Abstract
Objective
Racial health disparities persist among black and white women for colorectal cancer. Understanding racial differences in the gut microbiota and related covariates (e.g., stress) may yield new insight into unexplained colorectal cancer disparities.
Methods
Healthy non-Hispanic black or white females (age ≥19 years) provided survey data, anthropometrics, and stool samples. Fecal DNA was collected and isolated from a wipe. PCR was used to amplify the V4 region of the 16SrRNA gene and 250 bases were sequenced using the MiSeq platform. Microbiome data was analyzed using QIIME. OTU data were log transformed and normalized. Analyses were conducted using linear models in R Package “limma’.
Results
Fecal samples were analyzed for 80 females (mean age = 39.9 (SD= 14.0) years; 47 black, 33 white). Blacks had greater average BMI (33.3 vs. 27.5 kg/m2; p<0.01) and waist circumference (98.3 vs. 86.6 cm; p=0.003) than whites. Whites reported more stressful life events (p=0.026) and greater distress (p=0.052) than Blacks. Final models accounted for these differences. There were no significant differences in dietary variables. Unadjusted comparisons revealed no racial differences in alpha diversity. Racial differences were observed in beta diversity and abundance of top-10 OTUs. Blacks had higher abundances than whites of Faecalibacterium (p=0.034) and Bacteroides (p=0.038). Stress was associated with abundances of Bifidobacterium. The association between race and Bacteroides (logFC=1.72; 0=0.020) persisted in fully adjusted models.
Conclusions
Racial differences in the gut microbiota were observed including higher Bacteroides among blacks. Efforts to cultivate an ‘ideal’ gut microbiota may help reduce colorectal cancer risk.
Keywords: gut microbiota, disparities, stress, race, colorectal cancer
Introduction
Racial health disparities persist among black and white women for many chronic health conditions including colorectal cancer (1–5). While some determinants that contribute to health disparities have been well documented and even intervened upon to reduce disparities, differences in health outcomes of blacks and whites diagnosed with colorectal cancer are still not fully understood. For example, progress has been made in reducing colorectal cancer incidence and mortality as a result of prevention and early detection through screening(6, 7); however, colorectal cancer disparities remain, in part, due to access to and utilization of screening along with unknown contributors (7). Even after accounting for differences in the distribution of known risk factors of colorectal cancer (e.g., diet, body weight, physical activity, smoking, alcohol use, family history, screening practices, personal health history, socioeconomic status, age), black women remain at 48% greater risk of diagnosis than white women (1) suggesting that an unidentified or unmeasured risk factor is contributing to this disparity.
Recent research has revealed associations between the gut microbiota and colorectal cancer. For example, in a case-control study reported by Ahn et al., participants with colorectal cancer displayed lower overall microbial diversity in the gut, lower abundance of Clostridium, and higher abundances of Fusobacterium and Porphyromonas compared to cancer-free individuals (8). Indeed, Fusobacterium is one of the most documented microbial taxa to be linked to colorectal cancer (8–12). However, several others including lower abundances of Lactobacillus (13, 14) and Bifidobacterium; higher abundances of Bacteroides (15), Ruminococcus (15), and Porphyromonas (8); and mixed findings of both significantly higher and lower abundances of Clostridium (15) have also been associated with colorectal cancer. While discovery of these associations are provocative and generate numerous research hypotheses, the exact mechanisms and pathophysiology for most of the observed associations have not been fully defined. Research has suggested that metabolic processes, i.e., the interaction of diet and colonization of the gut, may explain the role of the gut microbiota and colorectal cancer risk. Ou et al. reported that the consumption of certain foods and the composition of the gut microbiota result in the production of either health-promoting metabolites like short-chain fatty acids or inflammatory and potentially carcinogenic metabolites (e.g., secondary bile acids) (14). Additional research has also shown that high diversity in the gut microbiota is generally associated with better health including lower risk for colorectal cancer as suggested by multiple case-control studies in which cases displayed lower diversity than cancer-free controls (8, 16–19). Given this evidence, one may hypothesize that racial differences in the composition of the gut microbiota may help explain observed differences in the incidence of colorectal cancer (20).
Increased sequencing capability made possible through technological advances over the past several years have led researchers to investigate possible links and mechanisms between the composition of the gut microbiota and chronic diseases including irritable bowel syndrome (inflammatory bowel disease), obesity, and colorectal cancer (21–23). However, there is a paucity of research examining whether the gut microbiota of blacks and whites differ which may ultimately contribute to observed disparities in colorectal cancer; and if so, whether psychosocial factors like stress, which has been shown to affect the gut microbiota in animal studies (24), is associated with the gut microbiota in humans. Furthermore, previous research has not specifically tested for racial differences in colorectal cancer-associated taxa in the gut microbiota. Hester and colleagues reported that microbial profiles and microbial metabolites differed by racial/ethnic group among a sample of healthy Hispanic and non-Hispanic AAs, American Indians, and Whites (25). However, this study was limited by a very small sample size (n=20; 5 per race/ethnic group), limited age range (all participants over 50 years), and failure to collect certain sociodemographic information that may be relevant.
