Abstract
Exposures to ionizing radiation (IR) may either be accidental or intentional, for medical purposes or even through terrorist actions. As certain populations emerge to be more radiosensitive than others, it is imperative to assess those individuals and treat them accordingly. To demonstrate the feasibility of rapid identification of such cases, we utilized the highly radiosensitive mouse model Atm−/− in the C57BL/6 background, and evaluated the urinary responses in 8–10 wk old male mice at early time points (4, 24, and 72h) after exposure to their respective LD50/30 doses [4 Gy for Atm−/−, and 8 Gy for wild type (WT)]. Urinary profiles from heterozygous animals exhibited remarkably similar responses to WT before and after radiation exposure. However, genotypic differences (WT or Atm−/−) were the primary driver to responses to radiation. Putative metabolites were validated through tandem mass spectrometry and included riboflavin, uric acid, D-ribose, D-glucose, pantothenic acid, taurine, kynurenic acid, xanthurenic acid, 2-oxoadipic acid, glutaric acid, 5′-deoxy-5′-methylthioadenosine, and hippuric acid. These metabolites mapped to several interconnected metabolic pathways which suggest that radiosensitive mouse models have underlying differences significantly impacting overall metabolism. This was further amplified by ionizing radiation at different time points. This study further emphasizes that genetically based radiosensitivity is reflected in the metabolic processes, and can be directly observed in urine. These differences in turn can potentially be used to identify individuals that may require altered medical treatment in an emergency radiological situation or modification of a regimen during a radiotherapy session.
Keywords: Metabolomics, Ataxia telangiectasia mutated, urine, ionizing radiation, biodosimetry
1. Introduction
Exposure to ionizing radiation, whether intentional or accidental, has increased the need for methods of rapid identification of exposed individuals. Incidents such as nuclear plant explosions (i.e. Chernobyl) or contamination of populations from mishandling of medical equipment (i.e. Goiânia, Brazil) [IAEA, 1988] highlight the importance for early and correct determination of the type and amount of radiation exposure one has received in order to rapidly and effectively provide appropriate medical treatment [NCRP, 2005]. This is particularly important in the case of a nuclear detonation, where thousands to millions of individuals will have to be rapidly assessed for radiation exposure [DiCarlo et al., 2011]. In addition, radiotherapy treatments, whether receiving partial or whole body irradiation, can lead to large delivered doses. During such treatments certain populations have emerged that exhibit higher than normal radiosensitivity. It is estimated that 2–4 % of humans have a variable degree of sensitivity to ionizing radiation (IR) that can also increase their risk of developing cancer after IR exposures [National Academy of Sciences, 2006], with particular importance for children who are in a more radiosensitive population. Radiotoxicity in these populations remains a primary concern for both short and long term. Identifying such individuals for biodosimetry or radiotherapy purposes is important for determination of effective treatment, whether through cytokine administration, transfusions, or modification of radiation regimen. The Health and Human Services (HHS) and the National Institute of Allergy and Infectious Diseases (NIAID) have designated under the biodosimetry program that rapid assessment of individuals after a radiological incident should begin at the earliest 24 h after the incident and continue in the first week, in order to provide immediate medical treatment that can save individuals from radiation-induced hematopoietic death, among other acute radiation exposure symptoms.
Metabolomics for radiation biodosimetry has been recognized as a powerful –omics approach to identify and quantify biomarkers through analysis of biofluids (urine, blood, saliva) and tissues in a minimally invasive manner [Coy et al., 2011; Pannkuk et al., 2017]. The collective assessment of small molecules (<1kDa) with technologies such as liquid chromatography (LC) and/or gas chromatography (GC) coupled with mass spectrometry (MS) can provide important information on the overall metabolic phenotype of an individual and responses to genotoxic agents. Studies for various radiation scenarios (dose, dose rate, external vs. internal emitters, radiation quality) have painted a picture of metabolic differences that can be used to investigate early outcome and possible delayed effects (reviewed in Pannkuk et al. [Pannkuk et al., 2017]). In addition, genotypic differences, such as in the Parp1−/− mouse model with defects in base excision repair, have shown a dramatic alteration to baseline metabolism and after exposure to IR [Laiakis et al., 2016]. Alterations in other signaling pathways involved in processes such as DNA repair are therefore expected to have unique metabolic fingerprints that can differentiate them from unirradiated or other genotypes based on construction of signatures, as has been recently demonstrated on a gene expression level in blood [Rudqvist et al., 2017].
Here, we extend our previous studies with metabolomics that explored the role of particular gene mutations in responses to ionizing radiation to include responses in the signaling pathway associated with ataxia telangiectasia mutated (Atm). Deficiency in ATM is associated with severe radiosensitivity in the human population and constitutes an extreme case of investigating genetic involvement to radiation injury. Atm has a multifaceted role in radiation responses through activation of DNA repair pathways, cellular metabolism [Shiloh and Ziv, 2013], and mitochondrial homeostasis [Valentin-Vega et al., 2012; Valentin-Vega and Kastan, 2012]. Atm patients are characterized by radiosensitivity, neurodegeneration, defects in T-cell maturation, and infertility. Although Atm homozygotes are well characterized, Atm heterozygotes are generally uncharacterized and carriers are undetermined, as they are phenotypically normal. Meta-analysis of data from carriers estimates that they have an increased risk of cancer and ischemic heart disease [van Os et al., 2016] and even modestly increased cellular radiosensitivity [National Academy of Sciences, 2006].
In this study, we analyzed urine from Atm−/− mice exposed to IR to identify metabolomic responses for the purpose of radiation biodosimetry, with time points relative to NIAID’s objectives (day 1 and 3 post irradiation). A very early time point (4h) provided further information on the kinetics of select biomarkers. LD50/30 doses were used for each genotype, based on previously published reports (8 Gy for WT and 4 Gy for Atm−/−). Baseline metabolomic profiles differed significantly between WT and Atm−/−, while heterozygotes resembled WT. Metabolic differences were identified as early as 4 h post IR and a combination of 5 metabolites was able to classify LD50/30 exposed WT from Atm−/− with high sensitivity and specificity. Utilizing extreme rare cases such as this for radiation biodosimetry can provide the basis for construction of biosignatures that can include results from genetic variants for a more comprehensive screening of individuals in a radiological event. Furthermore, this highlights the power of metabolomics to identify phenotypic differences based on small molecule analysis.
