Abstract
Electron crystallography is widespread in material science applications, but for biological samples its use has been restricted to a handful of examples where two-dimensional (2D) crystals or helical samples were studied either by electron diffraction and/or imaging. Electron crystallography in cryoEM, was developed in the mid-1970s and used to solve the structure of several membrane proteins and some soluble proteins. In 2013, a new method for cryoEM was unveiled and named Micro-crystal Electron Diffraction, or MicroED, which is essentially three-dimensional (3D) electron crystallography of microscopic crystals. This method uses truly 3D crystals, that are about a billion times smaller than those typically used for X-ray crystallography, for electron diffraction studies. There are several important differences and some similarities between electron crystallography of 2D crystals and MicroED. In this review, we describe the development of these techniques, their similarities and differences, and offer our opinion of future directions in both fields.
Keywords: MicroED, Electron diffraction, cryoEM (electorn cryo-microscopy), electron crystallography
1. Introduction
Electron cryo-microscopy (cryoEM) has revolutionized structural biology. CryoEM includes a set of techniques to investigate cryogenically cooled specimens using a transmission electron microscope (TEM) [1]. These techniques are divided into those that focus a scattered beam of electrons into images of the sample and those that utilize the scattered electron beam (diffraction). Cryo electron tomography [2] and single particle cryoEM [3,4] are two techniques that use the imaging mode. Tomography reconstructs the three-dimensional structure of single objects by stitching together multiple high-resolution images of the same object taken at many different angles. Single particle cryoEM takes images that have many copies of a single protein in multiple orientations. The three-dimensional protein structure is modeled by classifying thousands of individual protein images (projections) into many different views to reconstruct a three-dimensional representation of the specimen. Biomolecules resolved in cryotomography use sub-tomogram averaging to solve individual structures in much the same way as single particle reconstructions [5]. Though these methods utilize the same mode of the electron microscope, tomography typically investigates a single specimen of interest utilizing fixed tilt angles, whereas single particle stochastically samples random orientations of different copies of the same protein. The cryoEM diffraction methods can be similarly divided into two major methodologies: electron crystallography [6] and MicroED [7••]. Electron crystallography collects diffraction data or images from two-dimensional crystals using still images over a range of tilt angles to solve structures. A two-dimensional protein crystal is an array of regularly arranged protein in X and Y (two dimensions) but is only a single protein layer thick. Each 2D crystal is oriented differently on the grid and images of a thousand 2D crystals collected at different tilt angles. The data is then combined to a single dataset that is then used for structure determination. Phases come from images or by molecular replacement [8]. In sharp contrast, MicroED is electron diffraction from three-dimensional crystals up to a micrometer thick [9••], not 2D crystals, and a single 3D nanocrystal is sometimes sufficient for the collection of an entire dataset [10,11•]. The crystal is continuously diffracted and under continuous rotation [10]. The data is collected on a fast camera as a movie and processed using standard X-ray diffraction software [12]. In this way, electron crystallography of 2D crystals is a more stochastic methodology, whereas MicroED is prescribed to a single 3D crystal sample. These differences are reminiscent to the differences between single particle analysis and cryotomography, respectively. There are clearly major differences between electron crystallography of 2D crystals and MicroED and here we discuss such differences. We give a brief overview of the ongoing developments in electron crystallography and MicroED, and discuss exciting avenues to future development.
2. Milestones in electron crystallography of 2D crystals
Investigation on biological specimens by electron crystallography arguably began when Parsons and Martius used electron diffraction to investigate the structure of muscle fibers in 1964 [13]. This same year Klug and Berger used electron microscopy images to corroborate optical diffraction data [14]. However, this study did not utilize the signal from diffracted electrons, nor did either study elucidate a novel structure. The first report of electron crystallography that used Fourier transforms from images to visualize a three-dimensional density and gain insight into a novel biological structure was done by DeRosier and Klug in 1968 on the structure of the T4 bacteriophage [15]. Hoppe et al. followed, discussing the possibility of solvent replacement and phasing in electron crystallography, and correlated the diffraction amplitudes of Myoglobin from X-ray and electron diffraction in 1968 [16]. Glaeser and Thomas demonstrated electron diffraction from a valine crystal to 1.06 Å resolution, and explored radiation sensitivity of biological specimens in 1969 [17]. Electron microscopy was used by Labaw and Rossmann in 1969 to observe the lattice spacing in lactate dehydrogenase (LAD) from dogfish muscle [18].
The first investigations of two-dimensional crystalline assemblies were done by Matricardi in 1972 [19], followed by Taylor and Glaeser [20], and Unwin and Henderson in 1975 [21]. However, it was not until Henderson et al. used the naturally occurring purple membrane protein bacteriorhodopsin in 1990 that a near atomic model was elucidated by electron diffraction at a resolution of 3.5 Å (Fig. 1) [22••]. These studies used the transforms of high magnification images to calculate the phases of the crystalline structure, and the square-root of the intensities from electron diffraction for the amplitudes. These were combined at many tilt angles to create projection maps of the structure being studied, and combined to form a fully featured 3D reconstruction. From this work, many others using similar methodology followed. Among these included work on light harvesting complex II by Kuhlbrandt in 1994 [23], a higher resolution bacteriorhodopsin structure by Kimura et al. in 1997 [24], the structure solution of tubulin from two-dimensional crystals by Nogales et al. in 1998 [25] and the first structure of a water channel aquaporin-1 by Walz, Engel and Fujiyoshi [26].