The purpose of the current study was to build upon a small but growing evidence base to further characterize and compare the gut microbiota of blacks and whites among a sample of generally healthy women 19 years and older and to specifically test for differences in taxa that have been associated with colorectal cancer. We hypothesized that black and white women would display differences in gut microbial composition with black women having lower diversity; higher abundances of Fusobacterium, Porphyromonas, Bacteroides and Ruminococcus; and lower abundances of Lactobacillus and Bifidobacterium in which case additional research would be needed to determine the mechanism of any differences, i.e., biological or non-biological factors associated with race. Thus, as secondary analyses, we also explored whether perceived psychological stress contributed to any observed differences. Currently, much of what is known about the impact of stress on the gut microbiota has been demonstrated only in animal models. Previous studies have shown that exposure to a social stressor led to perturbation of the gut microbiota among mice (26). Translational work remains to be validated in humans. Given the evidence from animal studies, we hypothesized that stress would be associated with gut microbial composition among our human sample with higher stress being associated with higher abundances of Fusobacterium, Porphyromonas, Bacteroides and Ruminococcus; and lower abundances of Lactobacillus and Bifidobacterium. Associations between stress and the gut microbiota are supported the body of evidence surrounding the brain-gut axis that suggests a bi-directional relationship between the central nervous system and the gastrointestinal system. For this study, we focused on perceived psychological chronic stress based on research indicating that this type of stress is associated with adverse health outcomes (27). Better characterization of the gut microbiota and contributing factors of healthy black and white women may provide valuable insights into the nearly 50% increased risk of colorectal cancer among black women after controlling for currently known risks.
Materials and Methods
For this cross-sectional study, generally healthy volunteers were recruited using flyers, word of mouth, and small media between March 2014 and August 2014. Flyers were posted throughout the University and the surrounding communities inviting non-Hispanic black or white, non-pregnant females who were interested in participating in a study to examine gut bacteria to contact the study coordinator. Similar recruitment messages were also deployed using small media including website postings for clinical trials and other local circulations (e.g., newsletters) throughout the community. Study participants were also encouraged to tell others who may be interested about the study. Participants were at least 19 years old and self-identified as non-Hispanic black or white. Exclusion criteria were the following: 1) current tobacco use, 2) current pregnancy, 3) prior cancer diagnosis, or 4) use of antibiotics or other medications known to alter the gut in the previous 90 days (28–32). All study-related protocols and questionnaires received approval from The University of Alabama at Birmingham Institutional Review Board for human subjects. Participants provided written informed consent and were compensated $50 for their time.
Participants completed two study visits during which demographics, anthropometrics, survey data, and biospecimens were collected. During visit 1, participants completed the following:
Demographics Survey – A demographic data collection survey was used to gather the following: age; race and ethnicity; education (less than high school, high school/GED, some college, Associates, Bachelors, Masters, Doctoral); household income (< $10,000, $10,000–$19,999, $20,000–$29,999, $30,000–$39,999, $40,000–$49,999 or $50,000 or more); and number of individuals in the household.
Anthropometric Measures – Weight and height were measured in indoor clothing, without shoes, using a calibrated digital measuring station (Seca 284 measuring station, Hanover, MD). BMI was calculated as weight (kg)/height (m2). Waist circumference was measured to the nearest 0.10 centimeter using the Gulick II tension spring measuring tape (model 67020).
Perceived Stress Scale – 10 (PSS-10) - Participants completed the PSS-10, a validated ten-item scale used to assess the degree to which situations in one’s life are appraised as stressful (33). Participants provided a response (0=Never, 1=Almost Never, 2=Sometimes, 3=Fairly Often and 4=Very Often) to a series of ten statements about the occurrence of stressful events. The PSS-10 overall score is obtained by reversing subjects’ responses (0=4, 1=3, 2=2, 3=1 and 4=0) of four positively stated items (PSS 4, 5, 7 and 8) and then summing across all 10 scale items. Higher scores on the PSS-10 indicate greater perceived stress (possible range 0–40) (33). This survey was selected to capture participants’ global perception of stress over the previous month. The PSS has good internal consistency (Cronbach’s alpha = 0.86) and is positively correlated with other indices of stress among adults (33, 34).
Weekly Stress Inventory – Short Form (WSI-SF) Participants also completed the WSI-SF, a validated 25-item instrument that assesses the number of minor life and associated distress in the previous week (35). Items were ranked on an 8-point Likert scale from which an event score and an impact score were calculated. The event score (WSI-SFE) is the total number of events endorsed by summing the counts of events. The impact sore (WSI-SFI) is the sum of the subjective ratings of distress by summing each of the values (35). This survey measured minor chronic stressors in the previous week. The instrument has good internal consistency (WSI-SFE Cronbach’s alpha = 0.92; WSI-SFI Cronbach’s alpha = 0.91) and has been validated in diverse populations (35).
At the end of the first study visit, participants were given a stool collection kit along with verbal and written instructions for proper sample collection at home. Participants were asked to collect one sample and bring to the next study visit, which was scheduled within the following 5–7 days. During visit 2, participants returned stool samples, completed a participant satisfaction survey and a provided dietary information using the National Cancer Institute Automated Self-administered 24-hour Dietary Recall (ASA-24) which includes multi-level food probes and cues to assess food types and amounts (36, 37).
Sample Processing and Analysis
Sample collection
Participants were asked to collect a first wipe using a moist towelette provided by the study following defecation no more than 48 hours prior to their second clinic visit. Wipes were then placed in a sealed sandwich bag and frozen until delivered to the microbiome lab for DNA extraction and processing.
DNA extraction and Illumina MiSeq DNA sequencing
Microbial genomic DNA was isolated using the Fecal DNA isolation kit from Zymo Research following the manufacturer’s instructions. Once the sample DNA was prepared, PCR was used with unique barcoded primers to amplify the variable region 4 (V4) region of the 16S rDNA gene to create an amplicon library from individual samples. The PCR product was ~255 bases from the V4 segment of the 16S rDNA gene, and we sequenced 251 bases single end reads using Illumina MiSeq (38, 39).