2. Materials and methods
2.1 Chemicals
All chemicals and solvents utilized were of the highest purity available. Water and acetonitrile were Optima LC/MS grade (Fisher Scientific Inc). Debrisoquine sulfate, 4-nitrobenzoic acid, and chlorpropamide were utilized as internal standards (Sigma-Aldrich®,St. Louis, MO). Riboflavin, kynurenic acid, pantothenic acid, uric acid, 2-oxoadipic acid, D-ribose, D-glucose, hippuric acid, glutaric acid, xanthurenic acid, and taurine were also purchased from Sigma-Aldrich® (St. Louis, MO). 5′-deoxy-5′-methylthioadenosine was purchased from Santa Cruz Biotechnology Inc (Dallas, TX).
2.2 Animal studies, irradiations, and sample collection
All initial breeders for WT C57BL/6 and Atm−/− (B6.129S6-Atmtm1Awb/J; Stock number 008536) were obtained from Jackson Laboratory and bred at Georgetown University. Mice for the experiments were obtained from Atm+/− x Atm+/− breeding pairs, therefore mice were littermates. All mice were male and 8–10 weeks old at the time of irradiation and all experiments were conducted according to approved protocols by the Georgetown University Institutional Animal Care and Use Committee (GUACUC). Whole body irradiations were conducted with gamma rays from a 137Cs source at a dose rate of ~1.45 Gy/min. Equitoxic LD50/30 doses were used for each genotype as previously published [Barlow et al., 1996; Budach et al., 1992; Laiakis et al., 2014], with 8 Gy for WT and 4 Gy for Atm. WT and Atm+/− (het) were also exposed to 4 Gy for equidose comparisons. Urine samples were collected with metabolic cages, as previously described [Laiakis et al., 2012], at 4 h, days 1 and 3 post irradiation, and stored at −80°C until analysis. The n of each group of mice is shown in Supplementary Table 1.
2.3 Sample processing and biomarker validation
Urine samples were prepared as previously described in detail in 50% acetonitrile: 50% water [Laiakis et al., 2012, 2016] with internal standards for monitoring of retention time drift. Quality control (QC) samples consisted of pooled urine samples, prepared in a similar manner, and run every 21 samples. Two μl of sample were injected into a Waters Acquity Ultra Performance Liquid Chromatography (UPLC) system coupled to a Xevo G2 time-of-flight mass spectrometry system (Waters Corporation, UK). A BEH C18 column, 130 Å was used for chromatography at 40° C. The chromatographic and MS conditions are presented in Supplementary Table 2. MS data were acquired in centroid mode (50 to 1200 Da) in both positive (ESI+) and negative (ESI-) ionization modes in MSE function, with intermittent injections of leucine enkephalin as Lockspray® for mass accuracy correction. For validation of putative biomarkers with tandem mass spectrometry (MS/MS), pure chemicals were diluted in 50% acetonitrile: 50% water to a concentration of 50 μM and fragmented with ramping collision energy. Fragmentation patterns from urine QC samples were compared to those from the pure standards and cross-referenced to MS/MS spectra in the METLIN library [Smith et al., 2005].
2.4 Data processing and analysis
Peak alignment and data deconvolution were performed with the software MarkerLynx® (Waters), and the ion intensities for each detected peak were normalized against the sum of the peak intensities within that sample. Normalization for each sample to account for changes in glomerular filtration rates was conducted to its respective creatinine levels [M+H]+=114.0667 with retention time 0.35 min. Multidimensional scaling plots (MDS) and heatmaps were generated with the machine learning algorithm Random Forests [Breiman, 2001] through R [Team, 2016] from the top 100 ranked ions. Twenty-five independent random forests were constructed with each random forest utilizing 10,000 trees to rank the ions according to importance.
Potential biomarkers were also identified with the statistical software MetaboLyzer [Mak et al., 2014], an informatics pipeline for analyzing untargeted metabolomics data. Briefly, complete-presence ions, which are defined as ions having a presence in at least 80% of the samples in both control and experimental groups, were analyzed via the non-parametric Mann-Whitney U test. Outliers were removed via 1.5 interquartile range (IQR) based filtering, and zero abundance values were also excluded during analysis. Partial-presence ions, which are defined as ions having an 80% presence in only one group, were analyzed categorically via the Barnard’s test. All p-values were corrected via the Benjamini-Hochberg step-up false discovery rate (FDR) procedure, and only ions with an FDR corrected p-value of < 0.10 were further scrutinized. Putative identities were obtained through the databases Kyoto Encyclopedia of Genes and Genomes (KEGG) [Kanehisa et al., 2014; Kanehisa and Goto, 2000], Human Metabolome Database (HMDB) [Wishart et al., 2013], and LIPID Metabolites and Pathways Strategy (LIPID MAPS®) [Sud et al., 2007], via neutral mass elucidation from the MS1 m/z values with a ppm error threshold of 10. Volcano plots (−log10 FDR corrected p-value vs. log2 fold change) were constructed from analysis of the complete-presence ions.
Graphical representation was conducted with the software Prism 6 (GraphPad Software, Inc.). The non-parametric Mann-Whitney U test (p<0.05 considered significant) through the same software was used to calculate the p-values reported in the figures, after outlier identification and removal with the ROUT testing and Q=0.1%, which is considered aggressive for removal of definitive outliers. Intergroup statistical analysis was performed with one-way ANOVA and Tukey’s multiple correction test (p<0.05 considered significant). Error bars are shown as standard error of the mean (SEM).
Receiver operating characteristic (ROC) curves and area under the curve (AUC) calculations were conducted through the software MetaboAnalyst [Xia et al., 2015; Xia and Wishart, 2011] by either including the full panel of identified and positively validated metabolites or a smaller subset. Features with >50% missing values were excluded, the data was Pareto scaled, and Random Forests were selected as the classification method and feature ranking method. An AUC value of >0.9 was considered excellent, while 0.8–0.9 was considered a good classification model and <0.8 fair.
2.5 Fold changes and statistically significant ion comparisons
No trend alterations were considered when fold changes were calculated between the limits of 0.85 and 1.15. These numbers were chosen to reflect at least a 15% change of levels (increased or decreased relative to control). Fold changes with increases between 1.15 and 2 were represented with ↑ and >2 with ↑↑. Fold changes with decreases between 0.85 and 0.5 were represented with ↓ and <0.5 [corresponding to −2 as calculated through –(1/fold change of 0.5)] with ↓↓. Statistically significant ion comparisons were based on the results from MetaboLyzer with the Mann-Whitney U test and Barnard’s test. Only ions that had at least one putative identification through HMDB, and therefore potential biological significance, were included in the analysis.