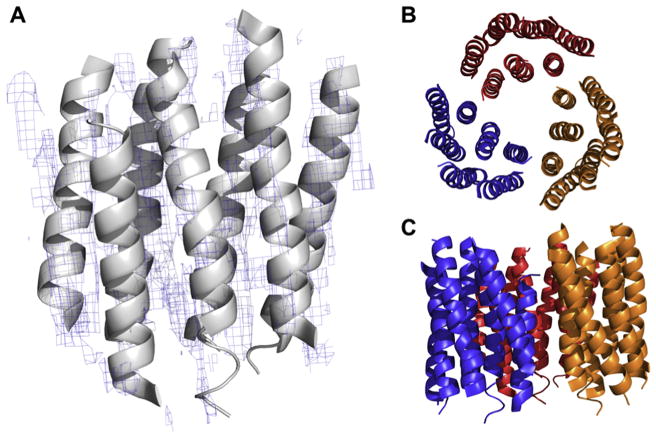
Fig. 1.
(A) Structure solution of Bacteriorhopsin at 3.5 Å resolution determined by electron crystallography of 2D crystals. Density contoured at 1.5σ level. (B, C) Biological assembly showing the bacteriorhodoprin trimer arrangement in the membrane.
The progress in the early 1990s of electron crystallography was largely fueled by the development of rapid freezing of thin layers of ice (or in glucose or trehalose as embedding media), resulting in vitrification rather than crystallization of the ice layer [27–29]. This came after work by Taylor and Glaeser in 1974, that created frozen hydrated specimens by sandwiched hydrophilic support films rather than plunge freezing [20]. In early 2000s a number of additional water channel structures were determined at ~3 Å resolution [26,30,31].
In 2005, Gonen et al. used the electron microscope operating exclusively in diffraction mode to collect thousands of diffraction data from two-dimensional aquaporin-0 (AQP0) crystals producing a structure of the channel in a closed conformation at 1.9 Å (Fig. 2) [32•]. It was at this point that electron diffraction of protein samples entered the atomic age. This was the first time that water was seen by cryoEM as well as the first structure of a eukaryotic membrane. It was also the first study where diffraction data alone was used for structure solution rather than coupling phases originating from images. Here protocols for molecular replacement in electron crystallography were established [33•].
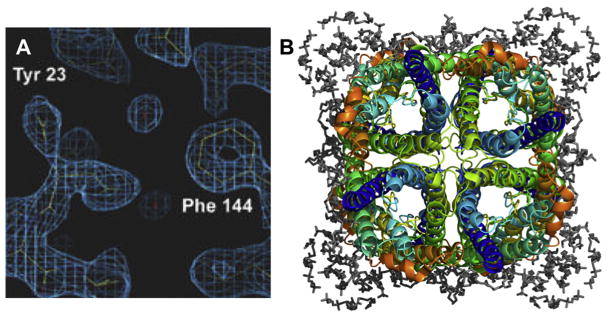
Fig. 2.
The structure solution of Aquaporin-0 from electron crystallography at 1.9 Å resolution adapted from [35]. (A) Density for waters in the pore area, and (B) the full biological assembly showing resolved lipids (grey) surrounding the channel tetramer.
A major reason for the slow progress in membrane protein structure solution from electron crystallography were often prohibitive sample requirements, lengthy and laborious data collection and analysis procedures. Many milligrams of purified membrane protein are necessary for 2D crystallization assays. Only 17 plane groups are allowed [34] while for 3D crystals 65 packing symmetries are allowed. This means it is more difficult to coax proteins to form a 2D crystal rather than a 3D crystal. Once 2D crystals are obtained, data collection and analysis are laborious and time consuming. Issues with phase retrieval further curbed the achievable resolution by electron crystallography. Solving a structure requires both amplitudes and phases, where the amplitudes were taken from diffraction patterns, and phases were collected from the Fourier transforms of high-resolution images. Where phases are available from a known structure, one only needs the amplitudes, as was done in the case of Aquaporin-0 [32•]. Using a known structure or homologue to solve the phase problem is known as molecular replacement [35]. This is a procedure that attempts to find a model that fits the experimentally measured intensities among the known structures scanning the model over the possible rotational and translational possibilities. Molecular replacement was used in electron crystallography only in the mid-2000s [30,32•] but traditionally phases originated from images of the 2D crystals. Collection of high resolution images from 2D crystals at multiple tilt angles is much more technically challenging than diffraction patterns: diffraction is translationally invariant, making the effects of drift and charging essentially unimportant. In 2011, Wisedchaisri and Gonen demonstrated that only low resolution images are needed if high resolution diffraction patterns are also present, and that the low-resolution phases can be extended to higher resolution to ultimately solve the 3D molecular structure to atomic resolution (Fig. 3) [36].
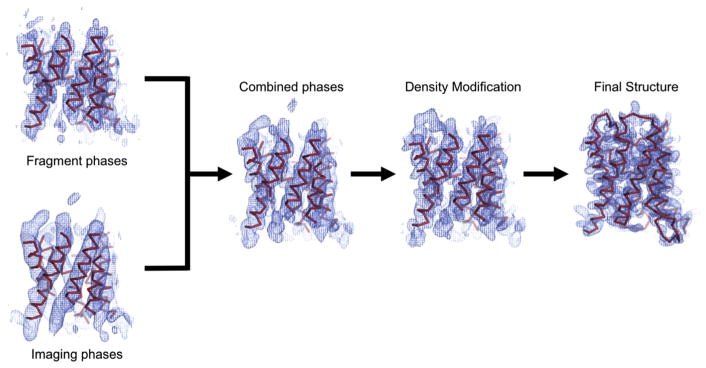
Fig. 3.
