman correlation analysis of phenotypes measured in BL21 cells and sequence covariates in a set of 190 GFP variants. The size and color of circles represents the correlation coefficient; crosses indicate nonsignificant correlations.
Several recent studies examined the effects of synonymous mutations on fitness in bacteria, either in endogenous genes or in overexpressed heterologous genes (2, 12–16). Fitness had been found to correlate with the CAI, GC content, RNA folding, protein expression level, a codon ramp near the start codon, and measured or predicted translation initiation rates. We quantified these variables in a set of 190 synonymous variants of GFP and analyzed their impact on fitness. We also considered two candidate toxic RNA fragments (GFP_170, nucleotides 514–645, and GFP_155, nucleotides 490–720), both of which were common to several constructs and appeared to negatively influence fitness (Fig. 3 C and D). High protein expression was previously shown to correlate with slow growth (14), whereas we found positive correlations of fitness with total protein yield or protein yield per cell. These correlations presumably reflect reduced protein yields and cell growth after the induction of toxic RNAs. As seen previously, growth rate and optical density were positively correlated with CAI, and GC content was correlated with optical density (2, 16). However, in a multiple regression analysis aimed to disentangle the effects of these covariates, we found that the presence of candidate toxic RNA fragments predicted slow growth in both BL21 and DH5α cells, whereas CAI and GC3 did not (SI Appendix). This suggests that the apparent correlation of CAI or GC content with fitness, observed in this and previous studies (2, 16), might result from the confounding effect of toxic RNA fragments. Consistently, an experiment with 22 new, unrelated synonymous GFP constructs spanning a wider range of GC content showed no correlation between GC content and bacterial growth (SI Appendix, Fig. S6). To further test whether toxicity could be explained by unusually high expression of certain GFP variants, we measured the mRNA abundance of 79 toxic and nontoxic RNAs by Northern blots, and correlated GFP mRNA abundance per cell with optical density (OD). Although we observed differences in mRNA abundance, mostly related to mRNA folding (2), we find no significant correlation between RNA abundance and toxicity (Spearman’s ρ = 0.12, P = 0.29). Furthermore, we detected no consistent differences in plasmid abundance between toxic and nontoxic variants.
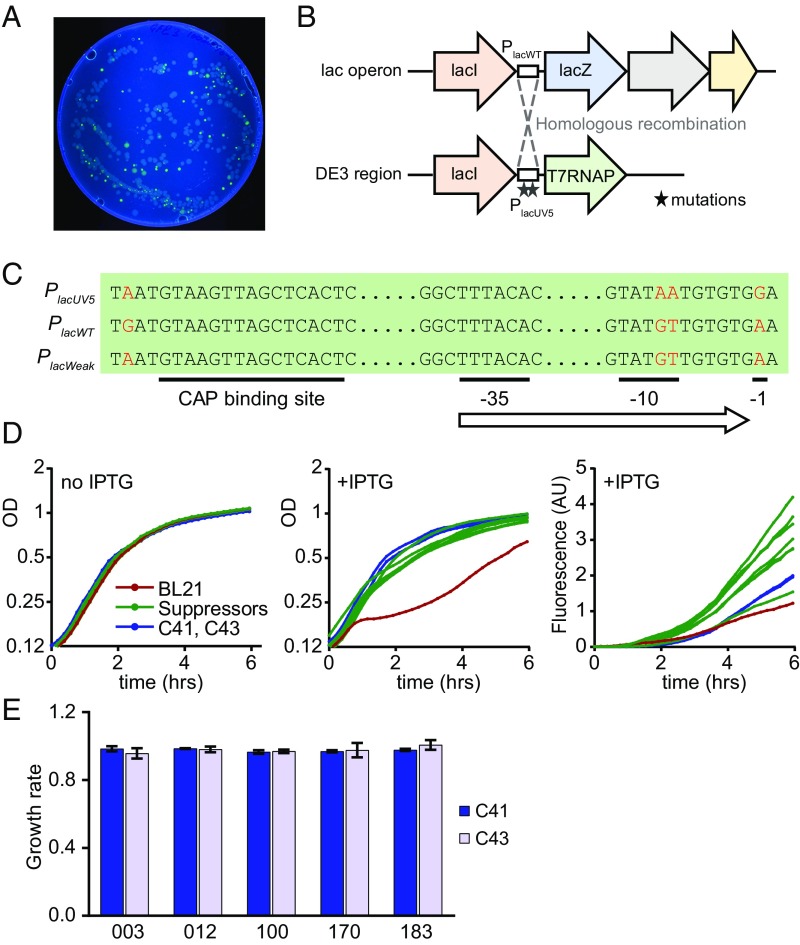
To study the molecular mechanisms of toxicity caused by mRNA overexpression, we aimed to evolve genetic suppressors of this phenotype. We selected several GFP constructs that showed both strong toxicity and moderate or high GFP fluorescence, and plated bacteria containing these constructs on LB agar plates with IPTG and ampicillin. We observed a number of large white colonies that apparently expressed no GFP, and smaller bright green colonies producing high amounts of the GFP protein (Fig. 4A). We hypothesized that the green colonies have acquired a genomic mutation that allowed cells to survive while expressing toxic RNAs. To support this, we cured the evolved strains of their respective plasmids and retransformed the cured strains with the same plasmid. The retransformed strains readily formed bright green colonies on IPTG+ampicillin plates, and exhibited faster growth rates in IPTG medium compared with the parental strain. This supported our hypothesis that the mutations were located on the chromosome and not the plasmid. We therefore selected 22 evolved strains and the parental strain for genome sequencing, and used the GATK pipeline for calling variants (SI Appendix).