3. Results
MS1 data for each sample was normalized to their respective endogenous creatinine level. No statistically significant differences were observed between creatinine levels at 4 h, day 1 (D1), and day 3 (D3). Supplementary Figure 1 shows the creatinine levels for D1 between the different groups, as biomarker identification was based on this time point and expanded to the other two.
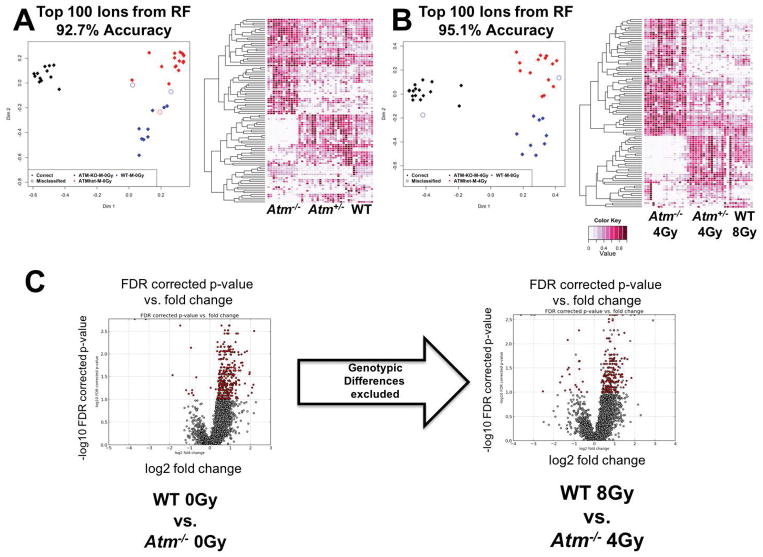
Random Forests analysis ranked the deconvoluted ions according to importance. Baseline analysis of metabolic differences between WT, Atm+/ − (het), and Atm−/− showed a clear separation on dimension 1, indicating that absence of a functional ATM is disruptive of the overall metabolism. The het group had a more similar metabolic pattern to WT (Figure 1A), with this pattern persisting after exposure to IR (Figure 1B). The similarities between the WT and het groups were further highlighted in depiction of the levels of the top 100 ranked ions with heatmaps (Figures 1A and 1B), where it appears that heterozygosity of this particular gene did not contribute to significant metabolomic changes, at least as determined in the urine. For this reason, het samples were not further analyzed. To graphically represent the most impactful putative metabolite changes in a two-group comparison, volcano plots were constructed, plotting the statistical significance of FDR corrected p-values vs. fold change (Figure 1C). WT vs. Atm−/− showed a shift towards a high statistical significance and fold change of metabolic levels, whether at basal levels or after exposure to equitoxic doses 4 Gy for Atm−/− and 8 Gy for WT.
Figure 1.
Multivariate data analysis at D1. Panels A and B: Multidimensional scaling plot (MDS) for basal levels (A) and post irradiation at day 1 (B). Blue indicates WT, red Atm+/−, and black Atm−/−. Both MDS plots were constructed through Random Forests of the top 100 ranked ions. These same ions were used to construct heatmaps, with darker colors indicating higher normalized abundance per sample. Panel C: Volcano plot of basal levels and responses to semi-lethal doses, following removal of basal genotypic differences. Statistically significant ions (FDR < 0.1, Mann-Whitney U test) are highlighted in red.
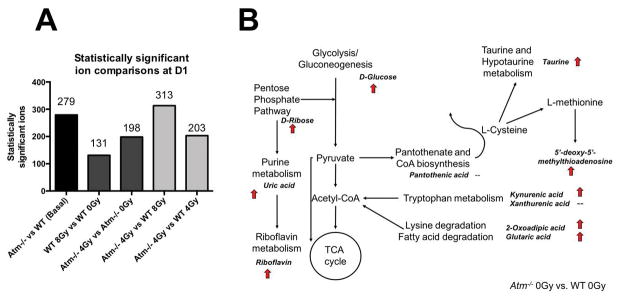
Univariate analysis was conducted via MetaboLyzer, an informatics pipeline for metabolomics, focusing on MS1 ions with HMDB assigned putative identities in order to elucidate biologically meaningful interpretations to the statistically significant results. A total of 279 putatively identified metabolites were found to be perturbed between WT and Atm−/−. Radiation responses in the two genotypes showed a clear higher response on the Atm−/− genotype (198 ions) compared to WT (131 ions). Following exposure to LD50/30 doses or doses that do not lead to death and initial removal of the baseline genotypic differences from the univariate analysis, it was evident that responses were primarily influenced by the underlying genotype instead of the stressor itself. Equitoxic doses revealed 313 statistically significant ions with a putative HMDB identity that were differentially excreted in urine, while equidose exposures showed 203 differentially excreted ions. The results are shown for comparison in Figure 2A.
Figure 2.
Panel A: Comparison of statistically significant ions with HMDB putative identifications, at basal levels and following IR at D1, from combination of results with the statistical tests Mann-Whitney U test and Barnard’s. Atm−/−, with the gene and protein defined as master regulators in signaling and metabolism, showed high basal metabolic differences from WT. Exposure to IR and removal of the basal levels of differential metabolites still resulted in a significant number of perturbed metabolites. Panel B: Identification of select metabolites and depiction in metabolic pathways is consistent with overall metabolic perturbations in Atm−/− compared to WT. Arrows are consistent with data presented in Table 2, representing an increase in fold change in Atm−/− of at least 1.15.