Bioinformatics
FASTQ conversion of the raw data files was performed following de-multiplexing using MiSeq reporter. Quality assessment of the FASTQ files was performed as previously described (38, 39). Briefly, sequences were grouped into operational taxonomic units (OTUs) using the clustering program UCLUST at a similarity threshold of 97%. The Ribosomal Database Program (RDP) classifier trained using the Greengenes (v13.8) 16S rRNA database was used to make taxonomic assignments for all OTUs at confidence threshold of 80% (0.8). The resulting OTU table included all OTUs, their taxonomic identification, and abundance information. OTUs whose average abundance was less than 0.005% were filtered out. OTUs were then grouped together to summarize taxon abundance at different hierarchical levels of classification (e.g. phylum, class, order, family, genus, and species) from which comparisons are reported at the genus level.
Statistical Analysis
Sample demographic & Correlation
The overall and race-stratified descriptive statistics were created using R package “tableone”. Means and standard deviations (SD) were calculated for continuous variables. Frequencies and percentages were calculated for categorical variables. Between-group differences were tested using two-sided independent sample t-tests for continuous variables, and Fisher’s exact test for categorical variables. In addition, we assessed correlations between all numeric variables using the Pearson correlation coefficient.
Diversity measurements
Alpha diversities of the gut microbiota were summarized into mean and standard deviation (SD) for two race-stratified groups. Two-sample t-tests were conducted for each of the five alpha diversity indexes: Chao1, Observed species, PD whole tree, Shannon and Simpson. For beta diversity, PERMANOVA test (40) was performed to test for gut microbiota differences between whites and blacks in three beta diversity distance metrics: Bray Curtis, Weighted UniFrac, and Unweighted UniFrac (41).
OTU Analysis
The .biom file generated by QIIME was converted into OTU matrix with count numbers at genus level by using R Package “phyloseq”. Log2 transformation was performed after adding 1 to each count numbers of OTUs. The data were normalized by sample mean centering. Linear models in R Package “limma’ were used to test statistical significance differences. Linear models were also adjusted for age and waist circumference to account for group differences in the distribution of these potential confounding factors. We first tested 7 candidate OTUs identified as associated with colorectal cancer based on the literature and top 10 most abundant OTUs. For candidate OTU tests, the nominal p-value was used because of our interest in the result of each one instead of selection. We also conducted global tests on all the OTUs to identify OTUs associated with study subject attributes. In this case, false discovery rate (FDR) was used for selection. The threshold for statistical significance level was set at the 0.05 level.
Graphics were generated using R basic graphics, ggplot2, or “vegan” and analyses were conducted using R (Version 3.3.2).
Results
Description of the Study Sample
Fecal samples were analyzed for 80 females (47 black, 33 white) described in table 1. Mean age and BMI of participants were 39.9 years and 30.9 kg/m2, respectively. Blacks had a higher average BMI than whites (33.3 vs. 27.5 kg/m2; p=0.003) and larger waist circumference (98.3 vs. 86.6 cm; p=0.006). Participants were well-educated and 37.5% of participants were married. There were no statistically significant differences in dietary factors including fiber (p=0.60) and percentage of calories from fat (p=0.63).
Table 1.
Participant Characteristics (N=80)
| Characteristic | Mean (SD) | P-value | ||
|---|---|---|---|---|
|
| ||||
| Race | ||||
|
|
||||
| Black (N=47) | White (N=33) | Total (N=80) | ||
| Age (year) | 43.70 (14.38) | 34.52 (11.59) | 39.91 (13.98) | 0.003 |
|
| ||||
| BMI (kg/m2) | 33.26 (9.50) | 27.48 (6.65) | 30.88 (8.88) | 0.003 |
|
| ||||
| Waist Circumference (cm) | 98.28 (19.30) | 86.62 (15.80) | 93.44 (18.73) | 0.006 |
|
| ||||
| HH Individuals* | 2.28 (1.31) | 2.21 (1.14) | 2.25 (1.24) | 0.820 |
|
| ||||
| Perceived Stress Scale Score | 15.09 (6.46) | 16.16 (7.05) | 15.52 (6.69) | 0.488 |
|
| ||||
| Weekly Stress Inventory (# events) | 10.86 (5.92) | 13.90 (5.34) | 12.14 (5.85) | 0.026 |
|
| ||||
| Weekly Stress Inventory (impact) | 25.63 (22.08) | 35.74 (21.18) | 29.87 (22.14) | 0.052 |
|
| ||||
| Total Fiber (g) | 17.05 (10.73) | 18.31 (10.29) | 17.56 (10.51) | 0.604 |
|
| ||||
| % of Calories from Fat | 38.21 (9.56) | 37.15 (9.80) | 37.78 (9.61) | 0.634 |
|
| ||||
| N (%) | ||||
|
| ||||
| Marital Status | 0.211 | |||
| Single | 16 (34.8) | 14 (42.4) | 30 (37.5) | |
| Married | 16 (34.8) | 14 (42.4) | 30 (37.5) | |
| Separated | 2 (4.3) | 0 (0.0) | 2 (2.5) | |
| Divorced | 7 (15.2) | 4 (12.1) | 11 (13.8) | |
