Abstract
It was demonstrated in previous studies that cysteine-rich angiogenic inducer 61 (Cyr61) plays vital roles in hematological disorders, and we have already reported that the Cyr61 protein is a tumor promoter in acute myeloid leukemia (AML). Here, we investigated the association between CYR61 gene polymorphisms and susceptibility to AML.
We genotyped 2 single-nucleotide polymorphisms (rs2297141 and rs6576776) in the region of the CYR61 gene by improved multiplex ligase detection reaction genotyping assays in a total of 275 samples, including samples from 137 AML patients and 138 healthy controls. Chi-squared tests and logistic regression analysis were performed to compare the different distributions of the genotypes and alleles between patients and healthy controls.
The rs2297141 A allele was associated with lower risk of AML compared with the G allele (odds ratio [OR] = 0.704, 95% confidence interval [CI] = 0.503–0.985, P = .04) in both the dominant (OR = 0.447, 95% CI = 0.22–0.909, P = .025, AA vs GG) and recessive inheritance models (OR = 0.419, 95% CI = 0.23–0.763, P = .004, AA vs GA + GG). Although the distribution of the rs6576776 alleles was not different between patients with AML and normal controls, the CC genotype significantly increased the risk of AML in the dominant inheritance model (OR = 6.064, 95% CI = 1.303–28.216, P = .01, CC vs GG) and the recessive inheritance model (OR = 5.937, 95% CI = 1.291–27.306, P = .01, CC vs GC + GG). Additionally, it was shown that the rs2297141 and rs6576776 genotypes were associated with AML-M5 and AML-M2, respectively.
Our findings indicated that genetic polymorphisms in the CYR61 gene may be considered potential AML risk factors in the Han Chinese population.
Keywords: AML, Cyr61, polymorphism
1. Introduction
Acute myeloid leukemia (AML) is a type of hematopoietic stem cell tumor. Genomic changes play vital roles in AML; mutations in AML inform the disease classification and prognostic stratification,[1,2] and gene analysis has significantly improved the diagnostic criteria and the classification of myeloid neoplasms (World Health Organization [WHO] 2016 edition).[3]
Cysteine-rich 61 (Cyr61), is a member of the connective tissue growth factor, cysteine-rich protein, which consists of 6 members. Cyr61 has diverse functions in several types of tumors, for example, cervical cancer,[4] breast cancer,[5] pancreatic cancer,[6] malignant melanoma,[7] esophageal squamous cell carcinoma,[8] and hepatocarcinogenesis.[9] Moreover, in recent years, there has been a focus on the association between polymorphisms of the Cyr61 gene and diseases; the Cyr61 rs12756618 variant increases the risk of Graves’ ophthalmopathy,[10] the Cyr61 p.R47W variant is related to atrial septal defect,[11] and the Cyr61 rs3753793 variant might contribute to prostate cancer risk.[12] In our previous study, we reported that Cyr61 is overexpressed in AML cell lines and patient samples and is a tumor promoter in AML through the MEK/ERK pathway.[13]
The results from ANLN clinical and RNA-seq data showed that there were diverse CYR61 expression patterns in different AML subtypes, and the expression of CYR61 was associated with FTL3 mutation, PML/RAR-fusion, or RAS activation (Fig. 1). In this study, to further understand the roles played by Cyr61 in AML, we analyzed the distributions of Cyr61 polymorphism in patients with AML and healthy controls to explore the relationship between Cyr61 polymorphisms and the risk of AML. It has been demonstrated that CYR61 polymorphism rs3753793 was related to Graves’ ophthalmopathy,[10] prostate cancer risk,[12] and HDL-cholesterol levels in obese individuals.[14] And, the data from population of CHB and CHS in 1000 genome database[15] show strong linkage relationship among single-nucleotide polymorphisms (SNPs) rs2297141, rs6576776, and rs3753793 in CYR61 gene through Haploview analysis. Therefore, theses SNPs were chosen for current investigation.
Figure 1.
