Abstract
PURPOSE
On computed tomography (CT), intrahepatic cholangiocarcinomas (ICC) are a visibly heterogeneous group of tumors. The purpose of this study was to investigate the associations between CT imaging phenotypes, patient survival and known genetic markers.
METHODS
A retrospective study was performed with 66 patients with surgically resected ICC. Pre-surgical CT images of ICC were assessed by radiologists blinded to tumor genetics and patient clinical data. Associations between qualitative imaging features and overall survival (OS) and disease free survival (DFS) were performed with Cox proportional hazards regression and visualized with Kaplan-Meier plots. Associations between radiographic features and genetic pathways (IDH1, Chromatin and RAS-MAPK) were assessed with Fisher's Exact test and the Wilcoxon Rank sum test where appropriate and corrected for multiple comparisons within each pathway using the False Discovery Rate correction.
RESULTS
Three imaging features were significantly associated with a higher risk of death: necrosis (hazard ratio (HR):2.95 – 95%CI:1.44–6.04, p=0.029), satellite nodules (HR:3.29, 95% confidence interval (CI):1.35–8.02, p=0.029), and vascular encasement (HR:2.63, 95%CI:1.28–5.41, p=0.029). Additionally, with each increase in axial size, the risk of death increased (HR:1.14, 95%CI:1.03–1.26, p=0.029). Similar to findings for OS, satellite nodules (HR:3.81, 95%CI:1.88–7.71, p=0.002) and vascular encasement (HR:2.25, 95%CI:1.24–4.06, p=0.019) were associated with increased risk of recurrence/death. No significant associations were found between radiographic features and genes in the IDH1, Chromatin or RAS-MAPK pathways (p=0.63–84).
CONCLUSION
This preliminary analysis of resected ICC suggests associations between CT imaging features and OS and DFS. No association was identified between imaging features and currently known genetic pathways.
Keywords: cholangiocarcinoma, radiomics, radiogenomics, survival
Introduction
Intrahepatic cholangiocarcinoma (ICC) belongs to a subset of bile duct tumors that are mass-forming and arise distal to the second order bile ducts [1–3]. It is the second most common primary liver cancer worldwide after hepatocellular carcinoma (HCC), with a rising incidence [4]. Unresectable cholangiocarcinoma has a poor prognosis, without treatment yielding a median survival of only 3.9 months [5]. Resection is the only known effective cure and even then recurrence is frequent with a reported 58% recurrence rate in a cohort of 189 patients, most commonly occurring in the first 24 months [6]. The role of the radiologist pre-resection is to describe imaging features contributing to stage and prognosis including tumor size, vascular invasion, periductal infiltration and presence of regional metastatic adenopathy or distant metastases [7].
While radiologists play a central role in diagnosing and staging liver tumors, recent efforts have shifted towards extracting more prognostic information from medical imaging [8,9]. High-throughput extraction of quantitative features can be performed to convert images into mineable data to be analyzed for prognostic biomarkers, a process termed radiomics [9]. Alternatively, qualitative evaluations by radiologists using a standard lexicon may also be used to annotate images prospectively and contribute to mineable databases, for example, by using terminology developed by the Liver Imaging Reporting and Data System (LI-RADS) [10].
Among ICCs, a range of imaging appearances are evident, from small hypervascular tumors mimicking HCC, to larger tumors with desmoplastic centers, as well as infiltrative and necrotic appearing ICC [2]. Qualitative descriptors of ICC may have prognostic value, as a number of studies have identified the potential of tumoral vascularity in differentiating short and long term survivors [11–14].
Given the imaging heterogeneity of ICC observable on CT and MRI, and recent advances in our genetic understanding of this tumor [15–18], we hypothesized that qualitative descriptors of ICC may correlate with different genetic subtypes of ICC. The purpose of our study was to correlate radiologic phenotypes of ICC on contrast enhanced CT with known genetic pathways and survival outcomes in surgical patients.