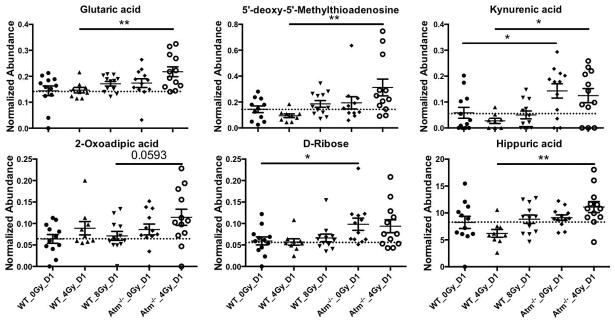
Further validation through MS/MS led to the positive identification of twelve metabolites. Table 1 describes the identified m/z and retention time, in addition to the major fragments from MS/MS and the metabolic pathways that are generally associated with each metabolite. In particular, riboflavin, kynurenic acid, xanthurenic acid, 5′-deoxy-5′-methylthioadenosine, and pantothenic acid were identified on ESI+. Uric acid, 2-oxoadipic acid, D-ribose, D-glucose, hippuric acid, glutaric acid, and taurine were identified on ESI-. These biomarkers are intermediates in various metabolic pathways that form an interconnected network, as depicted in Figure 2B. Baseline comparison clearly showed that the metabolism in Atm−/− animals is perturbed, with higher trends of excretion in those mice compared to WT. Calculated fold changes for genotypic differences and responses to radiation, trends, and FDR corrected p-values of univariate statistical analysis with the Mann-Whitney U test are shown in Table 2. Results showed that Atm−/− mice (with and without IR) exhibited a generalized higher response for all metabolites at D1 post IR when compared to WT. Kynurenic acid was the only biomarker that showed >2 fold change with or without IR. Interestingly, although trend analysis showed similar patterns between the equitoxic analysis and equidose analysis, it was the equidose analysis that revealed the highest statistically significant differences in these markers with p<0.05 at both D1 and D3. Some graphed examples of biomarkers at D1 are shown in Figure 3.
Table 1.
Validated biomarkers and MS/MS fragments
| Metabolite | Adduct | Ret. time | m/z Found (Precursor) | m/z Predicted | ppm error | MS/MS fragments | Major Metabolic Pathway Involvement | ||
|---|---|---|---|---|---|---|---|---|---|
| Fragment 1 | Fragment 2 | Fragment 3 | |||||||
| Riboflavin | [M+Na]+ | 2.91 | 399.1283 | 399.1275 | 2.0 | 359.1323 | 243.088 | 158.9626 | Riboflavin metabolism |
| Kynurenic acid | [M+H]+ | 1.45 | 190.0504 | 190.0499 | 2.6 | 172.0407 | 144.0456 | 116.0511 | Tryptophan metabolism |
| Xanthurenic acid | [M+H]+ | 1.19 | 206.0453 | 206.0448 | 2.5 | 188.0336 | 160.0382 | 132.0435 | Tryptophan metabolism |
| 5′-Deoxy-5′-Methylthioadenosine | [M+H]+ | 1.32 | 298.0981 | 298.0968 | 4.4 | 136.0616 | 119.0354 | - | Cysteine and methionine metabolism |
| Pantothenic acid | [M+H]+ | 0.79 | 220.1184 | 220.1180 | 2.0 | 202.1071 | 184.0965 | 90.0553 | beta-alainine metabolism, pantothenate and CoA biosynthesis |
| Uric acid | [M−H]− | 0.35 | 167.0208 | 167.0211 | 1.8 | 124.0147 | 96.0203 | 69.0097 | Purine metabolism |
| 2-Oxoadipic acid | [M−H]− | 0.39 | 159.0302 | 159.0299 | 1.9 | 115.0404 | 87.0459 | 59.015 | Lysine biosynthesis, tryptophan metabolism |
| D-Ribose | [M-H]− | 0.34 | 149.0451 | 149.045 | 0.7 | 89.0239 | 71.0143 | 59.0137 | Pentose phosphate pathway |
| D-Glucose | [M-H]− | 0.34 | 179.0552 | 179.0561 | 5.0 | 127.0377 | 92.0394 | 85.0281 | Glycolysis/ Gluconeogenesis |
| Hippuric acid | [M-H]− | 1.97 | 178.0501 | 178.051 | 5.1 | 134.0607 | 77.0399 | - | Phenylalanine metabolism |
| Glutaric acid | [M-H]− | 0.79 | 131.0345 | 131.035 | 7.6 | 113.0244 | 87.0458 | 69.0358 | Fatty acid degradation, lysine degradation |
| Taurine | [M-H]− | 0.32 | 124.0069 | 124.0074 | 4.0 | 79.9567 | - | - | Primary bile acid biosynthesis, taurine and hypotaurine metabolism |
Table 2.
Fold changes between genotypes and radiation doses.
| Fold changes - 4h time point | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | ATM−/ − 0Gy/ WT 0Gy | p-value | WT 8Gy/ WT 0Gy | p-value | ATM−/ − 4Gy/ ATM−/ − 0Gy | p-value | ATM−/ − 4Gy/ WT 8Gy | p-value | ATM−/ − 4Gy/ WT 4Gy | p-value | |||||
| Riboflavin | 0.78 | 0.66 | ⬇ | 0.99 | 0.87 | - | 1.36 | 0.52 | ⬆ | 1.08 | 0.92 | - | 1.87 | 0.03 | - |
| Kynurenic acid | 0.21 | 0.04 | ⬇ ⬇ | 0.51 | 0.08 | ⬇ | 13.73 | 0.07 | ⬆ ⬆ | 5.51 | 0.08 | ⬆ ⬆ | 2.42 | 0.84 | ⬆ ⬆ |
| Xanthurenic acid | 1.26 | 0.58 | ⬆ | 0.63 | 0.05 | ⬇ | 0.85 | 0.53 | - | 1.71 | 0.04 | ⬆ | 1.28 | 0.28 | ⬆ |