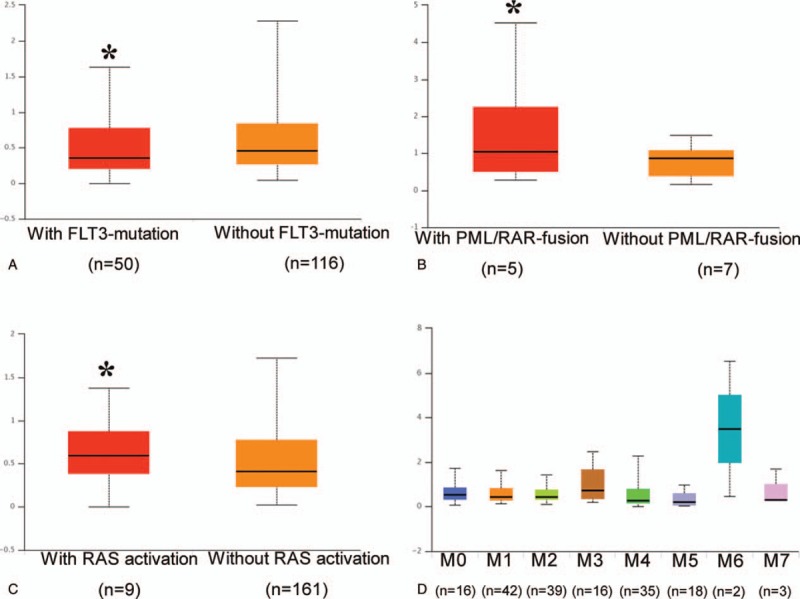
Comparison of CYR61 RNA-seq expression level based on FLT3 mutation status (A), PML/RAR-fusion status (B), RAS activation status (C), and French–American–British classification (D) in the Cancer Genome Atlas samples. ∗P < .05.
2. Methods
2.1. Patient cohorts and healthy controls
Bone marrow samples from AML patients (n = 137) and peripheral blood samples of healthy controls (n = 138) were obtained from Chongqing General Hospital and Chongqing Xinqiao Hospital from November 2016 to September 2017. All participants were of Chinese Han ethnicity. The patients were diagnosed according to the French–American–British (FAB) and WHO classifications, and the patients who received hematopoietic stem cell transplantations were excluded. Healthy subjects who had no self-reported cancer history or known blood disorders were enrolled as the controls. The patients with AML and the healthy donors provided informed consent, and the study was approved by the Ethics Committee of Chongqing General Hospital. In addition, the authors did not have access to information that could identify individual participants during or after data collection.
2.2. Molecular genetic analysis
Genomic DNA was extracted with the Tiangen kit (Beijing, China) according to the manufacturer's instructions. Samples were stored at −80°C. The CYR61 genotypes were determined by improved multiplex ligase detection reaction, which was developed by Genesky Biotechnologies Inc. (Shanghai, China) as previously reported.[16] The sequences of the probes are as follows:
rs2297141:
5′-TTCCGCGTTCGGACTGATATGCAGCCTTCCGAGGTGGACG-3′, 5′- TACGGTTATTCGGGCTCCTGTGCAGCCTTCCGAGGTGGACA-3′, 5′-GGCTGGACGAGATCAGAGGCTTTTTTT-3′;
rs6576776:
5′- TTCCGCGTTCGGACTGATATGCAAAGGAATGCAAGGAATTTCTAG -3′, 5′-TACGGTTATTCGGGCTCCTGTGCAAAGGAATGCAAGGAATTTCTAC-3′, 5′-TACTTTTCCAATAGCGTGAGGGCTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′.
The mutations in AML genes (c-kit, FLT3-TKD, CEBPA, NPM1, DNMT3A, U2AF1, SRSF2, ASXL1), which play crucial roles in the diagnosis, stratification, and treatment of AML,[2,3,17,18] were analyzed with Sanger sequencing: DNA was amplified for the genes (c-kit, FLT3-TKD, CEBPA, NPM1, DNMT3A, U2AF1, SRSF2, ASXL1) with S1000 Thermal Cycler, then amplified products were analyzed with 3730xl DNA Analyzer (Thermo Fisher Scientific, Waltham).
2.3. Statistics
ANLN clinical and RNA-seq data were downloaded from the Cancer Genome Atlas (https://portal.gdc.cancer.gov/) and analyzed by using the online software UALCAN (http://ualcan.path.uab.edu/analysis.html) following their policies. Transcripts per million expression values employed for the generation of box plots were also used to estimate the significance of difference in gene expression levels between groups. The Student t test was performed using a PERL script with Comprehensive Perl Archive Network module.