| Widowed | 5 (10.9) | 0 (0.0) | 5 (6.3) | |
| Domestic Partner | 0 (0.0) | 1 (3.0) | 1 (1.3) | |
|
| ||||
| Education Level | 0.112 | |||
| Less than High School | 1 (2.1) | 0 (0.0) | 1 (1.3) | |
| High School/GED | 6 (12.8) | 1 (3.0) | 7 (8.8) | |
| Some College | 12 (25.5) | 3 (9.1) | 15 (18.8) | |
| Associates | 5 (10.6) | 2 (6.1) | 7 (8.8) | |
| Bachelors | 14 (29.8) | 16 (48.5) | 30 (37.5) | |
| Masters | 9 (19.1) | 10 (30.3) | 19 (23.8) | |
| Doctoral | 0 (0.0) | 1 (3.0) | 1 (1.3) | |
|
| ||||
| Household Income | 0.123 | |||
| Less than 10,000 | 11 (23.4) | 6 (18.2) | 17 (21.3) | |
| 10,000–19,000 | 3 (6.4) | 2 (6.1) | 5 (6.3) | |
| 20,000–29,000 | 2 (4.3) | 1 (3.0) | 3 (3.8) | |
| 30,000–39,000 | 10 (21.3) | 3 (9.1) | 13 (16.3) | |
| 40,000–49,000 | 11 (23.4) | 4 (12.1) | 15 (18.8) | |
| 50,000 or over | 10 (21.3) | 17 (51.5) | 27 (33.8) | |
SD, standard deviation; HH Individuals, number of individuals are in a household;
Bolded p-values are significant at the 0.05 level of significance
The Composition of the Gut Microbiota
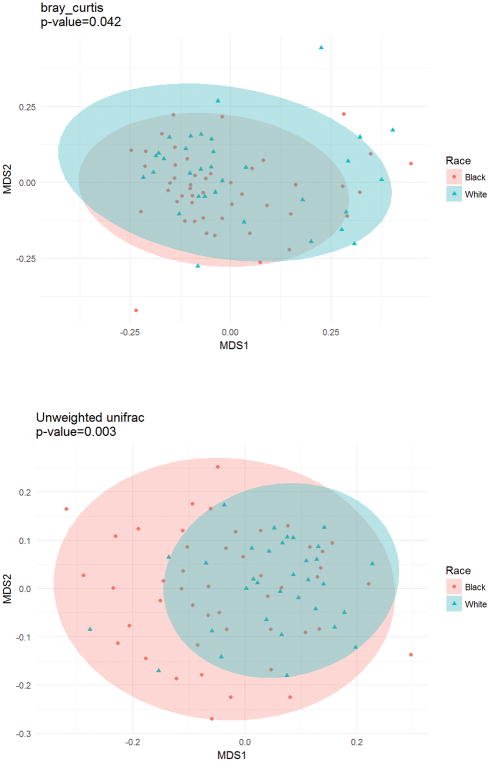
Measures of alpha diversity (shown in table 2) indicated similar within-sample gut microbial diversity for black and white women. Beta diversity, which compares the overall microbial distribution between groups, indicated racial differences using the Bray Curtis (p=0.042) and Unweighted Unifrac (p=0.003) methods (figure 1), but not Weighted Unifrac (p=0.12).
Table 2.
Measures of alpha diversity by race
| Measures | Mean (SD) | P-value | |
|---|---|---|---|
| Race | |||
| Black (N=55) | White (N=45) | ||
| Chao1 | 329.00 (52.33) | 337.53 (49.51) | 0.465 |
| Observed species | 274.96 (45.08) | 285.67 (37.23) | 0.265 |
| PD whole tree | 21.09 (2.74) | 21.89 (2.73) | 0.199 |
| Shannon | 5.19 (0.62) | 5.16 (0.61) | 0.801 |
| Simpson | 0.93 (0.04) | 0.92 (0.06) | 0.348 |
Figure 1.
Schematic of a Non-metric multidimensional scaling (NMDS) plot. NMDS is an indirect gradient analysis approach which produces an ordination based on a distance and attempts to represent, as closely as possible, the pairwise dissimilarity between objects in a low-dimensional space. Points represent individual participants’ gut microbiota. Participants with gut microbiota that are more similar to one another are ordinated closer together. Circles are confidence ellipses.
Racial comparisons of top 10 OTUs and 7 colorectal cancer-associated Genera
The average relative abundance of the top 10 OTUs at the genus level stratified by race is shown in figure S1 (Supplemental Digital Content 1). To formally compare blacks and whites for these OTUs, we first transformed and normalized the data and then used linear models. Results from our linear model indicated a significant racial difference in Bacteroides (p=0.035), and Faecalibacterium (p=0.032) where blacks had higher abundances of both.
Figure S2 (Supplemental Digital Content 2) illustrates racial comparisons of genera that were identified for examination a priori based on evidence in the literature of an association with colorectal cancer. Unadjusted analyses indicated significant racial differences for Bacteroides (p=0.035), but not for Bifidobacterium, Clostridium, Fusobacterium, Lactobacillus, Porphyromonas or Ruminococcus (shown in Table 3). The Y-axis represents normalized to the largest total counts (Bacteroides) size on the log scale.
Table 3.
Racial comparisons of average abundance of selected colorectal cancer-associated genera
| OTUs | Average Abundance (Black) | Average Abundance (White) | Log(B/W)* | P Value |
|---|---|---|---|---|
| Bacteroides | 7.59 | 6.38 | 1.205 | 0.035 |
| Bifidobacterium | 3.55 | 2.72 | 0.828 | 0.244 |
| Clostridium | 2.91 | 3.45 | −0.536 | 0.368 |
| Fusobacterium | −1.56 | −1.64 | 0.082 | 0.775 |
| Lactobacillus | 4.08 | 4.39 | −0.311 | 0.601 |
| Porphyromonas | 1.45 | 1.65 | −0.195 | 0.752 |
| Ruminococcus | 5.87 | 5.66 | 0.211 | 0.620 |
B, Black; W, White
Bolded p-values are significant at the 0.05 level of significance
Examination of associations between race, selected phenotypic traits and the gut microbiota
As shown in table 1, racial stratification revealed differences in other phenotypic traits of our study sample including age, BMI, and waist circumference which necessitated additional analyses to account for potential confounding of results. Due to the high correlation between BMI and waist circumference (r=0.94, p<0.001), we decided to focus on a single measure of body size. Waist circumference was chosen due to literature indicating that waist circumference is a better indicator of actual body composition than BMI and may account for observed racial differences in body composition for black and white females with similar BMIs. When controlling for waist circumference, race was significantly associated with Bacteroides (p=0.040), where black race was associated with a greater abundance (table 4). There was no significant waist-by-race interaction (table 4).