| 5′-Deoxy-5′-Methylthioadenosine | 0.74 | 0.21 | ⬇ | 1.73 | 0.26 | ⬆ | 2.54 | 0.14 | ⬆ ⬆ | 1.08 | 0.85 | - | 3.05 | 0.014 | ⬆ ⬆ |
| Pantothenic acid | 0.69 | 0.07 | ⬇ | 1.15 | 0.69 | - | 1.14 | 0.66 | - | 0.68 | 0.09 | ⬇ | 1.17 | 0.39 | ⬆ |
| Uric acid | 1.48 | 0.31 | ⬇ | 4.50 | 0.14 | ⬆ ⬆ | 1.03 | 0.99 | - | 0.34 | 0.28 | ⬇ ⬇ | 0.98 | 0.61 | - |
| 2-Oxoadipic acid | 0.84 | 0.39 | ⬇ | 0.96 | 0.97 | - | 0.76 | 0.74 | ⬇ | 0.67 | 0.23 | ⬇ | 0.57 | 0.07 | ⬇ |
| D-Ribose | 1.40 | 0.34 | ⬆ | 0.81 | 0.38 | ⬇ | 0.61 | 0.31 | ⬇ | 1.05 | 0.78 | - | 1.11 | 0.87 | - |
| D-Glucose | 1.36 | 0.47 | ⬆ | 0.96 | 0.52 | - | 0.64 | 0.47 | ⬇ | 0.90 | 0.96 | - | 1.01 | 0.87 | - |
| Hippuric acid | 1.02 | 0.96 | - | 0.90 | 0.69 | - | 0.73 | 0.59 | ⬇ | 0.83 | 0.67 | ⬇ | 0.70 | 0.6 | ⬇ |
| Glutaric acid | 0.91 | 0.89 | - | 1.06 | 0.87 | - | 0.68 | 0.31 | ⬇ | 0.59 | 0.28 | ⬇ | 0.68 | 0.07 | ⬇ |
| Taurine | 1.42 | 0.14 | ⬆ | 0.94 | 0.69 | - | 0.56 | 0.07 | ⬇ | 0.85 | 0.76 | ⬇ | 0.89 | 0.85 | - |
| Fold changes - D1 time point | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | ATM−/ − 0Gy/ WT 0Gy | p-value | WT 8Gy/ WT 0Gy | p-value | ATM−/ − 4Gy/ ATM−/ − 0Gy | p-value | ATM−/ − 4Gy/ WT 8Gy | p-value | ATM−/ − 4Gy/ WT 4Gy | p-value | |||||
| Riboflavin | 1.24 | 0.03 | ⬆ | 1.12 | 0.20 | - | 0.97 | 0.96 | - | 1.07 | 0.70 | - | 1.20 | 0.11 | ⬆ |
| Kynurenic acid | 2.44 | 0.03 | ⬆ ⬆ | 0.86 | 0.93 | - | 0.87 | 0.75 | - | 2.49 | 0.08 | ⬆ ⬆ | 4.57 | 0.03 | ⬆ ⬆ |
| Xanthurenic acid | 0.94 | 0.96 | - | 0.92 | 0.70 | - | 1.09 | 0.79 | - | 1.11 | 0.54 | - | 0.92 | 0.17 | - |
| 5′-Deoxy-5′-Methylthioadenosine | 1.38 | 0.60 | ⬆ | 1.32 | 0.31 | ⬆ | 1.61 | 0.10 | ⬆ | 1.68 | 0.18 | ⬆ | 3.33 | 0.0013 | ⬆ ⬆ |
| Pantothenic acid | 1.07 | 0.87 | - | 1.18 | 0.58 | - | 1.05 | 0.62 | - | 1.45 | 0.62 | ⬆ | 1.45 | 0.03 | ⬆ |
| Uric acid | 1.27 | 0.82 | ⬆ | 1.30 | 0.40 | ⬆ | 0.80 | 0.69 | ⬇ | 0.78 | 0.45 | ⬇ | 0.61 | 0.04 | ⬇ ⬇ |
| 2-Oxoadipic acid | 1.34 | 0.20 | ⬆ | 1.11 | 0.66 | - | 1.33 | 0.25 | ⬆ | 1.61 | 0.06 | ⬆ | 1.29 | 0.21 | ⬆ |
| D-Ribose | 1.66 | 0.04 | ⬆ | 1.12 | 0.74 | - | 0.95 | 0.66 | - | 1.41 | 0.18 | ⬆ | 1.64 | 0.11 | ⬆ |
| D-Glucose | 1.57 | 0.20 | ⬆ | 1.17 | 0.58 | ⬆ | 0.93 | 0.54 | - | 0.97 | 0.26 | - | 1.52 | 0.11 | ⬆ |
| Hippuric acid | 1.11 | 0.47 | - | 1.07 | 0.66 | - | 1.22 | 0.09 | ⬆ | 1.25 | 0.16 | ⬆ | 1.79 | 0.0018 | ⬆ |
| Glutaric acid | 1.21 | 0.34 | ⬆ | 1.19 | 0.37 | ⬆ | 1.25 | 0.18 | ⬆ | 1.27 | 0.09 | ⬆ | 1.48 | 0.0033 | ⬆ |
| Taurine | 1.23 | 0.37 | ⬆ | 1.18 | 0.26 | ⬆ | 0.93 | 0.74 | - | 0.97 | 0.68 | - | 0.92 | 0.5 | - |
| Fold changes - D3 time point | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | ATM−/ − 0Gy/ WT 0Gy | p-value | WT 8Gy/ WT 0Gy | p-value | ATM−/ − 4Gy/ ATM−/ − 0Gy | p-value | ATM−/ − 4Gy/ WT 8Gy | p-value | ATM−/ − 4Gy/ WT 4Gy | p-value | ||||||
| Riboflavin | 0.93 | 0.52 | - | 0.96 | 0.82 | - | 1.29 | 0.23 | ⬆ | 1.25 | 0.44 | ⬆ | 1.38 | 0.07 | ⬆ | |
| Kynurenic acid | 1.68 | 0.52 | ⬆ | 1.76 | 0.53 | ⬆ | 0.58 | 0.34 | ⬇ | 0.55 | 0.50 | ⬇ | 1.28 | 0.99 | ⬆ | |
| Xanthurenic acid | 1.14 | 0.60 | - | 0.96 | 0.96 | - | 1.35 | 0.29 | ⬆ | 1.61 | 0.10 | ⬆ | 1.52 | 0.18 | ⬆ | |
| 5′-Deoxy-5′-Methylthioadenosine | 2.74 | 0.68 | ⬆ | 2.95 | 0.46 | ⬆ | 0.31 | 0.52 | ⬇ ⬇ | 0.29 | 0.29 | ⬇ ⬇ | 1.18 | 0.99 | ⬆ | |
| Pantothenic acid | 1.25 | 0.30 | ⬆ | 1.05 | 0.74 | ⬆ ⬆ | 1.17 | 0.23 | ⬆ | 1.40 | 0.31 | ⬇ | 2.00 | 0.0087 | ⬆ ⬆ | |
| Uric acid | 1.25 | 0.07 | ⬆ | 0.67 | 0.29 | ⬇ | 2.17 | 0.36 | ⬆ ⬆ | 1.88 | 0.62 | ⬆ | 1.82 | 0.39 | ⬆ | |
| 2-Oxoadipic acid | 1.21 | 0.30 | ⬆ | 0.57 | 0.05 | ⬇ | 0.95 | 0.71 | - | 2.11 | 0.04 | ⬆ ⬆ | 1.27 | 0.18 | ⬆ | |
| D-Ribose | 0.92 | 0.29 | - | 0.75 | 0.25 | ⬇ | 1.05 | 0.92 | - | 1.23 | 0.52 | ⬆ | 1.49 | 0.02 | ⬆ | |
| D-Glucose | 0.66 | 0.05 | ⬇ | 0.83 | 0.25 | ⬇ | 0.92 | 0.60 | - | 0.79 | 0.43 | ⬇ | 0.98 | 0.9 | - | |
| Hippuric acid | 1.07 | 0.25 | - | 0.68 | 0.34 | ⬇ | 0.88 | 0.36 | - | 1.57 | 0.36 | ⬆ | 1.29 | 0.3 | ⬆ | |
| Glutaric acid | 1.53 | 0.02 | ⬆ | 0.90 | 0.96 | - | 1.04 | 0.92 | - | 1.71 | 0.04 | ⬆ | 1.63 | 0.02 | ⬆ | |
| Taurine | 0.70 | 0.45 | ⬇ | 1.19 | 0.25 | ⬆ | 0.84 | 0.23 | ⬇ | 0.58 | 0.02 | ⬇ | 0.54 | 0.0022 | ⬇ | |
Figure 3.