Fig. 4.
Isolation and characterization of genetic suppressors of toxicity. (A) Fluorescence image of LB+Amp+IPTG Petri dish with BL21 cells expressing GFP_003 variant. (B) Genetic organization of lac and DE3 loci in BL21 cells. Dashed lines indicate homologous recombination between the loci in suppressor strains. (C) Sequence variation between the three types of promoters found in the suppressor strains. Substitutions are marked in red. (D) Growth curves and fluorescence of strains carrying the GFP_003 variant: parental BL21 strain (red), suppressors strains (n = 7, green), C41 and C43 strains (blue). (E) Growth rates of C41 and C43 cells expressing several GFP variants. GFP_003, GFP_100 and GFP_170 are toxic in the BL21 strain; GFP_012 and GFP_183 are not. Growth curves are averages of three replicates.
In all green suppressor strains, we found a single cluster of mutations in the Plac promoter of the T7 polymerase gene that explains the suppressor phenotype (Fig. 4 B and C and SI Appendix, Table S1). The parental BL21 strain contains two alleles of the Plac promoter: the wild-type allele PlacWT controls the lac operon, and a stronger derivative allele PlacUV5 controls T7 RNA polymerase. In the suppressor strains, recombination between these two loci associates PlacWT promoter with T7 polymerase, leading to reduced levels of polymerase and presumably to reduced transcription of GFP. The same Plac promoter mutations were recently observed in the C41(DE3) and C43(DE3) strains of E. coli (the “Walker strains”), and were responsible for the reduced T7 RNA polymerase expression, high-level recombinant protein production, and improved growth characteristics of those strains (22–24). Similar to our suppressor strains, C41(DE3) and C43(DE3) allowed high protein expression of toxic GFP variants, and little toxicity was observed in these strains (Fig. 4D). Taken together, these results support our conclusion that high levels of RNA, rather than RNA translation or protein, are responsible for toxicity.
To test whether translation-independent RNA toxicity might affect genes other than GFP, we turned to the ogcp gene, which encodes a membrane protein oxoglutarate-malate transport protein (OGCP) believed to be toxic for E. coli. OGCP overexpression was originally used to derive the C41(DE3) strain, now commonly used for recombinant protein expression (22). As expected, we found that expression of OGCP was toxic to BL21 but not to C41(DE3) cells. In agreement with our observations for GFP, a translation-incompetent variant of OGCP lacking the Shine–Dalgarno sequence was just as toxic to BL21 cells as a translation-competent variant (SI Appendix, Fig. S7). A translation-competent, codon-optimized variant of OGCP retained toxicity in BL21 cells. These experiments suggest that translation-independent RNA toxicity might be a widespread phenomenon associated with heterologous gene expression in E. coli.
Heterologous protein expression is known to inhibit growth of E. coli. Toxicity is typically attributed to the foreign protein itself, and it is often remedied by lowering expression, reducing growth temperature, or using special strains of E. coli, such as C41(DE3). Here we demonstrate that the same strategies and strains also prevent toxicity when RNA, rather than protein, is the toxic molecule. We speculate that other cases of toxicity, previously attributed to proteins, may in fact be caused by RNA. Although the molecular mechanisms of RNA toxicity are presently unclear, we identified several GFP and OGCP variants with similar phenotypes, suggesting that the phenomenon may be common. Interestingly, induction of wild-type APE_0230.1 in E. coli inhibits growth, but a codon-optimized variant does not inhibit growth despite increased protein yield (25). In addition, several recent high-throughput studies found unexplained cases of slow growth or toxicity upon the expression of various random sequences in E. coli (14, 26, 27). Our results point to RNA toxicity as a possible cause of these observations.