SPSS 20.0 software was used for the statistical analysis. The quality control was assessed with Hardy–Weinberg equilibrium by the differences in the distributions of the patients and the healthy donors. Chi-squared tests or/and Fischer exact tests were used to analyze the genotype and allele frequency data. The association between CYR61 gene polymorphisms and AML risk was evaluated by odds ratio (OR) with 95% confidence intervals (CIs). Logistic regression analysis was used to analyze the independent factors. P < .05 indicated a statistically significant result.
3. Results
3.1. The information of AML patients and healthy controls
There were no statistically significant differences between the groups with regard to gender (77/60 vs 76/62, P = .850) or age (mean 39.4 ± 15.6, 37.8 ± 17.2, P = .445) in the total 275 samples (137 AML patients and 138 healthy controls) (Table 1). The distribution of the AML subtypes based on the FAB classification was as follows: AML-M1, 9 (6.6%); AML-M2, 44 (32.1%); AML-M3, 10 (7.3%); AML-M4, 35 (25.6%); AML-M5, 29 (21.2%); AML-M6, 4 (2.9%); and AML-other type, 6 (4.4%). We analyzing the AML 8 gene mutations of AML samples (from 137 AML patients) with Sanger sequencing, and the results showed that the distribution of the AML 8 genes mutations were as follows: c-Kit, 6 (4.4%); FLT3-TKD, 16 (11.7%); CEBPA, 10 (7.3%); NPM1, 8 (5.8%); DNMT3A, 5 (3.7%); U2AF1, 1 (0.7%); SRSF2, 2 (1.5%); and ASXL1, 2 (1.5%).
Table 1.
Demographics and clinical characteristics of the patients with AML and the healthy controls.

3.2. The distribution of CYR61 gene polymorphisms in the patients with AML
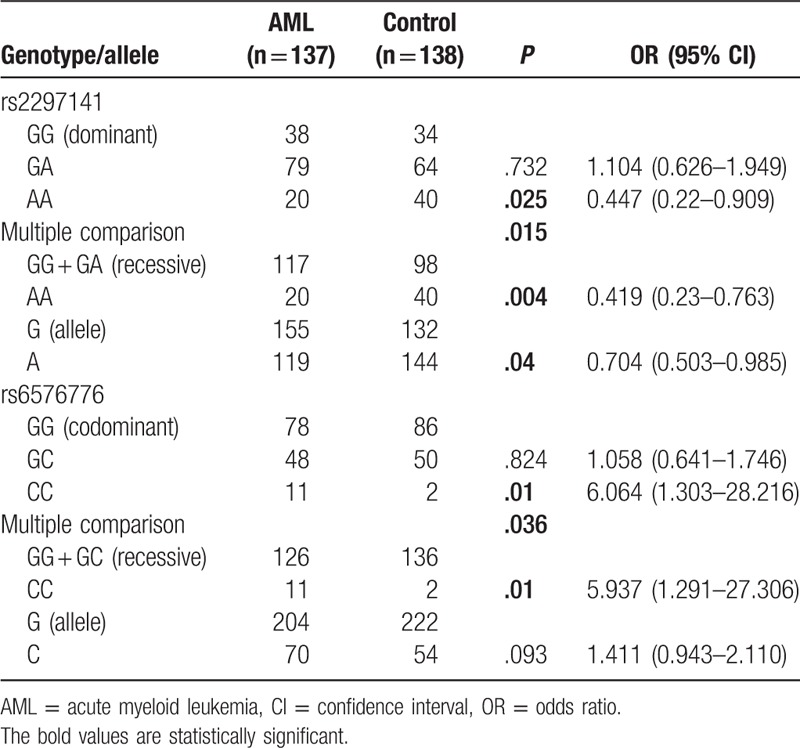
As shown in Table 2, the rs2297141 A allele significantly decreased the risk of AML compared with the G allele (OR = 0.704, 95% CI = 0.503–0.985, P = .04) in both the dominant (OR = 0.447, 95% CI = 0.22–0.909, P = .025, AA vs GG) and recessive inheritance models (OR = 0.419, 95% CI = 0.23–0.763, P = .004, AA vs GA + GG). Although the G allele of rs6576776 was more frequent in AML cases (25.5%) than in controls (19.6%), it was not a statistically significant difference (P = .093). The genotype CC of rs6576776 significantly increased the risk of AML, whether in the dominant (OR = 6.064, 95% CI = 1.303–28.216, P = .01, CC vs GG) or the recessive inheritance model (OR = 5.937, 95% CI = 1.291–27.306, P = .01, CC vs GC + GG).