Normalized abundance levels shown in scatter plot format of glutaric acid, 5′-deoxy-5′-methylthioadenosine, kynurenic acid, 2-oxoadipic acid, D-ribose, and hippuric acid at D1 post IR. The dotted line is based on the mean of WT 0 Gy for easy visualization of the differences. All data is presented as mean ± SEM. Asterisks signify statistical significance, with * <0.05 and ** <0.01.
ROC curve analysis (Figure 4) with the combination of all twelve metabolites as a signature gave a high degree of confidence in identifying Atm−/− and WT both exposed to 4 Gy, with high sensitivity and specificity at all time points (AUC for 4h=0.903, AUC for D1=0.893, AUC for D3=0.942). However, the full signature did not fare well when equitoxic doses were compared (AUC for 4h=0.69, AUC for D1=0.662, AUC for D3=0.753), indicating that based on the collective biomarkers the signal that can distinguish between the two different genotypes may be weakened. By constructing ROC curves for the individual metabolites (data not shown), we selected the top five based on their individual values. For 4 h uric acid, 2-oxoadipic acid, kynurenic acid, xanthurenic acid, and pantothenic acid were combined to give an AUC of 0.7. At D1 2-oxoadipic acid, D-Ribose, hippuric acid, glutaric acid, and kynurenic acid were combined to give an AUC of 0.8. Finally, at D3, 2-oxoadipic acid, glutaric acid, taurine, xanthurenic acid, and 5′-deoxy-5′-methylthioadenosine were combined to give an AUC of 0.902. Therefore, by limiting the biosignature to a small number of metabolites we can increase the accuracy and sensitivity and specificity.
Figure 4.
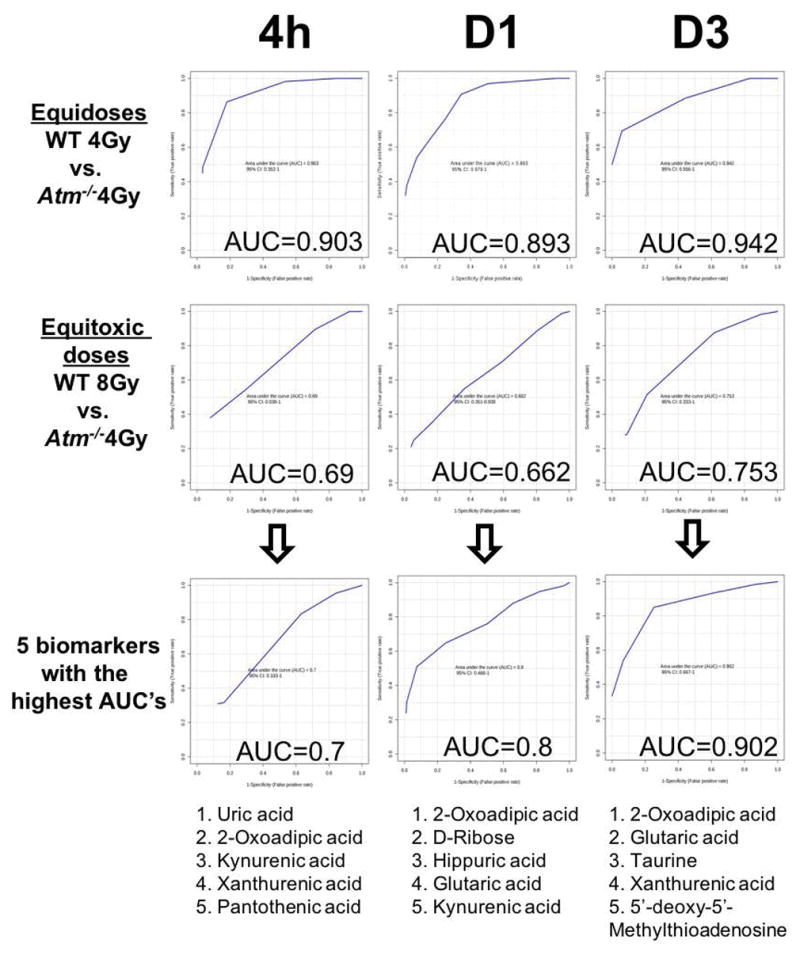
ROC curves and AUC values of equidose or equitoxic exposures with twelve identified metabolites over three time points. Equidose graphs showed a high sensitivity and specificity, while equitoxic doses showed an improvement by restricting the signature to the highest qualified metabolites (n=5).
4. Discussion
In this study we investigated the urinary metabolomic responses of Atm−/−, a genetic model that is well characterized for its increased radiosensitivity compared to WT mice. This represents an extreme model and although individuals exist in the population that carry such mutations, carriers are more prevalent. The radiosensitivity associated with the carriers however, remains largely unknown. However, increased chromosomal aberrations in heterozygotes have been documented following radiation exposure [Tchirkov et al., 1997] together with an increased risk of developing contralateral breast cancer [Bernstein et al., 2010, 2017]. For the purposes of biodosimetry or radiotherapy, a metabolic signature will need to be inclusive of all degrees of radiosensitivity. Therefore, here we demonstrate the effects a genetic mutation can have on metabolism and the variable responses compared to an equitoxic dose in WT mice. We previously analyzed urine from Parp1−/− mice [Laiakis et al., 2016] with defects in base excision repair, where we showed possible systemic perturbations in the tricarboxylic acid (TCA) cycle and other metabolites associated with energy metabolism, as observed in urine. We extended our studies to analyze urine from Atm−/− mice with a generalized defect in signaling post IR and Atm+/− to investigate whether heterozygosity manifests in an intermediate phenotype.