Table 4.
Examining waist circumference as an independent predictor of OTUs and a potential confounder of the relationship between race and OTUs
| OTUs | Unadjusted | Model 1 | Model 2 | |||
|---|---|---|---|---|---|---|
| logFC | P Value | logFC | P Value | logFC | P Value | |
| Bacteroides | 0.003 | 0.866 | 1.283 | 0.040 | 0.018 | 0.610 |
| Bifidobacterium | 0.006 | 0.744 | 1.103 | 0.150 | −0.036 | 0.408 |
| Clostridium | 0.005 | 0.776 | −0.554 | 0.390 | −0.055 | 0.133 |
| Fusobacterium | 0.005 | 0.471 | −0.056 | 0.833 | −0.004 | 0.779 |
| Lactobacillus | −0.015 | 0.355 | −0.347 | 0.586 | −0.028 | 0.433 |
| Porphyromonas | 0.010 | 0.521 | −0.370 | 0.569 | −0.068 | 0.065 |
| Ruminococcus | 0.011 | 0.327 | 0.153 | 0.732 | 0.013 | 0.600 |
Unadjusted, linear model for interested OTUs based on waist circumference without controlling for anything;
Model 1, linear model for interested OTUs based on race while controlling for waist circumference;
Model 2, linear model for interested OTUs based on interaction between race and waist circumference
Examination of associations between race, stress measures and the gut microbiota
We compared racial groups on measures of stress (table 1) based on preliminary evidence that exposure to stress may affect gut microbial composition. There were no differences in scores on the PSS-10 which appraises an individual’s overall stress in the previous month (p=0.49). When comparing the WSI-SF which measures the number of and impact of minor stressors in the previous week, white women reported a higher number of stressful events than black women (13.9 vs. 10.9; p=0.026). White women also reported a higher perceived impact, i.e. level of distress from stressful events, compared to black women (35.7 vs. 25.6; p=0.052). For each stress measure, our first set of models examined the crude relationships between the individual stress measure (i.e., PSS-10, WSI-SFE, WSI-SFI) and each pre-specified colorectal cancer-associated genus group. Then, consistent with our overall hypothesis of exploring racial differences in the gut microbiota and possible contributors to observed differences, the next set of models examined the relationships between race and pre-specified colorectal cancer-associated genus groups while controlling for stress. Lastly, we tested for race-by-stress interactions as a predictor of taxa abundances.
Results of analyses examining the relationship between race, the PSS-10 and our pre-specified colorectal cancer associated genera are shown in Table S1 (Supplemental Digital Content 3). Unadjusted analysis revealed no significant associations between PSS-10 scores and any of the pre-specified genus groups. When controlling for PSS-10 score, race was significantly associated with Bacteroides (p=0.041) where black race was associated with a higher abundance. Analyses also revealed a significant race-by-stress (as indicated by PSS-10 score) interaction as associated with Clostridium (p=0.021), and Ruminococcus (p=0.014). Among whites, as PSS-10 score increased, the abundances of Clostridium and Ruminococcus also increased whereas there was a slight decrease in abundances for blacks with higher PSS-10 scores.
Table S1 also shows the results of analyses examining the relationship between race, the WSI-SF and our pre-specified colorectal cancer associated genera. Unadjusted analysis indicated that the WSI-SFE score (# of stressful events) was associated with Bifidobacterium (p=0.010). After controlling for WSI-SFE scores, race was significantly associated with Bacteroides (p=0.021) with blacks having a higher abundance after controlling for the number of stressful events. Race-by-stress (as indicated by WSI-SFE score) interactions were associated with abundances of Fusobacterium (p=0.017) such that among blacks, Fusobacterium increased with higher WSI-SFE score, but decreased with higher WSI-SFE score among whites.
Relationships between race, the WSI-SFI (perceived impact of stressful events), and our pre-specified colorectal cancer associated genera are also shown in table S1. A significant association between WSI-SFI score and Bifidobacterium (p=0.012) was observed in unadjusted analysis where the increased impact of stress was associated with lower abundance of Bifidobacterium. After controlling WSI-SFI scores, race was significantly associated with Bacteroides (p=0.032). Significant race-by-stress (as indicated by WSI-SFI) interactions were also observed for Fusobacterium (p=0.036). Among blacks, as WSI-SFI increased, abundances of Fusobacterium also increased. Among whites, increased WSI-SFI scores were associated with lower abundances of Fusobacterium.
Examination of associations between race, age, waist, stress measures, dietary measures and the gut microbiota
In the full model examining the relationship between race and our pre-specified colorectal cancer associated genera while controlling for age, waist, dietary and stress measures, an association was observed between race and a single genus group – Bacteroides (logFC= 1.79; p=0.019). Among our study sample, black race was associated with a greater abundance of Bacteroides.