Materials and Methods
Patient Population
The study was approved by our Institutional Review Board and was compliant with the Health Insurance Portability and Accountability Act. A retrospective review was performed to identify patients with resected mass-forming ICC from January 1993 to December 2014, who had genetic mutation profiling and available pre-surgical CT imaging available for evaluation on a picture archiving and communications system (PACS – GE Centricity, Milwaukee, WI). Pathologic specimens included surgical tissue resections (n = 63 cases) and core biopsy samples (n = 3 cases). Before treatment, patients had given informed consent for review of medical records, imaging, pathologic and genetic data for correlated research. 61 of 66 (92%) patients had no prior therapy prior to tumor sampling, with the remaining five receiving chemotherapy.
CT Image Acquisition
The CT studies selected for review were the most recently acquired study prior to the specimen date, including both CTs performed at our own institution (n=51, 77%) and those performed at other institutions (n=15, 23%), all uploaded to PACS as complete Digital Imaging and Communication in Medicine (DICOM) imaging set. These CT studies were performed on a variety of scanner models all with multidetector CT (16 to 64 slices) and included single phase (n=12, 18%) and multi-phase post contrast imaging (n=54, 82%). Arterial phase imaging was available in 52/66 CTs. Delayed phase imaging was available in 6 CTs. All scans had maximal axial slice thicknesses of 5 mm.
Image Analysis
Imaging features were reviewed in consensus by [blinded for review] and [blinded for review], who were blinded to clinical and genomic data. The radiologists were aware that each patient had a pathologically proven ICC.
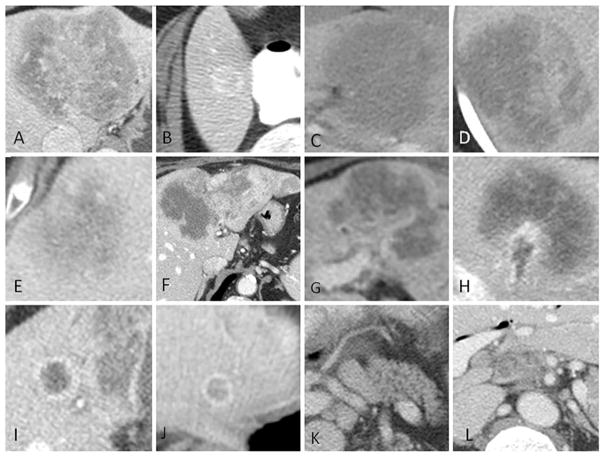
Lesion size was measured on the imaging phase on which the lesion was most clearly delineated both as the largest axial size and the perpendicular axial size (or short axis). The presence or absence of the following LI-RADS imaging features (v 2014) were assessed: washout appearance, mosaic architecture, disproportionate biliary obstruction, central necrosis, liver surface retraction, tumor in vein. Other imaging features included in the analysis were: lesion border (well defined or infiltrative), enhancement pattern (homogeneous or heterogeneous), central fibrosis, vascular encasement without thrombus (present or absent), hepatolithiasis (present or absent), peri-lesional perfusional abnormality (present or absent), satellite nodules - defined as tumors within 1 cm of the primary tumor border (present or absent), intrahepatic metastases - defined as tumors >1 cm from the primary tumor (present or absent) and adenopathy (absent - short axis < 1.0 cm, present - short axis > 1.0 cm and non-specific, or present and suspicious for nodal metastases) (Figure 1).
Fig. 1.
Imaging features of intrahepatic cholangiocarcinoma visually assessed on CT Examples of ICC with A) central fibrosis, B) well-defined borders, C) homogeneous enhancement and infiltrative borders, D) heterogeneous enhancement, E) central necrosis, F) liver surface retraction, G) vascular encasement without thrombus, H, tumor in vein, I) satellite nodules, J) intrahepatic metastasis, K) non-specific adenopathy, L) suspicious adenopathy.
Genetic Analysis
Genetic mutation profiling was completed using the [blinded for review], a customized array of 341 cancer-associated genes with subsequent expansion to 410 cancer-associated genes [reference blinded for review]. The most commonly occurring genetic alterations were identified and grouped by previously identified cancer pathways and families as follows: IDH1, chromatin-remodeling gene family (BAP2, ARID1A, PBRM1), and KRAS/RASA1 [19–21].