Overall baseline metabolic profiles levels of Atm−/−, Atm+/−, and WT and of irradiated mice revealed that the urinary profiles of the heterozygous mice were closer to WT and responded to radiation in a similar manner. Unlike cytogenetic parameters and epidemiological data that have identified a clear radiosensitivity of Atm carriers, this was not evident in urinary metabolomic data. Further research with other biofluid or tissue analysis may however reveal such a relationship. For our purposes however, no further analysis was conducted on Atm+/−.
Metabolites that were identified in this study mapped to a variety of metabolic pathways, and in a simplified way, shown in Figure 2B, exhibited an interconnected network of metabolic changes. Compared to our previously published study on Parp1−/− mice [Laiakis et al., 2016], alterations in the canonical energy metabolism pathway (TCA cycle) were not observed in urine. Therefore, it is possible to discriminate between genotypes associated with severe radiosensitive syndromes and their responses to IR with the use of global metabolomics and construction of highly targeted signatures. However, it should be emphasized that the responses to radiation are largely driven by the underlying genetics at the early time points post IR. Interestingly, when dissecting the kinetics for each biomarker in each time point, seen through fold changes in Table 2, there was a considerable shift to increased excretion of the majority of the biomarkers as time progressed, highlighting that Atm−/− mice show considerable metabolic dysregulation. This was also evident in the equitoxic comparison and in the equidose comparison, further indicating that the Atm−/− genotype is a significant contributing factor in the responses to IR. It is also possible that the inability to effectively mount a response to the radiation exposure, as Atm is a key regulator of signaling pathways that initiate processes such as DNA repair, may lead to increased apoptotic levels and elimination of such products in urine.
Limited metabolomic studies have investigated responses to radiation in these mouse models, and particularly in easily accessible biofluids such as urine. Two studies in ATM deficient isogenic cell lines [Jung et al., 2011; Varghese et al., 2010] showed perturbations in purine metabolism, linoleic acid metabolism, arginine and proline metabolism, and fructose and mannose metabolism amongst the most prominent altered metabolic pathways. The connection between ATM deficiency, radiation, and metabolic alterations are well known, including direct phosphorylation of key mitochondrial proteins and reactive oxygen species (ROS) regulation following exposure to genotoxic stressors [Shiloh and Ziv, 2013]. Such significant metabolic alterations were also indicated in our study, with increases in metabolites such as uric acid (anti-oxidant) from purine catabolism [Tasaki et al., 2017] and glutaric acid (pro-oxidant) from fatty acid degradation [Latini et al., 2007].
One metabolite, 5′-deoxy-5′-methylthioadenosine (MTA) has been implicated in processes such as apoptosis, differentiation, proliferation and even gene expression changes, as it can be metabolized to 5-methylthioribose-1-phosphate and adenine, further associated with both methionine and purine metabolism [Avila et al., 2004; Mills and Mills, 1985]. Elevated levels in urine post IR in the early time points (4h and D1) may be directly related to downstream deficient DNA repair, as cleavage of MTA provides adenine endogenously [Fitchen et al., 1986] and lowering of the endogenous pool due to loss may contribute to errors. The increased levels of this metabolite at the early time points, as compared to the levels in the equidose WT group, indicate that apoptosis-related metabolites are perturbed to a significant level in the Atm−/− genotype compared to WT. As apoptosis is initiated by high IR exposure as one of the steps in the IR response, products of such can be reflected in urine at time points as early as 4 h post IR.
Finally, although the overall endogenous metabolism in highly perturbed in the Atm−/− mice, the pattern is clear that there is elevated excretion of important metabolic intermediates. Although in some cases, i.e. equidose exposures, a large panel of biomarkers may offer high classification between the groups, in other cases a smaller panel may provide increased classification power, as seen in our study. Given that metabolomic analysis is a rapid and high throughput method, the incorporation of multiple biomarkers in a radiation signature is feasible and could be accurately used in an emergency situation to effectively provide medical specialized treatments. In another scenario, monitoring of radiation responses in radiotherapy patients that receive high doses may lead to modifications of radiotherapy regimens to protect normal tissue. Such investigation will be of particular importance in individuals receiving whole body irradiation prior to hematopoietic stem cell transplantation, particularly in pediatric populations. The genotype chosen in this study constitutes an extreme of radiosensitivity and therefore it remains to be answered whether genetics that confer intermediate radiosensitivity may also be identified through metabolomics methods in other biofluids or tissues.
Supplementary Material
Creatinine levels at all time points. No statistically significant differences were observed except for the intergroup comparison at 4h of WT 4 Gy and Atm−/− 0 Gy, therefore creatinine was used for normalization.
Acknowledgments
Funding
This work was funded by the National Institutes of Health (National Institute of Allergy and Infectious Diseases) grant U19 AI067773 (P.I. David J. Brenner, performed as part of Columbia University Center for Medical Countermeasures against Radiation). The project described above was also supported by Award Number P30 CA051008 (P.I. Louis Weiner) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
We would like to thank Ms. Kirandeep Gill from the Georgetown University Lombardi Comprehensive Cancer Center Metabolomics Shared Resource for data acquisition.
Footnotes
Author contribution
ECL and AJF were responsible for designing the experiment. PA was responsible for mouse breeding and mouse handling. BHM performed the genotyping and assisted with irradiations. ECL, SJS, YWW, and PA performed the irradiations and collected the samples. TDM was involved with the statistical analysis. ECL interpreted the MS data and wrote the manuscript. All authors provided comments and approved the final submitted version.
Conflict of interest statement
The authors declare no competing financial interest.