Discussion
In our examination of the composition of the gut microbiota of sample of generally healthy black and white females, we observed no differences in the amount of microbial diversity, i.e., alpha diversity, of black and white women. We did, however, observe racial differences in the overall composition of the gut microbiota indicated by tests of beta diversity. While many of the racial differences that emerged in our unadjusted comparisons of the top 10 OTUs and 7 pre-specified colorectal cancer-associated genera were explained by other factors, Bacteroides remained as statistically different after controlling for other measured correlates in this study. Also, racial differences in multiple other genus groups were identified in exploratory post hoc analyses (Tables S2 – S5, Supplemental Digital Content 3).
Our study is consistent with previous work that has demonstrated racial differences in the gut microbiota overall. For example, Yazici and colleagues reported that blacks had greater abundances of sulfidogenic bacteria than whites (42). Other research has also shown differences in the gut microbiota at the phylum level including a report by Hester et al. suggesting significantly higher Firmicutes among blacks compared to whites (25). While our study did not necessarily compare the same taxonomic groups as these previous studies, our study contributes to a small body of literature demonstrating a general pattern of racial differences in the gut microbiota. When looking at specific taxa, we identified a higher abundance of Bacteroides among black women in our study which is consistent with our hypothesis that blacks would have a greater abundance of colorectal cancer-associated bacteria. When comparing Bacteroides between native Africans, a group with a very low incidence of colorectal cancer, and African Americans, the abundance of Bacteroides was much higher among African Americans. To our knowledge, our study is among the first to directly compare Bacteroides of black and white women. We also observed that the Weekly Stress Inventory measure, a measure of ongoing chronic stress was inversely associated with Bifidobacterium. We also observed racial differences in the WSI with white women reporting a higher number of stressful events and a greater impact. After controlling for stress, the racial difference in Bacteroides persisted with black women having a higher abundance which is of note based on previous research where colorectal cancer patients display higher abundances of Bacteroides than cancer-free individuals. Other research, primarily animal models (24, 43), but more recently human studies (44, 45), has also suggested a relationship (46–48)—potentially bi-directional—between stress and the gut microbiota. While the pathways linking the gut microbiota to health outcomes like colorectal cancer and to upstream correlates like stress remain to be elucidated, stress-induced alterations in neuro-endocrine-immune pathways may provide one possible mechanism for further exploration (27). The interplay of stress, inflammation, and the gut microbiota (49)may provide another pathway for investigation given the association between inflammation and cancer risk.
This study does not come without limitations. First, the cross-sectional design only allows for reporting of observed associations and does not explain mechanisms by which observed differences may exist. Next, given the sample size, it was not possible to simultaneously investigate all of the potential correlates of the gut microbiota. Indeed, in a 2016 paper, Falony et al reported that 69 different factors were significantly associated with variation in the gut microbiota among a cohort of 1106 Flemish participants (50). While our study narrowed the focus of correlates being investigated, the racial diversity of our sample still allows for novel information to be determined about the gut microbiota among a wider range of individuals. Also, medication data was not included in this analysis because it was not collected in a systematic quantifiable manner. While individuals who reported any antibiotic use were excluded from the study, use of other medications like anti-hypertensives was qualitatively recorded. Future studies should rigorously collect medication data so that more can be learned about how a range of medication usage may affect the gut microbiota. Also, while we opted to use waist circumference to estimate central adiposity to account for racial/ethnic differences in body fat distribution, future studies might consider more definitive measures of body fat and body composition such as bioelectrical impedance analysis (BIA). Finally, there is limited generalizability to groups other than middle-class non-Hispanic black and white females.
This study also has several strengths. First, the racially diverse sample allows for the examination of the gut microbiota across racial groups which may be a key factor in better understanding and addressing health disparities for a number of chronic diseases including colorectal cancer. This study also included a well-phenotyped sample for examination. Thus, we were able to examine and account for demographic or phenotypic differences that may contribute to differences in the gut microbiota. Finally, the inclusion of psychological measures of stress further enhances the impact of this study Taken together, these factors build on the current human microbiome research by moving the field closer to exploring the microbiome using a more comprehensive approach by considering the microbiome within the context of the immediate and proximal environments (51). One framework, the biopsychosocial ecological model, suggests that a variety of factors at the immediate, proximal, distal, and intermediate levels may influence the gut microbiota (51). Hence, future research should consider inclusive frameworks like to more rigorously examine this emerging area.
In summary, racial differences in the overall composition of the gut microbiota were observed although only one of our 7 a priori identified candidate genera revealed any racial differences. While we fully acknowledge that findings may include false positives as a result of a relatively small, self-selected sample, our research suggests that additional investigation is warranted. Additionally, we do not assert that these racial differences were due to race alone, but more likely due to behavioral, environmental and psychosocial factors that are related to race that were unmeasured in this study. Given that temporality has not been established between microbial dysbiosis and colorectal cancer, the absence of significant racial differences among our pre-specified genera is meaningful information. First, it provides some direction for future hypotheses regarding temporality. Also, it suggests that modifiable factors, not biological factors associated with race, are the main drivers of OTU abundance for those 7 genera.
Nevertheless, while only a few racial differences in OTUs emerged for the top10 OTUs or our a priori groups of interest, racial differences for many other groups emerged in exploratory analysis suggesting that understanding the gut microbiota is complex and must be fully explored rather than focusing on a limited number of microbes. This study has completed an important first step of establishing that there are, in fact, racial differences in the gut microbiota. It remains unclear whether these differences are driven by genetics, behavior, or a combination. Future research in this area should expand racial comparisons to include individuals with and without colorectal cancer. Additionally, longitudinal follow-up studies of racially diverse, well-phenotyped cohorts will allow us to better understand the role of the gut microbiota in the etiology of colorectal cancer.