Statistical Analysis
Radiologic characteristics were summarized, and continuous characteristics and categorical characteristics with at least 10 patients in each category were considered for analysis. The associations between radiologic features and genetic pathways were assessed with Fisher's Exact test and the Wilcoxon Rank Sum test, where appropriate, and corrected for multiple comparisons within each pathway using the False Discovery Rate correction. Adjusted p-values less than 0.05 were considered for multivariate analysis.
We also compared radiographic features to overall (OS) and disease free (DFS) survival with Cox proportional hazards regression. OS was calculated from the time of resection until death. Patients alive at last follow up were censored. DFS was calculated from resection until recurrence or death. Patients alive and disease free at last follow up were censored. Univariate p-values were corrected using the false discovery rate approach (FDR), and adjusted p-values less than 0.05 were considered for multivariate analysis. Multicollinearity was examined. The multivariate model discrimination ability was assessed with Gonen and Heller's K, also known as the Concordance Probability Estimate. All analyses were performed with SAS 9.4 (The SAS Institute, Cary, NC).
Results
Patient Characteristics and Radiologic Features
Sixty-six patients (41 female, 25 male) met the inclusion criteria with a median age at diagnosis of 64.5 years (range 29 – 87 years). Tumor sizes ranged from 1.6 to 15.9 cm in maximal axial dimension (Table 1). A majority of ICC had well defined borders (n=55, 83%) with heterogeneous enhancement patterns (n=60, 90.9%), and lacked evidence of central necrosis (n=41, 62.1%) (Table 1). A minority of our patients had evidence of advanced disease, such as adenopathy (n=23, 34.9%), satellite nodule (n= 12, 18.2%), intrahepatic metastasis (n=4, 6.1%), and tumor in vein (n=1, 1.5%). Of the 23 patients with adenopathy, 11 were assessed as suspicious for nodal metastases (16.7%) and 12 were deemed non-specific (18.2%).
Table 1.
Imaging Features of ICC
| CT Feature | N (%) | |
|---|---|---|
| Adenopathy | Present-non-specific | 12 (18.2) |
| Present-Suspicious | 11 (16.7) | |
| Absent | 43 (65.2) | |
| Disproportionate Biliary Obstruction | Present | 12 (18.2) |
| Absent | 54 (81.8) | |
| Border | Well-Defined | 55 (83.3) |
| Infiltrative | 11 (16.7) | |
| Central Fibrosis | Present | 25 (37.9) |
| Absent | 41 (62.1) | |
| Enhancement | Homogeneous | 6 (9.1) |
| Heterogeneous | 60 (90.9) | |
| Hepatolithiasis | Present | 4 (6.1) |
| Absent | 62 (93.9) | |
| Intrahepatic Metastasis | Present | 4 (6.1) |
| Absent | 62 (93.9) | |
| Liver Surface Retraction | Present | 36 (54.5) |
| Absent | 30 (45.5) | |
| Central necrosis | Present | 25 (37.9) |
| Absent | 41 (62.1) | |
| Perilesion Perfusion Abnormality | Present | 20 (30.3) |
| Absent | 46 (69.7) | |
| Satellite Nodule | Present | 12 (18.2) |
| Absent | 54 (81.8) | |
| Largest Axial Size, cm | Median (range) (N=66) | 5.90 (1.60–15.9) |
| Perpendicular Axial Size, cm | Median (range) (N=66) | 4.35 (1.00–13.3) |
| Tumor in Vein | Present | 1 (1.5) |
| Absent | 65 (98.5) | |
| Vascular Encasement | Present | 20 (30.3) |
| Absent | 46 (69.7) | |
| Mosaic Architecture | Absent | 66 (100) |
| Washout | Absent | 52 (78.8) |
| NA | 14 (21.2) |
Numbers represent frequency with percent of total sample in parentheses, unless otherwise specified
Correlation with Gene Pathways
Median time between CT and IMPACT was 0.6 months (range: 0.1–6.2 months). The most commonly identified families and pathways, which were expressed in 10 or more patients in our study population, were IDH1, Chromatin remodelling gene family and KRAS/RASA1 pathways (manuscript in preparation by (blinded for review)). No significant associations were found between radiologic imaging features and genes in the IDH1 pathway (p=0.70–>0.95), Chromatin remodelling Pathway (p=0.59–>0.95) or KRAS/RASA1 pathway (p=0.63–84) (Table 2).