Table 2.
The allelic and genotypic frequencies of CYR61 polymorphisms in AML patients and controls.
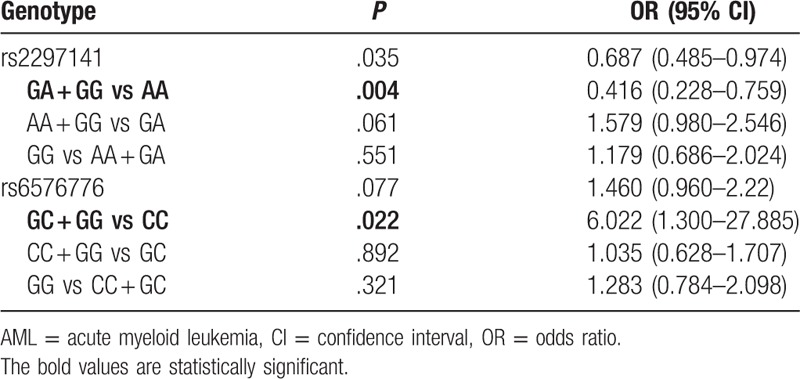
Furthermore, logistic regression analyses adjusted for gender and age were used to analyze the association of 2 CYR61 polymorphisms with the risk of AML. As shown in Table 3, the CC genotype of rs6576776 in patients with AML was associated with a higher risk of AML, but the genotype AA of rs2297141 in patients with AML was associated with a lower risk of AML.
Table 3.
Logistic regression analysis for the association of rs2297141 and rs6576776 polymorphisms with AML.
3.3. Patient characteristics in relation to CYR61 polymorphisms
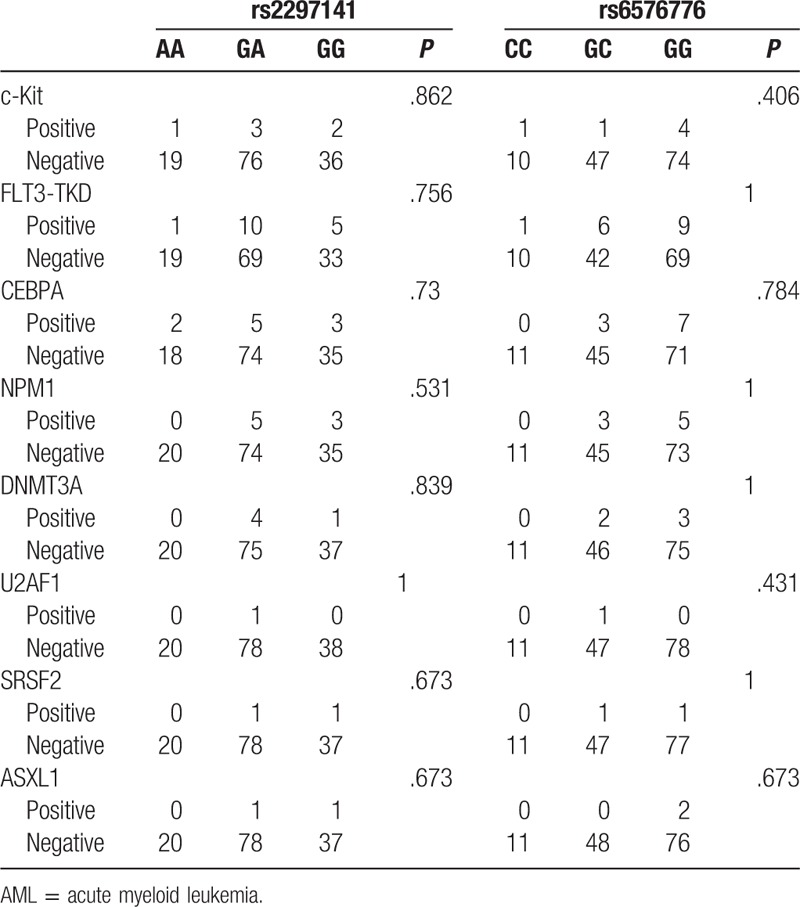
Eight AML gene mutations were analyzed in the patients with AML, and the mutations were found in 39 patients. We also analyzed the relationship of rs2297141 and rs6576776 with the mutation of 8 AML genes, and the data showed that there were no associations (Table 4).