Supplementary Material
Acknowledgments
Source of Funding: This work was supported by the UAB Comprehensive Cancer Center Young Supporters Board Young Investigator Award (T.L.C.), the UAB Center for Clinical and Translational Science Grant Number UL1TR001417 from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH), and the National Cancer Institute (K01CA190559 to T.L.C.).
The following are also acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30 CA013148), Center for AIDS Research (5P30AI027767), Center for Clinical Translational Science (UL1TR001417) and Heflin Center for Genomic Sciences.
Abbreviations
- OTU
Operational Taxonomic Unit
- PSS
Perceived Stress Scale
- WSI-SFE
Weekly Stress Inventory-Short Form Event
- WSI-SFI
Weekly Stress Inventory-Short Form Impact
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Ollberding NJ, Nomura AMY, Wilkens LR, Henderson BE, Kolonel LN. Racial/ethnic differences in colorectal cancer risk: The multiethnic cohort study. International Journal of Cancer. 2011;129:1899–906. doi: 10.1002/ijc.25822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hertz RP, Unger AN, Cornell JA, Saunders E. Racial disparities in hypertension prevalence, awareness, and management. Archives of internal medicine. 2005;165:2098–104. doi: 10.1001/archinte.165.18.2098. [DOI] [PubMed] [Google Scholar]
- 3.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Executive Summary: Heart Disease and Stroke Statistics—2012 Update. A Report From the American Heart Association. 2012;125:188–97. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- 4.Golden SH, Brown A, Cauley JA, Chin MH, Gary-Webb TL, Kim C, Sosa JA, Sumner AE, Anton B. Health disparities in endocrine disorders: biological, clinical, and nonclinical factors--an Endocrine Society scientific statement. The Journal of clinical endocrinology and metabolism. 2012;97:E1579–639. doi: 10.1210/jc.2012-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017 doi: 10.3322/caac.21395. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 6.Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT. Long-term colorectal-cancer incidence and mortality after lower endoscopy. The New England journal of medicine. 2013;369:1095–105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desantis C, Naishadham D, Jemal A. Cancer statistics for African Americans, 2013. CA Cancer J Clin. 2013 doi: 10.3322/caac.21173. [DOI] [PubMed] [Google Scholar]
- 8.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. Journal of the National Cancer Institute. 2013;105:1907–11. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell host & microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PloS one. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, Shureiqi I, Toyota M, Kondo Y, Estecio MR, Issa JP. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer research. 2014;74:1311–8. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA. Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP) 2015 doi: 10.1097/CEJ.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 13.Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Applied and environmental microbiology. 1995;61:3202–7. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, Gaskins HR, O’Keefe SJ. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. The American journal of clinical nutrition. 2013;98:111–20. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PloS one. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett C, Knight R, Gordon JI. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heiman ML, Greenway FL. A healthy gastrointestinal microbiome is dependent on dietary diversity. Molecular Metabolism. 2016;5:317–20. doi: 10.1016/j.molmet.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, Wang J, Imhann F, Brandsma E, Jankipersadsing SA, Joossens M, Cenit MC, Deelen P, Swertz MA, Weersma RK, Feskens EJM, Netea MG, Gevers D, Jonkers D, Franke L, Aulchenko YS, Huttenhower C, Raes J, Hofker MH, Xavier RJ, Wijmenga C, Fu J. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–9. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. British journal of cancer. 2016;114:237–42. doi: 10.1038/bjc.2015.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Findley K, Williams DR, Grice EA, Bonham VL. Health Disparities and the Microbiome. Trends in Microbiology. 2016;24:847–50. doi: 10.1016/j.tim.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiBaise JK, Frank DN, Mathur R. Impact of the Gut Microbiota on the Development of Obesity: Current Concepts. Am J Gastroenterol Suppl. 2012;1:22–7. [Google Scholar]
- 22.Sartor RB, Mazmanian SK. Intestinal Microbes in Inflammatory Bowel Diseases. Am J Gastroenterol Suppl. 2012;1:15–21. [Google Scholar]
- 23.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Micro [Review] 2014;12:661–72. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 24.Galley JD, Bailey MT. Impact of stressor exposure on the interplay between commensal microbiota and host inflammation. Gut Microbes. 2014;5:390–6. doi: 10.4161/gmic.28683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hester CM, Jala VR, Langille MG, Umar S, Greiner KA, Haribabu B. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World journal of gastroenterology : WJG. 2015;21:2759–69. doi: 10.3748/wjg.v21.i9.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen R, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain, Behavior, and Immunity. 2011;25:397–407. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster JA, Rinaman L, Cryan JF. Stress & the gut-brain axis: Regulation by the microbiome. Neurobiology of Stress. 2017;7:124–36. doi: 10.1016/j.ynstr.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raymond F, Ouameur AA, Déraspe M, Iqbal N, Gingras H, Dridi B, Leprohon P, Plante P-L, Giroux R, Bérubé È, Frenette J, Boudreau DK, Simard J-L, Chabot I, Domingo M-C, Trottier S, Boissinot M, Huletsky A, Roy PH, Ouellette M, Bergeron MG, Corbeil J. The initial state of the human gut microbiome determines its reshaping by antibiotics. The ISME Journal. 2016;10:707–20. doi: 10.1038/ismej.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsen JA, Ptacek TS, Carter SJ, Liu N, Kumar R, Hyndman L, Lefkowitz EJ, Morrow CD, Rogers LQ. Gut microbiota composition associated with alterations in cardiorespiratory fitness and psychosocial outcomes among breast cancer survivors. Supportive Care in Cancer. 2017;25:1563–70. doi: 10.1007/s00520-016-3568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS biology. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. Journal of clinical microbiology. 2005;43:5588–92. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Medicine. 2016;8:39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. Journal of Health and Social Behavior. 1983;24:385–96. [PubMed] [Google Scholar]