Phase extension in electron crystallography from low resolution scheme adapted from [37]. Low resolution phases and amplitudes are collected from cryo-EM images and combined with phases from initially placed fragments. Density modification is done with phases from the combined phases and the electron diffraction intensities. Density modification then allows for final building and refinements.
3. Milestones in MicroED of 3D crystals
Microcrystal electron diffraction (MicroED) is a cryoEM method that was unveiled only recently [7••] to solve protein structures from three-dimensional crystals using a transmission electron microscope. Electron diffraction of three-dimensional crystals was thought to be intractable, as simulations suggest that the measured intensities in the diffraction spots would be far too inaccurate due to dynamical scattering [37–39]. Dynamical or multiple scattering is thought to occur when the electron undergoes multiple scattering events within the same sample instead of a single scattering event. This is thought more possible in electron scattering, because the interaction between electrons and matter is much greater than the interaction between light and matter, as is the case for X-rays. However, micrographs showing diffraction spots from three-dimensional crystals had been reported earlier from catalase crystals at room temperature as far back as 1930, with diffraction from catalase crystals at cryogenic conditions reported in 1974 by Taylor and Glaeser [20]. Unwin and Henderson then created a density projection map from three dimensional crystals of catalase in 1975 with a resolution of 9 Å [40]. Wau Chiu et al. observed the electron diffraction patterns from three dimensional crystals of tetanus toxin to create a general density from a single reciprocal lattice pattern to deduce that the toxin molecules bundle together in groups of 6 in each unit cell of the three-dimensional crystal [41], and then determined the P4222 space group and arrangement of crotoxin from thin, three-dimensional crystals using projection maps from 2.2 Å diffraction and 4.4 Å imaging resolutions [42]. After his work with Unwin on the acetylcholine receptor, Toyoshima and Stokes observed diffraction from Ca2+-ATPase crystals in the electron microscope, though ultimately solved the structure by X-ray diffraction [43].
Lysozyme was the first protein structure solved from a three-dimensional crystal using electron diffraction in 2013 (Fig. 4) [7••]. The method was named microcrystal electron diffraction, or MicroED, and distinguishes itself from electron crystallography in that it utilizes three-dimensional protein crystals up to 0.5 micrometer thick consisting of many protein layers arranged regularly in three dimensions. MicroED also differs in both data collection and processing methods. This first MicroED structure was solved by combining still-diffraction data from 3 different lysozyme crystals. Data from each crystal span up to a 90-degree wedge of reciprocal space, allowing proper indexing and integration of the partially recorded reflections. The data are collected at liquid nitrogen temperatures using an ultra-low dose rate of ~0.1 e−A−2 s−1. Diffraction spots were collected to a resolution of 1.7 Å with a final refinement to 2.9 Å. Each crystal yielded up to 90 degrees of rotation data due to the ultra-low dose, allowing multiple crystals to be merged by accurately knowing the positions of each crystal in reciprocal space relative to one another.
Fig. 4.
Lysozyme structure (A) and densities from MicroED with data collected from (B - 3J4G at 2.9 Å) stills, (C - 3J6K at 2.5 Å) continuous rotation, and (D - 5K7O at 1.8 Å) continuous rotation with fragmentation.
The MicroED method was improved by the development and implementation of a new mode of data collection called continuous rotation in 2014 [10]. Continuous rotation MicroED uses a high frame rate CMOS or a direct electron detector to collect diffraction data while the crystal is being continuously rotated in a single direction in the electron beam at a fixed rotation speed. In this way, the data is output as a movie where each frame corresponds to a fixed angular wedge of reciprocal space, rather than a stationary slice through it [9••]. Continuous rotation is thought to reduce the contributions of dynamic scattering via intensity redistribution similarly to the precession method used in materials science [44], and therefore yields much more accurately measured reflections [12]. Oscillation of the sample during diffraction seemingly averages the signal over the entirety of the rocking curve, where the contributions of multiple scattering essentially cancel out, as first described by Blackman in 1939 [45]. This method increases the amount of information gathered, since full reflections can be collected on a single image as the crystal is rotated through the diffracting condition for each Bragg plane. This method is analogous to the rotation method in X-ray crystallography [46] so MicroED data can be indexed, integrated and merged using modern X-ray crystallography software, such as iMOSFLM, DIALS, and XDS [47–49]. In this way, MicroED continuous rotation data was recorded, and processed in iMOSFLM to yield a fully refined model of lysozyme at 2.5 Å resolution from a single nanocrystal [10] and later to 1.5 Å resolution [50]. Apart from the first proof of principle work that was done with diffraction stills [7••] all subsequent studies use continuous rotation exclusively. Continuous rotation is the standard data collection method in current MicroED experiments.
As the first reported electron diffraction from three-dimensional crystals were from bovine liver catalase back in 1930 [51], for historical reasons this protein was chosen as the next structure studied by MicroED [11•]. Catalase is much larger than lysozyme (240 kDa compared with 14 kDa, respectively), has lower symmetry and adopts a preferred orientation on cryoEM grids. Nevertheless, a successful MicroED structure solution was found from a single nanocrystal only ~8 protein layers thick [52]. This was the second time that a single nano-crystal was sufficient for structure solution by MicroED. This structure showed difference density in the binding site of the NADP when it was not present in the replacement model, and the suggested conformational rotation of F197 upon protonation of NADP to NADPH. This study demonstrated that MicroED could be used to deduce new information from previously solved structures. Catalase was finally solved using electron diffraction nearly 100 years after the first electron diffraction pattern had been published.