Table 2.
Imaging Features and Genetic Pathways
| IDH1 Pathway Mutation | Chromatin Pathway Mutation | KRAS/RASA1 Pathway Mutation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No | Yes | p-value | No | Yes | p-value | No | Yes | p-value | ||
| Adenopathy | Absent | 34 (66.7) | 9 (60) | 0.94 | 25 (71.4) | 18 (58.1) | 0.72 | 32 (68.1) | 11 (57.9) | 0.63 |
| Non-specific | 8 (15.7) | 4 (26.7) | 7 (20) | 5 (16.1) | 6 (12.8) | 6 (31.6) | ||||
| Suspicious | 9 (17.6) | 2 (13.3) | 3 (8.6) | 8 (25.8) | 9 (19.1) | 2 (10.5) | ||||
| Disproportionate Biliary Obstruction | Absent | 43 (84.3) | 11 (73.3) | 0.70 | 28 (80) | 26 (83.9) | 0.93 | 40 (85.1) | 14 ( 73.7) | 0.63 |
| Present | 8 (15.7) | 4 (26.7) | 7 (20) | 5 (16.1) | 7 (14.9) | 5 (26.3) | ||||
| Border | Well-Defined | 42 (82.4) | 13 (86.7) | >0.95 | 28 (80) | 27 (87.1) | 0.82 | 42 ( 89.4) | 13 (68.4) | 0.63 |
| Infiltrative | 9 (17.6) | 2 (13.3) | 7 (20) | 4 (12.9) | 5 (10.6) | 6 (31.6) | ||||
| Central Fibrosis | Absent | 29 (56.9) | 12 (80) | 0.70 | 20 (57.1) | 21 (67.7) | 0.82 | 28 ( 59.6) | 13 (68.4) | 0.64 |
| Present | 22 (43.1) | 3 (20) | 15 (42.9) | 10 (32.3) | 19 (40.4) | 6 (31.6) | ||||
| Liver Surface Retraction | Absent | 20 (39.2) | 10 (66.7) | 0.70 | 16 (45.7) | 14 (45.2) | >0.95 | 19 (40.4) | 11 (57.9) | 0.63 |
| Present | 31 (60.8) | 5 (33.3) | 19 (54.3) | 17 (54.8) | 28 (59.6) | 8 (42.1) | ||||
| Necrosis | Absent | 31 (60.8) | 10 (66.7) | 0.94 | 20 (57.1) | 21 (67.7) | 0.82 | 40 (85.1) | 14 (73.7) | 0.63 |
| Present | 20 (39.2) | 5 (33.3) | 15 (42.9) | 10 (32.3) | 7 (14.9) | 5 (26.3) | ||||
| Perilesion Perfusion Abnormality | Absent | 35 (68.6) | 11 (73.3) | >0.95 | 24 (68.6) | 22 (71) | >0.95 | 42 (89.4) | 13 (68.4) | 0.63 |
| Present | 16 (31.4) | 4 (26.7) | 11 (31.4) | 9 (29) | 5 (10.6) | 6 (31.6) | ||||
| Satellite Nodule | Absent | 43 (84.3) | 11 (73.3) | 0.70 | 28 (80) | 26 (83.9) | 0.93 | 28 (59.6) | 13 (68.4) | 0.64 |
| Present | 8 (15.7) | 4 (26.7) | 7 (20) | 5 (16.1) | 19 (40.4) | 6 (31.6) | ||||
| Largest Axial Size, cm | Median (range) | 5.9 (1.6–15.9) | 5.3 (1.7–11.3) | 0.70 | 6.5 (1.6–15.9) | 5.2 (2.2–11.8) | 0.59 | 40 (85.1) | 14 (73.7) | 0.63 |