- 34.Sharp LK, Kimmel LG, Kee R, Saltoun C, Chih-Hung C. Assessing the Perceived Stress Scale for African American Adults with Asthma and Low Literacy. Journal of Asthma [Article] 2007;44:311–6. doi: 10.1080/02770900701344165. [DOI] [PubMed] [Google Scholar]
- 35.Brantley PJ, Bodenlos JS, Cowles M, Whitehead D, Ancona M, Jones GN. Development and validation of the Weekly Stress Inventory-Short Form. Journal of Pscyhopathology and Behavioral Assessment. 2007;29:55–60. [Google Scholar]
- 36.Subar AF, Kirkpatrick SI, Mittl B, Zimmerman TP, Thompson FE, Bingley C, Willis G, Islam NG, Baranowski T, McNutt S, Potischman N. The Automated Self-Administered 24-hour dietary recall (ASA24): a resource for researchers, clinicians, and educators from the National Cancer Institute. Journal of the Academy of Nutrition and Dietetics. 2012;112:1134–7. doi: 10.1016/j.jand.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirkpatrick SI, Subar AF, Douglass D, Zimmerman TP, Thompson FE, Kahle LL, George SM, Dodd KW, Potischman N. Performance of the Automated Self-Administered 24-hour Recall relative to a measure of true intakes and to an interviewer-administered 24-h recall. The American journal of clinical nutrition. 2014;100:233–40. doi: 10.3945/ajcn.114.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar R, Eipers P, Little RB, Crowley M, Crossman DK, Lefkowitz EJ, Morrow CD. Getting started with microbiome analysis: sample acquisition to bioinformatics. Current protocols in human genetics. 2014;82:18 8 1–8 29. doi: 10.1002/0471142905.hg1808s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar R, Maynard CL, Eipers P, Goldsmith KT, Ptacek T, Grubbs JA, Dixon P, Howard D, Crossman DK, Crowley MR, Benjamin WH, Jr, Lefkowitz EJ, Weaver CT, Rodriguez JM, Morrow CD. Colonization potential to reconstitute a microbe community in patients detected early after fecal microbe transplant for recurrent C. difficile. BMC microbiology. 2016;16:5. doi: 10.1186/s12866-015-0622-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson MJ, Walsh DCI. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecological Monographs. 2013;83:557–74. [Google Scholar]
- 41.Baselga A. Partitioning the turnover and nestedness components of beta diversity. Global Ecology and Biogeography. 2010;19:134–43. [Google Scholar]
- 42.Yazici C, Wolf PG, Kim H, Cross T-WL, Vermillion K, Carroll T, Augustus GJ, Mutlu E, Tussing-Humphreys L, Braunschweig C, Xicola RM, Jung B, Llor X, Ellis NA, Gaskins HR. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017 doi: 10.1136/gutjnl-2016-313321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bharwani A, Mian MF, Foster JA, Surette MG, Bienenstock J, Forsythe P. Structural & functional consequences of chronic psychosocial stress on the microbiome & host. Psychoneuroendocrinology. 2016;63:217–27. doi: 10.1016/j.psyneuen.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Michels N, Van de Wiele T, De Henauw S. Chronic Psychosocial Stress and Gut Health in Children: Associations With Calprotectin and Fecal Short-Chain Fatty Acids. Psychosomatic medicine. 2017;79:927–35. doi: 10.1097/PSY.0000000000000413. [DOI] [PubMed] [Google Scholar]
- 45.Hemmings SMJ, Malan-Muller S, van den Heuvel LL, Demmitt BA, Stanislawski MA, Smith DG, Bohr AD, Stamper CE, Hyde ER, Morton JT, Marotz CA, Siebler PH, Braspenning M, Van Criekinge W, Hoisington AJ, Brenner LA, Postolache TT, McQueen MB, Krauter KS, Knight R, Seedat S, Lowry CA. The Microbiome in Posttraumatic Stress Disorder and Trauma-Exposed Controls: An Exploratory Study. Psychosomatic medicine. 2017;79:936–46. doi: 10.1097/PSY.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golubeva AV, Crampton S, Desbonnet L, Edge D, O’Sullivan O, Lomasney KW, Zhdanov AV, Crispie F, Moloney RD, Borre YE, Cotter PD, Hyland NP, O’Halloran KD, Dinan TG, O’Keeffe GW, Cryan JF. Prenatal stress-induced alterations in major physiological systems correlate with gut microbiota composition in adulthood. Psychoneuroendocrinology. 2015;60:58–74. doi: 10.1016/j.psyneuen.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Gur TL, Bailey MT. Effects of Stress on Commensal Microbes and Immune System Activity. Advances in experimental medicine and biology. 2016;874:289–300. doi: 10.1007/978-3-319-20215-0_14. [DOI] [PubMed] [Google Scholar]
- 48.Qin HY, Cheng CW, Tang XD, Bian ZX. Impact of psychological stress on irritable bowel syndrome. World J Gastroenterol. 2014;20:14126–31. doi: 10.3748/wjg.v20.i39.14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rea K, Dinan TG, Cryan JF. The microbiome: A key regulator of stress and neuroinflammation. Neurobiology of Stress. 2016;4:23–33. doi: 10.1016/j.ynstr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D’Hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, Raes J. Population-level analysis of gut microbiome variation. Science. 2016;352:560–4. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 51.Maier KJ, al’Absi M. Toward a Biopsychosocial Ecology of the Human Microbiome, Brain-Gut Axis, and Health. Psychosomatic medicine. 2017;79:947–57. doi: 10.1097/PSY.0000000000000515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.