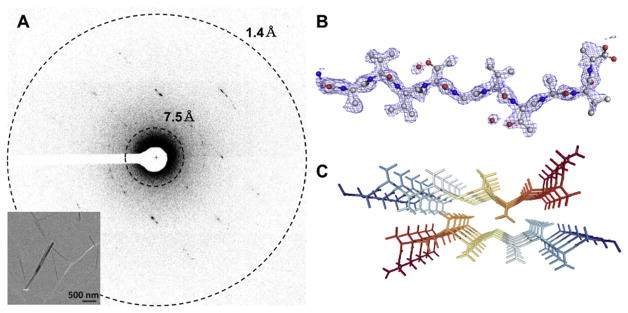
The first novel structure solved by MicroED was that of the toxic core of α-synuclein published in 2015 [53•]. α-Synuclein is the protein responsible for Parkinson’s disease [54]. α-Synuclein is a small protein and at its core lies a stretch of 10 amino acids that are both fibrillar and confer toxicity named the NACore. This fragment and its analogous non-cytotoxic form were the first novel structures to be solved by MicroED [53•]. NACore crystals were so small that they were dubbed “invisible” since they were smaller than the wavelength of visible light at only ~50 nm wide and thick (Fig. 5). These crystals were far too small for microfocus X-ray crystallography and were also not suitable for analysis by X-ray Free Electron Lasers (XFELS) although they were tested extensively. MicroED delivered structures of both oligopeptides at 1.4 Å resolution, marking a new record for the highest resolution protein structures solved using any form of cryoEM at the time. Furthermore, these structures explained why the crystals were unable to grow any larger and had been so evasive: the twist in the beta sheet stacking of these peptides disallowed long range growth along the c* direction of the lattice. This opened the door for other structures known to form beta-sheet aggregates, but were unable to create crystals sufficiently large for X-ray sources, to be solved using MicroED.
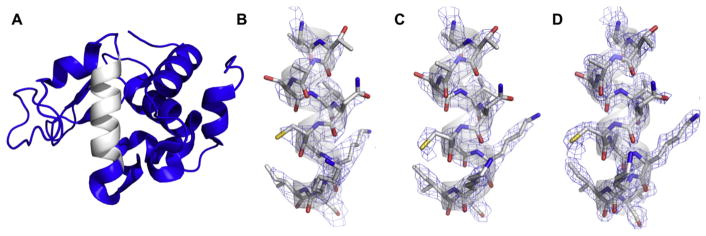
Fig. 5.
(A) MicroED pattern from nanocrystals of the NACore of α-synuclein, inset with low-dose micrograph of typical crystals; adapted from [49]. (B) The structure solution of the asymmetric unit, and (C) arrangement of the biological assembly.
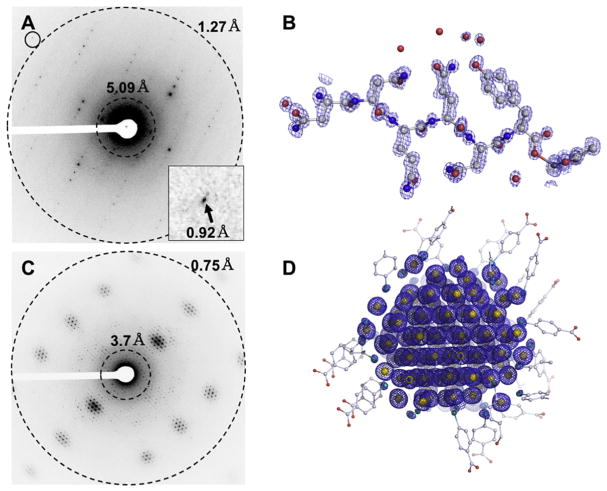
The first protein structures solved by MicroED at atomic resolution by ab initio direct methods were published in 2016 [55]. Up until that stage, all structures determined by MicroED were phased by molecular replacement. The protein fragments of the amyloid core of the Sup35 prion protein yielded MicroED data to ~1.1 Å resolution, and allowed the structures to be solved by ab initio direct methods [56]. This was the first demonstration that the intensities in MicroED experiments are accurate and maintain the relationship between phase and amplitude when the resolution extends beyond 1.2 Å – (the same resolution requirement as with X-ray crystallography for ab initio methods) [56]. These experiments also demonstrated that dynamical scattering did not affect the experimental phasing indicating that MicroED is a viable method for structure determination. In fact, direct methods have recently been used to solve a material hydrated gold nanoparticle novel structure in a frozen-hydrated state using MicroED to 0.8 Å resolution [57••] (Fig. 6).
Fig. 6.
(A) Sub-atomic resolution MicroED data breaking the 1 Å barrier from crystals of a segment of the Sup35 prior protein with (B) corresponding ab initio direct methods solution to the structure. (C) Diffraction from Au146 crystals extending beyond 0.8 Å, and (D) ab initio direct method solutions showing clear density for all gold and sulfur atoms. Figures adapted from [51,53•].