Table 4.
The relationship of rs2297141 and rs6576776 with the mutations of 8 AML genes.
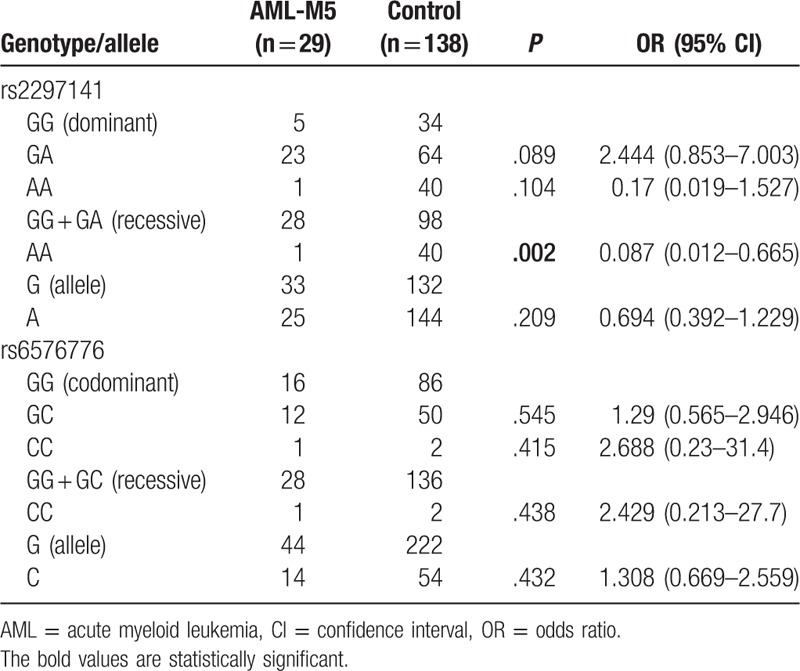
The association of Cyr61 polymorphisms with the AML FAB subtypes was also analyzed. Chi-squared tests showed that the rs2297141 genotype was associated with AML-M5 (P = .001), and the rs6576776 genotype was associated with AML-M2 (P = .005) (Table 5), compared with the healthy control. Furthermore, the dominant inheritance models, recessive inheritance models, and alleles were analyzed in the AML-M2 and AML-M5 subtypes. In addition, the results (Tables 6 and 7) showed that compared with the control, there was a lower risk of AML-M5 with the AA genotype (OR = 0.087, 95% CI = 0.012–0.665, P = .002, AA vs GG + GA) and a higher risk of AML-M2 with the CC genotype (OR = 12.286, 95% CI = 2.313–65.25, P = .002, CC vs GG; OR = 10.737, 95% CI = 2.082–55.36, P = .003, CC vs GG + GC; OR = 2.021, 95% CI = 1.184–3.4, P = .013, C vs G).
Table 5.
Chi-squared tests for the association of CYR61 polymorphisms with the AML FAB subtypes.
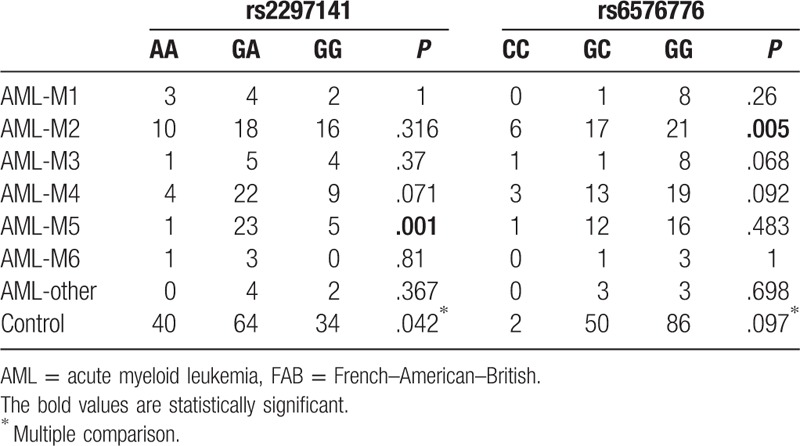
Table 6.