References
- Avila MA, García-Trevijano ER, Lu SC, Corrales FJ, Mato JM. Methylthioadenosine. Int J Biochem Cell Biol. 2004;36:2125–2130. doi: 10.1016/j.biocel.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Bernstein JL, Haile RW, Stovall M, Boice JD, Shore RE, Langholz B, Thomas DC, Bernstein L, Lynch CF, Olsen JH, et al. Radiation exposure, the ATM Gene, and contralateral breast cancer in the women’s environmental cancer and radiation epidemiology study. J Natl Cancer Inst. 2010;102:475–483. doi: 10.1093/jnci/djq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein JL, WECARE SCG, Concannon P. ATM, radiation, and the risk of second primary breast cancer. Int J Radiat Biol. 2017;93:1121–1127. doi: 10.1080/09553002.2017.1344363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Budach W, Hartford A, Gioioso D, Freeman J, Taghian A, Suit HD. Tumors arising in SCID mice share enhanced radiation sensitivity of SCID normal tissues. Cancer Res. 1992;52:6292–6296. [PubMed] [Google Scholar]
- Coy SL, Cheema AK, Tyburski JB, Laiakis EC, Collins SP, Fornace A. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol. 2011;87:802–823. doi: 10.3109/09553002.2011.556177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo AL, Maher C, Hick JL, Hanfling D, Dainiak N, Chao N, Bader JL, Coleman CN, Weinstock DM. Radiation injury after a nuclear detonation: medical consequences and the need for scarce resources allocation. Disaster Med Public Health Prep. 2011;5(Suppl 1):S32–44. doi: 10.1001/dmp.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchen JH, Riscoe MK, Dana BW, Lawrence HJ, Ferro AJ. Methylthioadenosine phosphorylase deficiency in human leukemias and solid tumors. Cancer Res. 1986;46:5409–5412. [PubMed] [Google Scholar]
- I.A.E.A. The radiological accident in Goiânia 1988 [Google Scholar]
- Jung M, Timofeeva O, Cheema AK, Varghese R, Ressom H, Dritschilo A. Human fibroblasts for large-scale “omics” investigations of ATM gene function. Adv Exp Med Biol. 2011;720:181–190. doi: 10.1007/978-1-4614-0254-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiakis EC, Hyduke DR, Fornace AJ. Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors γ radiation and lipopolysaccharide. Radiat Res. 2012;177:187–199. doi: 10.1667/rr2771.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiakis EC, Pannkuk EL, Diaz-Rubio ME, Wang YW, Mak TD, Simbulan-Rosenthal CM, Brenner DJ, Fornace AJ. Implications of genotypic differences in the generation of a urinary metabolomics radiation signature. Mutat Res. 2016;788:41–49. doi: 10.1016/j.mrfmmm.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiakis EC, Strassburg K, Bogumil R, Lai S, Vreeken RJ, Hankemeier T, Langridge J, Plumb RS, Fornace AJ, Astarita G. Metabolic phenotyping reveals a lipid mediator response to ionizing radiation. J Proteome Res. 2014;13:4143–4154. doi: 10.1021/pr5005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini A, Ferreira GC, Scussiato K, Schuck PF, Solano AF, Dutra-Filho CS, Vargas CR, Wajner M. Induction of oxidative stress by chronic and acute glutaric acid administration to rats. Cell Mol Neurobiol. 2007;27:423–438. doi: 10.1007/s10571-006-9134-9. [DOI] [PubMed] [Google Scholar]
- Mak TD, Laiakis EC, Goudarzi M, Fornace AJ. MetaboLyzer: a novel statistical workflow for analyzing Postprocessed LC-MS metabolomics data. Anal Chem. 2014;86:506–513. doi: 10.1021/ac402477z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GC, Mills JS. Urinary excretion of methylthioadenosine in immunodeficient children. Clin Chim Acta. 1985;147:15–23. doi: 10.1016/0009-8981(85)90005-1. [DOI] [PubMed] [Google Scholar]
- National Academy of Sciences. Health Risks from exposure to low levels of ionizing radiation BEIR VII Phase 2. 2006 [PubMed] [Google Scholar]
- NCRP. Commentary No. 19- Key elements of preparing emergency responders for nuclear and radiological terrorism 2005 [Google Scholar]
- Pannkuk EL, Fornace AJ, Laiakis EC. Metabolomic applications in radiation biodosimetry: exploring radiation effects through small molecules. Int J Radiat Biol. 2017:1–26. doi: 10.1080/09553002.2016.1269218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudqvist N, Laiakis EC, Ghandhi SA, Kumar S, Knotts JD, Chowdhury M, Fornace AJ, Jr , Amundson SA. Global gene expression response in mouse models of DNA repair deficiency after gamma irradiation. 2017 doi: 10.1667/RR14862.1. Submitted to Radiation Research journal (Accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Murphy RC, Raetz CR, Russell DW, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–32. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki E, Sakurai H, Nitao M, Matsuura K, Iuchi Y. Uric acid, an important antioxidant contributing to survival in termites. PLoS One. 2017;12:e0179426. doi: 10.1371/journal.pone.0179426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchirkov A, Bay JO, Pernin D, Bignon YJ, Rio P, Grancho M, Kwiatkowski F, Giollant M, Malet P, Verrelle P. Detection of heterozygous carriers of the ataxia-telangiectasia (ATM) gene by G2 phase chromosomal radiosensitivity of peripheral blood lymphocytes. Hum Genet. 1997;101:312–316. doi: 10.1007/s004390050634. [DOI] [PubMed] [Google Scholar]
- Team, R.C. R Foundation for statistical computing, 2015. Vienna, Austria: 2016. A language and environment for statistical computing. [Google Scholar]
- Valentin-Vega YA, Kastan MB. A new role for ATM: regulating mitochondrial function and mitophagy. Autophagy. 2012;8:840–841. doi: 10.4161/auto.19693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Maclean KH, Tait-Mulder J, Milasta S, Steeves M, Dorsey FC, Cleveland JL, Green DR, Kastan MB. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Os NJ, Roeleveld N, Weemaes CM, Jongmans MC, Janssens GO, Taylor AM, Hoogerbrugge N, Willemsen MA. Health risks for ataxia-telangiectasia mutated heterozygotes: a systematic review, meta-analysis and evidence-based guideline. Clin Genet. 2016;90:105–117. doi: 10.1111/cge.12710. [DOI] [PubMed] [Google Scholar]
- Varghese RS, Cheema A, Cheema P, Bourbeau M, Tuli L, Zhou B, Jung M, Dritschilo A, Ressom HW. Analysis of LC-MS data for characterizing the metabolic changes in response to radiation. J Proteome Res. 2010;9:2786–2793. doi: 10.1021/pr100185b. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Creatinine levels at all time points. No statistically significant differences were observed except for the intergroup comparison at 4h of WT 4 Gy and Atm−/− 0 Gy, therefore creatinine was used for normalization.