4. Data collection and processing in electron crystallography of 2D crystals
Traditional data collection in electron crystallography is typically a two-step process: collection of crystal images and collection of diffraction. Images contain information regarding both the phase and amplitude of the 2D protein crystal [15]. Amplitudes and phases may be extracted from the Fourier transform of the crystal image after motion and drift correction [58]. Structures can be entirely determined by images alone in this way. However, reliable high resolution imaging of 2D crystals remains difficult mainly because of charging and motion of the sample. Images provide accurate and high quality phase information but rather inaccurate amplitudes. The measured amplitudes are modulated by the contrast transfer function; affected by the microscope’s optical properties, and charging of the specimen during exposure causing movement and interference, ultimately lowering the signal to noise ratio [59]. The second step is typically the collection of diffraction data from 2D crystals. High resolution diffraction data is much easier to obtain than a high-resolution image, as the signal is translationally invariant and is not affected by sample charging or drift. Measured intensities from electron diffraction therefore tend to be much more accurate than those obtained by high resolution imaging.
Samples in electron crystallography must be truly two-dimensional, ideally only a single protein layer thick, and they must lay flat on the cryo grids. Data collection is laborious and time consuming and could benefit from automation. Each 2D crystal typically lasts for a single diffraction image. As a result, data collection typically involves the collection of 1000 diffraction images from 1000 different 2D crystals. Each crystal is oriented differently on the grid and one collects different tilt angles (0–70°). Each pattern is indexed and the data merged together. Assumptions are made regarding z* and the reflections interpolated using a cardinal sin function (sinc). In such a way, the reciprocal space is built up to and including 95% completeness if the sample is tilted to ±70° [60].
Data in 2D electron crystallography is typically processed in the MRC suite of software [61] and more recently images of 2D crystals can be processed by the GUI 2dx [62]. While many packages exist, the overall processing pipeline has remained relatively unchanged since the beginning of electron crystallography. Images or dose fractionated movies are recorded and corrected for distortions and drift [63], corrected for the contrast transfer function [64], and phases are extracted from the correct image Fourier transforms [14]. Intensities are recorded from diffraction patterns by summing the spot intensities and subtracting the local background. The phases and amplitudes for images at all tilt angles are used to model the crystal lattice lines that are contiguous along z*. This phase and amplitude set are inverse Fourier transformed to give a density map that has a model built into it. The model can be refined using standard X-ray crystallography refinement pipelines. New software has utilized maximum likelihood methods for reducing bias and making the most of lower resolution data [65].
Although most structures 2D electron crystallography have made use of the phase information from images, structures such as AQP0 [32•] and AQP4 [31] were solved using only diffraction data with the phases being solved by molecular replacement. Recent work also demonstrated that phases originating from images can be extended to the higher resolution of the electron diffraction [36].
5. Data collection and processing in MicroED
The largest difference in data collection between MicroED and 2D electron crystallography is that entire data sets can be collected in MicroED from a single nano-crystal [10,11•]. Crystals are typically less than ~0.5 micrometer thick, consisting of many protein layers, to maximize signal to noise while minimizing absorption artifacts. A single 3D nanocrystal in MicroED is continuously diffracted while it is rotating slowly, uni-directionally in the beam while diffraction data is collected on a fast camera as a movie [12]. This is referred to as continuous rotation MicroED [10]. Depending on the symmetry of the crystal, a single nanocrystal can yield >90% completeness allowing structure solution entirely on its own. MicroED data is similar to X-ray rotation data and therefore is processed exclusively using X-ray software [9••,12].
Indexing poses a unique challenge in electron diffraction experiments. An electron accelerated through a voltage of 200 kV has a wavelength of ~0.0251 Å, whereas a typical X-ray wavelength is ~1 Å [66]. The Ewald sphere in X-ray diffraction is curved sufficiently to allow indexing from one or two frames given good data [46]. The Ewald sphere in a MicroED experiment is essentially flat compared to X-ray experiments. Spots appear in rows and columns with individual spots flashing in and out of the diffraction condition along those rows and columns during the experiment. An individual frame can be visually inspected to obtain two cell axes relative to one another by the distance between spots in adjacent rows and columns, but the third dimension requires knowledge of when a spot reappears after some degrees of rotation. This leads to MicroED experiments only being indexed by using a suitably large wedge of data, typically ~20° of Phi is sufficient for accurate indexing [12]. Thus, far MicroED data has been processed in MOSFLM [47], DIALS [49], XDS [48], and SHELX [67]. Detailed protocols have been written recently outlining the data collection [9••] processing [12] in MicroED. MicroED data has been recorded on CMOS cameras such as oneview (Gatan), ceta (FEI) and F416 (TVIPS) as well as direct electron detectors such as the falcon (FEI), k2 (Gatan), Timepix, and Direct Electron.
6. Future directions in MicroED and electron crystallography
Electron crystallography is a well-developed technique with a rich history. However, there are still many possibilities for further advancement. The more obvious being automation and the use of newer hardware such as direct electron detectors and microscopes with advanced optics. Advances in hardware and software are likely to benefit both electron crystallography and MicroED. The potential future of electron crystallography likely lies in the ability to study membrane proteins in differing lipid environments, ultimately gleaming new, physiologically relevant information with greater context [68].