The dominant inheritance models, recessive inheritance models, and alleles were analyzed in the AML-M2 subtype.
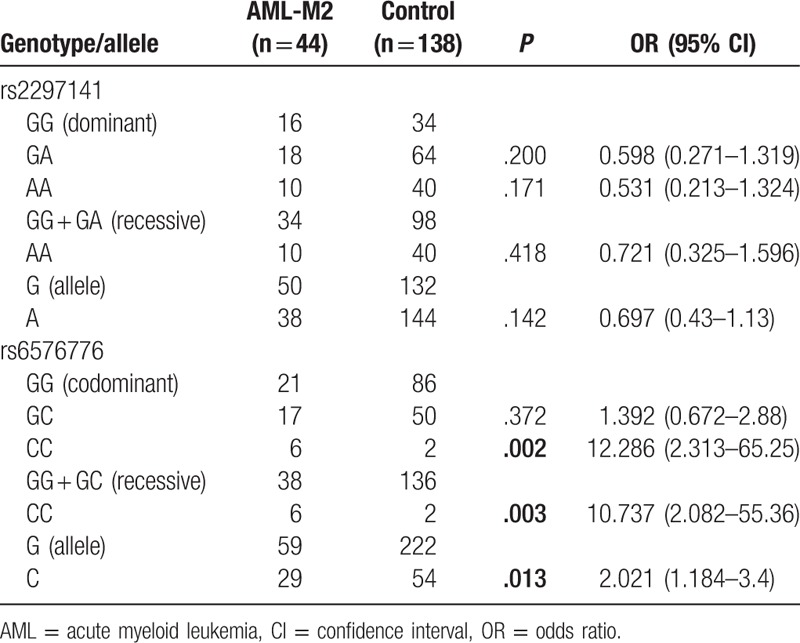
Table 7.
The dominant inheritance models, recessive inheritance models, and alleles were analyzed in the AML-M5 subtype.
4. Discussion
Genetic heterogeneity explains the variations in cancer predisposition.[19] In this study, we analyzed the distribution of CYR61 polymorphisms in patients with AML, and it was demonstrated that rs2297141 and rs6576776 genotypes were associated with the risk of AML in a Han Chinese population. Moreover, these 2 genotypes were associated with AML-M5 and AML-M2, respectively. Together with our previously finding,[13] this study confirmed the role of Cyr61 in the development of AML.
Cyr61 regulates hematological tumorigenesis by diverse mechanisms.[20] The Cyr61 gene is located on chromosome 1p22.3. It was demonstrated in this study that the rs2297141 genotype and the rs6576776 genotype are associated with the risk of AML. Rs2297141 is in intron 1 of CYR61, and rs6576776 is on the 3′-flanking region of Cyr61. Although the SNPs cannot directly affect the sequence of the Cyr61 protein, they may regulate the transcription of Cyr61 or other genes at many different levels.[21] That intron has been shown to regulate gene expression,[22,23] and it was reported that breast carcinogenesis was accompanied by a shift in intron 3 of the CYR61 gene moving toward a phenotype wherein intron 3 is missing from the mRNA transcript.[24] It was also demonstrated that the 5′-flanking region of the Cyr61 gene regulated its expression,[25,26] but little is known about the 3′-flanking region of the Cyr61 gene. Although this study provides a clue regarding the roles of CYR61 polymorphisms in AML, whether and how the roles of Cyr61 polymorphisms are associated with hematological tumorigenesis remain unclear and need further study.
It has been demonstrated that CYR61 polymorphisms (rs3753793, rs6682848, and rs12756618) were related to Graves’ ophthalmopathy,[10] and that a CYR61 polymorphism (rs3753793) was associated with prostate cancer risk[12]; however, these polymorphisms were not associated with AML in the Chinese population (data not shown). Based on these studies, we speculated that the distribution of CYR61 polymorphisms in AML may be different from the distribution in other diseases, which may be the result of the roles played by Cyr61 in different diseases or/and ethnic groups.