Our results are relevant to the phenomenon of synonymous site selection in microorganisms. Synonymous mutations can influence fitness directly (in cis), by changing the expression of the gene in which the mutation occurs (12, 13, 15), or indirectly (in trans), by influencing the global metabolic cost of expression (2, 14, 16, 28). Experiments with essential bacterial genes predominately uncover cis-effects, most of them mediated by changes of RNA structure or other properties that influence translation yield. For example, mutations in Salmonella enterica rpsT down-regulated the gene, and could be compensated by additional mutations in or around rpsT or by increase of the gene copy number (13). Similarly, mutations that disrupted mRNA structure of the E. coli infA gene, through local or long-range effects, explained much variation in fitness across a large collection of mutants (12). Protein abundance and RNA structure contribute to the observed trans-effect of mutations (14). Although our results are broadly consistent with a role of RNA structure, the specific structure is unknown, and the effects we uncovered are translation-independent, suggesting that a novel mechanism is involved. Toxic RNAs might interact with an essential cellular component, either nucleic acid or protein, and interfere with its normal function. Such interactions might be uncovered by pulldowns of toxic RNAs combined with sequencing or mass spectrometry. Alternatively, RNA phase transitions may be involved; such transitions have been shown to contribute to the pathogenicity of CAG-expansion disorders in Eukaryotes, providing a mechanistic explanation for this phenomenon (29). Further studies should address the mechanisms, biotechnology applications, and evolutionary consequences of RNA toxicity in bacteria.
Materials and Methods
We expressed a library of 347 synonymous variants of GFP in BL21-Gold(DE3) and DH5α strains of E. coli using IPTG-inducible multicopy plasmids. We monitored cell growth following IPTG induction using automated plate readers, fluorescence microscopy, and spot assays. Genetic suppressors of toxicity were isolated by plating BL21-Gold(DE3) cells carrying GFP variants onto LB plates containing 1 mM IPTG. Whole-genome sequencing of 22 suppressor strains was carried out on the Illumina HiSEq. 2500 platform (Edinburgh Genomics), and was analyzed using the GATK pipeline. A more detailed description of methods is available in SI Appendix.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP149903).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810022115/-/DCSupplemental.
References
- 1.Zhou Z, et al. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc Natl Acad Sci USA. 2016;113:E6117–E6125. doi: 10.1073/pnas.1606724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 4.Sørensen MA, Kurland CG, Pedersen S. Codon usage determines translation rate in Escherichia coli. J Mol Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 5.Akashi H. Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Presnyak V, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plotkin JB, Kudla G. Synonymous but not the same: The causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamary JV, Parmley JL, Hurst LD. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 12.Kelsic ED, et al. RNA structural determinants of optimal codons revealed by MAGE-seq. Cell Syst. 2016;3:563–571.e6. doi: 10.1016/j.cels.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knöppel A, Näsvall J, Andersson DI. Compensating the fitness costs of synonymous mutations. Mol Biol Evol. 2016;33:1461–1477. doi: 10.1093/molbev/msw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frumkin I, et al. Gene architectures that minimize cost of gene expression. Mol Cell. 2017;65:142–153. doi: 10.1016/j.molcel.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agashe D, et al. Large-effect beneficial synonymous mutations mediate rapid and parallel adaptation in a bacterium. Mol Biol Evol. 2016;33:1542–1553. doi: 10.1093/molbev/msw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghavan R, Kelkar YD, Ochman H. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci USA. 2012;109:14504–14507. doi: 10.1073/pnas.1205683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brandis G, Hughes D. The selective advantage of synonymous codon usage bias in Salmonella. PLoS Genet. 2016;12:e1005926. doi: 10.1371/journal.pgen.1005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stemmer WP. DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mairhofer J, Wittwer A, Cserjan-Puschmann M, Striedner G. Preventing T7 RNA polymerase read-through transcription-A synthetic termination signal capable of improving bioprocess stability. ACS Synth Biol. 2015;4:265–273. doi: 10.1021/sb5000115. [DOI] [PubMed] [Google Scholar]
- 21.Krzyzosiak WJ, et al. Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res. 2012;40:11–26. doi: 10.1093/nar/gkr729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 23.Kwon SK, Kim SK, Lee DH, Kim JF. Comparative genomics and experimental evolution of Escherichia coli BL21(DE3) strains reveal the landscape of toxicity escape from membrane protein overproduction. Sci Rep. 2015;5:16076. doi: 10.1038/srep16076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlegel S, Genevaux P, de Gier JW. De-convoluting the genetic adaptations of E. coli C41(DE3) in real time reveals how alleviating protein production stress improves yields. Cell Rep. 2015;10:1758–1766. doi: 10.1016/j.celrep.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 25.Boël G, et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016;529:358–363. doi: 10.1038/nature16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neme R, Amador C, Yildirim B, McConnell E, Tautz D. Random sequences are an abundant source of bioactive RNAs or peptides. Nat Ecol Evol. 2017 doi: 10.1038/s41559-017-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cambray G, Guimaraes JC, Arkin AP. Massive factorial design untangles coding sequences determinants of translation efficacy. bioRxiv. October 25, 2017 doi: 10.1101/208801. [DOI] [Google Scholar]
- 28.Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiol Rev. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.