MicroED is a new and exciting cryoEM method that has shown continued growth in solved structures year over year. MicroED experiments have typically been solved by molecular replacement and by ab initio phase retrieval. Molecular replacement is the most common method for solving the phase problem in crystallography [69], but requires the use of a search model with at least 30% sequence homology [35]. Ab initio methods are useful only when the diffraction extends to at least 1.2 Å resolution with high completeness and low noise [70]. Solutions of protein structures from ab initio methods are rare, as large globular or membrane proteins contain large regions of high flexibility and therefore low crystallinity, limiting the maximum resolution [71]. A robust, accurate method of phase retrieval for MicroED experiments represents the next phase of method development. This was recognized in X-ray crystallography by anomalous scattering wherein intensity mismatch between Friedel mates occurs due to elemental absorption edges of heavy atoms [72,73]. No similar absorption edges are known for any element at the wavelengths typical in electron diffraction so such anomalous methods are impractical in MicroED. A promising avenue is isomorphous replacement which requires significant exploration. Imaging 3D crystals is also a possibility. As with 2D crystals, images will carry accurate phase information and those low-resolution phases could be used for phase extension protocols [36].
Another emergent method to study nanocrystals are X-ray free electron lasers (XFELs) [74•]. These use very short, high intensity bursts of X-rays shot at a large number of nanocrystals to produce still diffraction patterns prior to obliteration of the crystal [75]. Each crystal provides only a single diffraction pattern and a million diffraction patters originating from a million crystals are merged together to yield complete data sets. The access to XFELs is rather limited but more importantly very large quantities of crystals are required and this can be quite prohibitive.
The true promise of MicroED comes from the use of protein nanocrystals to inform about the charged states in molecules. The structures studied by MicroED are built into maps of electrostatic potential, rather than electron density maps as with X-ray crystallography. This is because electrons are not scattered by the electrons and nuclei in the protein themselves, but by their summed interactions with the surrounding environment. With adequate modeling of scattering and accurate data, MicroED can begin to tell us where charges are moving in structures with different drugs or ligands bound, and go from a static picture of structural biology to a more nuanced understanding of biological function at the atomic level.
Acknowledgments
The Gonen laboratory is supported by the Howard Hughes Medical Institute.
References and recommended reading
• Of special interest.
•• Of outstanding interest.
- 1.Zeitler E. Cryo electron microscopy. Ultramicroscopy. 1982;10:1–5. doi: 10.1016/0304-3991(82)90181-4. [DOI] [PubMed] [Google Scholar]
- 2.Doerr A. Cryo-electron tomography. Nat Methods. 2017;14:34. doi: 10.1038/nmeth.4115. [DOI] [Google Scholar]
- 3.Cheng Y, Grigorieff N, Penczek PA, Walz T. A primer to single-particle cryo-electron microscopy. Cell. 2015;161:438–49. doi: 10.1016/j.cell.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell. 2015;161:450–7. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaeser RM. Review: electron crystallography: present excitement, a nod to the past, anticipating the future. J Struct Biol. 1999;128:3–14. doi: 10.1006/jsbi.1999.4172. [DOI] [PubMed] [Google Scholar]
- 7••.Shi D, Nannenga BL, Iadanza MG, Gonen T. Three-dimensional electron crystallography of protein microcrystals. Elife. 2013;2:e01345. doi: 10.7554/eLife.01345. This is the proof of principle paper outlining the principles of MicroED and showing that protein structure can be determined by electron diffraction. Before this paper it was widely believed that such an approach was not possible. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renault L, Chou HTT, Chiu PLL, Hill RMM, Zeng X, Gipson B, et al. Milestones in electron crystallography. J Comput Aided Mol Des. 2006;20:519–27. doi: 10.1007/s10822-006-9075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Shi D, Nannenga BL, de la Cruz MJ, Liu J, Sawtelle S, Calero G, et al. The collection of MicroED data for macromolecular crystallography. Nat Protoc. 2016;11:895–904. doi: 10.1038/nprot.2016.046. This is a protocol that describes all the steps in a typical MicroED experiment. The protocol begins with crystal detection and continues to cryoEM preparation, microscope setup and data collection. It also briefly talks about data processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nannenga BL, Shi D, Leslie AGW, Gonen T. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat Methods. 2014;11:927–30. doi: 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Nannenga BL, Shi D, Hattne J, Reyes FE, Gonen T. Structure of catalase determined by MicroED. Elife. 2014;3:e03600. doi: 10.7554/eLife.03600. This paper illustrates the wide applicability of MicroED as a method for determining protein structure. Catalase was the second protein determined by this method and is so far the largest at ~240 kDa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattne J, Reyes FE, Nannenga BL, Shi D, De La Cruz MJ, Leslie AGW, et al. MicroED data collection and processing. Acta Crystallogr A Found Adv. 2015;71:353–60. doi: 10.1107/S2053273315010669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons DF, Martius U. Determination of the α-helix configuration of poly-γ-benzyl-L-glutamate by electron diffraction. J Mol Biol. 1964;10:IN4–533. doi: 10.1016/S0022-2836(64)80071-1. [DOI] [PubMed] [Google Scholar]