CEBPA, c-Kit, FLT3, DNMT3A, NPM, ASXL1, U2AF1, and SRSF2 mutations are used in clinical practice and affect the diagnosis, stratification, and treatment of AML.[2,18] Combined with the data from ANLN clinical tests and RNA-seq, we speculated that there were relationships between these mutations and Cyr61 polymorphisms, but no correlations were found. Furthermore, we analyzed the distributions of rs2297141 and rs6576776 polymorphisms in the subtypes of AML. The genotype distributions of the polymorphisms with respect to the subtypes of AML revealed that the rs2297141 SNP was associated with AML-M5, and the rs6576776 polymorphism was associated with AML-M2; the results were also analyzed with the dominant inheritance models and recessive inheritance models. In recent years, gene analysis has played a vital role in the subclassification of AML, providing more information on which to base treatment and prognostic decisions,[3,27] and this study also showed the potential subclassification role of Cyr61 in AML. The role of the rs2297141 polymorphism in AML-M5 and the role of the rs6576776 polymorphism in AML-M2 contributed the most to their roles in AML. The finding that there are roles played by different polymorphisms in different subtypes of AML is consistent with the diversity of Cyr61 functions.[28]
Overall, our findings revealed a significant association between Cyr61 rs2297141 and rs6576776 polymorphisms and the risk of AML, especially the AML-M2 and AML-M5 subtypes. Whether and how the Cyr61 polymorphisms can affect CYR61 gene expression or the function of the CYR61 gene product should be studied further. Our observations provided a clue to guide further exploration of the leukemogenic roles of Cyr61.
Acknowledgments
The authors thank the Cancer Genome Atlas for providing the data, and online software UALCAN for analyzing, and thank for the support by the National Natural Science Foundation of China (81702075) and the Chongqing Basic and Frontier Research (cstc2016jcyjA0177), China.
Author contributions
Conceptualization: Chang-Chun Niu.
Data curation: Chang-Chun Niu, Ya-Fang Wan, Cheng Yang.
Formal analysis: Chang-Chun Niu, Ya-Fang Wan.
Investigation: Cheng Yang.
Methodology: Chang-Chun Niu, Cheng Yang, Tian Li.
Resources: Cheng Yang, Tian Li.
Writing – original draft: Chang-Chun Niu.
Writing – review and editing: Pu Liao.
Footnotes
Abbreviations: AML = acute myeloid leukemia, CI = confidence interval, Cyr61 = cysteine-rich angiogenic inducer 61, FAB = French–American–British, HWE = Hardy–Weinberg equilibrium, OR = odds ratio, SNP = single-nucleotide polymorphism, WHO = World Health Organization.
Chang-Chun Niu and Ya-Fang Wan have contributed equally to this work.
National Natural Science Foundation of China (81702075) and the Chongqing Basic and Frontier Research (cstc2016jcyjA0177), China.
The authors have no conflicts of interest to disclose.