- 14.Klug A, Berger JE. An optical method for the analysis of periodicities in electron micrographs, and some observations on the mechanism of negative staining. J Mol Biol. 1964;10:565-IN15. doi: 10.1016/S0022-2836(64)80081-4. [DOI] [PubMed] [Google Scholar]
- 15.De Rosier DJ, Klug A. Reconstruction of three dimensional structures from electron micrographs. Nature. 1968;217:130–4. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- 16.Hoppe W, Langer R, Knesch G, Poppe C. Protein-Kristallstrukturanalyse mit Elektronenstrahlen. Naturwissenschaften. 1968;65:333–6. doi: 10.1007/BF00600449. [DOI] [PubMed] [Google Scholar]
- 17.Glaeser RM, Thomas G. Application of electron diffraction to biological electron microscopy. Biophys J. 1969;9:1073–99. doi: 10.1016/S0006-3495(69)86437-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labaw LW, Rossmann MG. Electron microscopic observations of L-lactate dehydrogenase crystals. J Ultrastruct Res. 1969;27:105–17. doi: 10.1016/S0022-5320(69)90022-7. [DOI] [PubMed] [Google Scholar]
- 19.Matricardi VR, Moretz RC, Parsons DF. Electron diffraction of wet proteins: catalase. Science. 1972;177:268–70. doi: 10.1126/science.177.4045.268. [DOI] [PubMed] [Google Scholar]
- 20.Taylor KA, Glaeser RM. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186:1036–7. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 21.Henderson R, Unwin PNT. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 22••.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. High resolution structure of the membrane protein bacteriorhodopsin determined by electron crystallography of two-dimensional crystals. The crystals were isolated from native membranes. [DOI] [PubMed] [Google Scholar]
- 23.Kühlbrandt W, Wang DN, Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994;367:614–21. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 24.Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, et al. The structure of bacteriorhodopsin at 3. 0 Å resolution based on electron crystallography: implication of the charge distribution. J Mol Biol. 1999;286:861–82. doi: 10.1006/jmbi.1998.2529. [DOI] [PubMed] [Google Scholar]
- 25.Nogales E, Grayer Wolf S, Khan IA, Ludueña RF, Downing K, et al. Structure of tubulin at 6. 5 Å and location of the taxol-binding site. Nature. 1995;375:424–7. doi: 10.1038/375424a0. [DOI] [PubMed] [Google Scholar]
- 26.Walz T, Hirai T, Murata K, Heymann JB, Mitsuoka K, Fujiyoshi Y, et al. The three-dimensional structure of aquaporin-1. Nature. 1997;387:624–7. doi: 10.1038/42512. [DOI] [PubMed] [Google Scholar]
- 27.Dubochet J, Lepault J, Freeman R, Berriman JA, Homo J-C. Electron microscopy of frozen water and aqueous solutions. J Microsc. 1982;128:219–37. doi: 10.1111/j.1365-2818.1982.tb04625.x. [DOI] [Google Scholar]
- 28.Lepault J, Booy FP, Dubochet J. Electron microscopy of frozen biological suspensions. J Microsc. 1983;129:89–102. doi: 10.1111/j.1365-2818.1983.tb04163.x. [DOI] [PubMed] [Google Scholar]
- 29.Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall AW, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129. doi: 10.1017/S0033583500004297. [DOI] [PubMed] [Google Scholar]
- 30.Gonen T, Sliz P, Kistler J, Cheng Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004;429:193–7. doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- 31.Hiroaki Y, Tani K, Kamegawa A, Gyobu N, Nishikawa K, Suzuki H, et al. Implications of the aquaporin-4 structure on array formation and cell adhesion. J Mol Biol. 2006;355:628–39. doi: 10.1016/j.jmb.2005.10.081. [DOI] [PubMed] [Google Scholar]
- 32•.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, et al. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–8. doi: 10.1038/nature04775. This was the first demonstration that cryoEM can reach atomic resolution. The structure of aquaporin-0 at 1.9 Å resolution was the first to display atomicity, the first to identify water molecules and a eukaryotic membrane by cryoEM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Wisedchaisri G, Gonen T. Phasing electron diffraction data by molecular replacement: strategy for structure determination and refinement. Methods Mol Biol. 2013;955:243–72. doi: 10.1007/978-1-62703-176-9_14. This paper describes in detail the steps necessary to perform molecular replacement in an electron crystallography experiment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nannenga BL, Iadanza MG, Vollmar BS, Gonen T. Current protocols in protein science. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2013. Overview of electron crystallography of membrane proteins: crystallization and screening strategies using negative stain electron microscopy; pp. 17.15.1–17.15.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wisedchaisri G, Gonen T. Fragment-based phase extension for three-dimensional structure determination of membrane proteins by electron crystallography. J Struct Des. 2011;19:976–87. doi: 10.1016/j.str.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian G, Basu S, Liu H, Zuo JM, Spence JCH. Solving protein nanocrystals by cryo-EM diffraction: multiple scattering artifacts. Ultramicroscopy. 2015;148:87–93. doi: 10.1016/j.ultramic.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 38.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 39.Glaeser RM, Ceska TA. High-voltage electron diffraction from bacteriorhodopsin (purple membrane) is measurably dynamical. Acta Crystallogr A Found Crystallogr. 1989;45:620–8. doi: 10.1107/S0108767389004599. [DOI] [PubMed] [Google Scholar]
- 40.Unwin PNT, Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975;94:425–40. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- 41.John P. Characterization of crystalline filtrate tetanus toxin. 1982;293:285–93. doi: 10.1016/s0022-5320(82)90004-1. [DOI] [PubMed] [Google Scholar]