References
- [1].Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bullinger L, Dohner K, Dohner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol 2017;35:934–46. [DOI] [PubMed] [Google Scholar]
- [3].Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- [4].Mayer S, Gabriel B, Erbes T, et al. Cyr61 expression pattern and association with clinicopathological factors in patients with cervical cancer. Anticancer Res 2017;37:2451–6. [DOI] [PubMed] [Google Scholar]
- [5].Nguyen LT, Song YW, Cho SK. Baicalein inhibits epithelial to mesenchymal transition via downregulation of Cyr61 and LOXL-2 in MDA-MB231 breast cancer cells. Mol Cells 2016;39:909–14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [6].Sano M, Driscoll DR, DeJesus-Monge WE, et al. Activation of WNT/beta-catenin signaling enhances pancreatic cancer development and the malignant potential via up-regulation of Cyr61. Neoplasia 2016;18:785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen J, Liu Y, Sun Q, et al. CYR61 suppresses growth of human malignant melanoma. Oncol Rep 2016;36:2697–704. [DOI] [PubMed] [Google Scholar]
- [8].Wang P, Li L, Li T. Positive correlation of cysteine-rich 61 and target genes of Wnt/beta-catenin pathway in esophageal squamous cell carcinoma. J Cancer Res Ther 2016;12(suppl):19–22. [DOI] [PubMed] [Google Scholar]
- [9].Chen CC, Kim KH, Lau LF. The matricellular protein CCN1 suppresses hepatocarcinogenesis by inhibiting compensatory proliferation. Oncogene 2016;35:1314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Planck T, Shahida B, Sjogren M, et al. Association of BTG2, CYR61, ZFP36, and SCD gene polymorphisms with Graves’ disease and ophthalmopathy. Thyroid 2014;24:1156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Perrot A, Schmitt KR, Roth EM, et al. CCN1 mutation is associated with atrial septal defect. Pediatr Cardiol 2015;36:295–9. [DOI] [PubMed] [Google Scholar]
- [12].Tao L, Chen J, Zhou H, et al. A functional polymorphism in the CYR61 (IGFBP10) gene is associated with prostate cancer risk. Prostate Cancer Prostatic Dis 2013;16:95–100. [DOI] [PubMed] [Google Scholar]
- [13].Niu CC, Zhao C, Yang Z, et al. Inhibiting CCN1 blocks AML cell growth by disrupting the MEK/ERK pathway. Cancer Cell Int 2014;14:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bouchard L, Tchernof A, Deshaies Y, et al. CYR61 polymorphisms are associated with plasma HDL-cholesterol levels in obese individuals. Clin Genet 2007;72:224–9. [DOI] [PubMed] [Google Scholar]
- [15].Hou Z, Luo Y, Wang Z, et al. Inferring the dynamics of effective population size using autosomal genomes. Sci Rep 2016;6:20079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Huang L, Peng JH, Liang QH, et al. Insulin-like factor-2 receptor rs9456497 G genotype overrepresents in males of average population and its correlation with cardiovascular risks. Arch Gerontol Geriatr 2018;76:202–9. [DOI] [PubMed] [Google Scholar]
- [17].Komanduri KV, Levine RL. Diagnosis and therapy of acute myeloid leukemia in the era of molecular risk stratification. Annu Rev Med 2016;67:59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhu YM, Wang PP, Huang JY, et al. Gene mutational pattern and expression level in 560 acute myeloid leukemia patients and their clinical relevance. J Transl Med 2017;15:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shaw V, Bullock K, Greenhalf W. Single-nucleotide polymorphism to associate cancer risk. Methods Mol Biol 2016;1381:93–110. [DOI] [PubMed] [Google Scholar]
- [20].Wells JE, Howlett M, Cheung LC, et al. The role of CCN family genes in haematological malignancies. J Cell Commun Signal 2015;9:267–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA 2003;9:607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Carey KT, Wickramasinghe VO. Regulatory potential of the RNA processing machinery: implications for human disease. Trends Genet 2018;34:279–90. [DOI] [PubMed] [Google Scholar]
- [23].Shaul O. How introns enhance gene expression. Int J Biochem Cell Biol 2017;91:145–55. [DOI] [PubMed] [Google Scholar]
- [24].Hirschfeld M, zur Hausen A, Bettendorf H, et al. Alternative splicing of Cyr61 is regulated by hypoxia and significantly changed in breast cancer. Cancer Res 2009;69:2082–90. [DOI] [PubMed] [Google Scholar]
- [25].Schutze N, Rucker N, Muller J, et al. 5′ flanking sequence of the human immediate early responsive gene ccn1 (cyr61) and mapping of polymorphic CA repeat sequence motifs in the human ccn1 (cyr61) locus. Mol Pathol 2001;54:170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Seefried L, Muller-Deubert S, Krug M, et al. Dissection of mechanoresponse elements in promoter sites of the mechanoresponsive CYR61 gene. Exp Cell Res 2017;354:103–11. [DOI] [PubMed] [Google Scholar]
- [27].Vardiman JW. The World Health Organization (WHO) classification of tumors of the hematopoietic and lymphoid tissues: an overview with emphasis on the myeloid neoplasms. Chem Biol Interact 2010;184:16–20. [DOI] [PubMed] [Google Scholar]
- [28].Li J, Ye L, Owen S, et al. Emerging role of CCN family proteins in tumorigenesis and cancer metastasis (review). Int J Mol Med 2015;36:1451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]