- 42.Jeng TW, Chiu W. Low dose electron microscopy of the crotoxin complex thin crystal. J Mol Biol. 1983;164:329–46. doi: 10.1016/0022-2836(83)90080-3. [DOI] [PubMed] [Google Scholar]
- 43.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2. 6 Å resolution. Nature. 2000;405:647–55. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 44.Oleynikov P, Hovmöller S, Zou XD. Precession electron diffraction: observed and calculated intensities. Ultramicroscopy. 2007;107:523–33. doi: 10.1016/j.ultramic.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 45.Blackman M. On the intensities of electron diffraction rings. Proc R Soc Lond. 1939:68–82. Series A. [Google Scholar]
- 46.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J Appl Cryst. 1988;21:916–24. doi: 10.1107/S0021889888007903. [DOI] [Google Scholar]
- 47.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–81. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkhurst JM, Winter G, Waterman DG, Fuentes-Montero L, Gildea RJ, Murshudov GN, et al. Robust background modelling in DIALS. J Appl Crystallogr. 2016;49:1–10. doi: 10.1107/S1600576716013595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat Methods. 2017:1–6. doi: 10.1038/nmeth.4178. [DOI] [PMC free article] [PubMed]
- 51.Sumner JB, Dounce AL. Crystalline catalase. Science. 1937;85:366–7. doi: 10.1126/science.85.2206.366. [DOI] [PubMed] [Google Scholar]
- 52.Eventoff W, Tanaka N, Rossmann MG. Crystalline bovine liver catalase. J Mol Biol. 1976;103:799–801. doi: 10.1016/0022-2836(76)90210-2. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez JA, Ivanova MI, Sawaya MR, Cascio D, Reyes FE, Shi D, et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015:486–90. doi: 10.1038/nature15368. • [advance on] This was the first novel structure solved by MicroED. This toxic core resisted structure determination by all other methods including X-FELs but MicroED was successful in delivering a structure at 1.4Å resolution. [DOI] [PMC free article] [PubMed]
- 54.Spillantini MG, Schmidt ML, Lee VM-Y, Trojanowski JQ, Jakes R, Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–40. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 55.Sawaya MR, Rodriguez J, Cascio D, Collazo MJ, Shi D, Reyes FE, et al. Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1606287113. 201606287. [DOI] [PMC free article] [PubMed]
- 56.Sheldrick GM. A short history of SHELX. Acta Crystallogr A Found Crystallogr. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 57••.Vergara S, Lukes DA, Martynowycz MW, Santiago U, Plascencia-Villa G, Weiss SC, et al. MicroED structure of Au146(p-MBA)57 at subatomic resolution reveals a twinned FCC cluster. J Phys Chem Lett. 2017;146:5523–30. doi: 10.1021/acs.jpclett.7b02621. This paper describes the sub-atomic resolution structure of a material: hydrated gold cage that was determined by MicroED and corroborated by X-ray crystallography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klug A, Crowther RA. Three-dimensional image reconstruction from the viewpoint of information theory. Nature. 1972;238:435–40. doi: 10.1038/238435a0. [DOI] [Google Scholar]
- 59.Wade RH. A brief look at imaging and contrast transfer. Ultramicroscopy. 1992;46:145–56. doi: 10.1016/0304-3991(92)90011-8. [DOI] [Google Scholar]
- 60.Dorset DL. Filling the missing cone in protein electron crystallography. Microsc Res Tech. 1999;46:98–103. doi: 10.1002/(SICI)1097-0029(19990715)46:2<98::AID-JEMT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 61.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 62.Gipson B, Zeng X, Zhang ZY, Stahlberg H. 2dx-User-friendly image processing for 2D crystals. J Struct Biol. 2007;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 63.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–2. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Biyani N, Righetto RD, McLeod R, Caujolle-Bert D, Castano-Diez D, Goldie KN, et al. Focus: the interface between data collection and data processing in cryo-EM. 2017 doi: 10.1016/j.jsb.2017.03.007. bioRxiv. [DOI] [PubMed]
- 66.Warren BE. X-ray diffraction methods. J Appl Phys. 1941;12:375–84. doi: 10.1063/1.1712915. [DOI] [Google Scholar]
- 67.Sheldrick GM. Crystal structure refinement with SHELXL. Acta Crystallogr C. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fujiyoshi Y. Electron crystallography of soluble and membrane proteins. Totowa, NJ: Humana Press; 2013. Future directions of electron crystallography; pp. 551–68. [DOI] [PubMed] [Google Scholar]
- 69.Scapin G. Molecular replacement then and now. Acta Crystallogr D Biol Crystallogr. 2013;69:2266–75. doi: 10.1107/S0907444913011426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheldrick GM. SHELXT - integrated space-group and crystal-structure determination. Acta Crystallogr A Found Crystallogr. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–84. doi: 10.1016/S0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 72.Karle J. Some developments in anomalous dispersion for the structural investigation of macromolecular systems in biology. Int J Quantum Chem. 2009;18:357–67. doi: 10.1002/qua.560180734. [DOI] [Google Scholar]
- 73.Dauter Z. New approaches to high-throughput phasing. Curr Opin Struct Biol. 2002;12:674–8. doi: 10.1016/S0959-440X(02)00372-X. [DOI] [PubMed] [Google Scholar]
- 74•.Schlichting I. Serial femtosecond crystallography: the first five years. IUCrJ. 2015;2:246–55. doi: 10.1107/S205225251402702X. This paper highlights the recent advances and history of serial femptosecond crystallography at X-ray free electron laser sources. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lomb L, Barends TRM, Kassemeyer S, Aquila A, Epp SW, Erk B, et al. Radiation damage in protein serial femtosecond crystallography using an x-ray free-electron laser. Phys Rev B Condens Matter Mater Phys. 2011:84. doi: 10.1103/PhysRevB.84.214111. [DOI] [PMC free article] [PubMed]