| Perpendicular Axial Size, cm | Median (range) | 4.30 (1.0–13.3) | 4.4 (1.5–7.0) | 0.70 | 4.4 (1.0–13.3) | 3.9 (1.5–8.2) | 0.82 | 7 (14.9) | 5 (26.3) | |
| Vascular Encasement | Absent | 37 (72.5) | 9 (60) | 0.70 | 21 (60) | 25 (80.6) | 0.59 | 42 (89.4) | 13 (68.4) | 0.64 |
| Present | 14 (27.5) | 6 (40) | 14 (40) | 6 (19.4) | 5 (10.6) | 6 (31.6) | ||||
P-values were adjusted for multiple hypotheses using the false discovery rate correction
Correlation with Survival
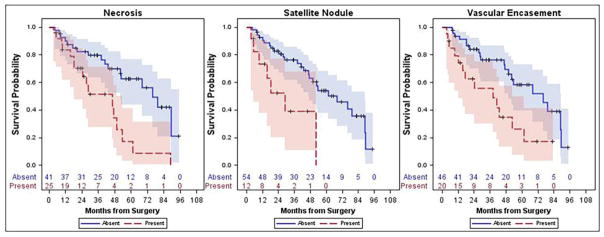
Overall, median OS was 53 months (95%CI: 43–79) with a 5-year OS of 48% (95%CI: 33–62%), and median DFS was 17 months (95%CI: 10–33) with a 5-year DFS of 26% (95%CI: 16–38%). In survivors, Median follow up was 41 months (range 5 – 95 months). Radiologic features were examined in association with both OS and DFS. Presence of central necrosis (hazard ratio (HR): 2.95, 95%CI: 1.44–6.04, p=0.029), satellite nodules (HR: 3.29, 95%CI: 1.35–8.02, p=0.029), and vascular encasement (HR: 2.63, 95%CI: 1.28–5.41, p=0.029) were all associated with a higher risk of death (Figure 2). Additionally, with each increase in axial size, the risk of death increased (HR: 1.14, 95%CI: 1.03–1.26, p=0.029). No other factors were significantly associated with OS (p=0.07–0.88); however perpendicular axial size approached significance (HR: 1.14, 95%CI: 1.01–1.28, p=0.07). In multivariate analysis, vascular encasement (HR: 2.52, 95%CI: 1.11–5.71, p=0.027) and necrosis (HR: 3.18, 95%CI: 1.29–7.86, p=0.012) remained significant independent predictors of OS. Presence of satellite nodules approach significance (HR: 2.57, 95%CI: 0.96–6.84, p=0.06) as well; however, axial size was not associated with OS in the multivariate model (HR: 0.97, 95%CI: 0.85–1.11, p=0.64).
Fig. 2.
Kaplan- Meier survival curves which illustrate that the presence of necrosis, satellite nodules and vascular encasement were all associated with decreased survival.
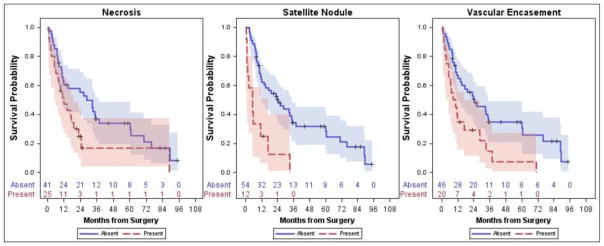
Similar to findings for OS, satellite nodules (HR: 3.81, 95%CI: 1.88–7.71, p=0.002) and vascular encasement (HR: 2.25, 95%CI: 1.24–4.06, p=0.019) were associated with reduced DFS (Figure 3). Additionally, with each increase in axial size, the risk of death increased (HR: 1.17, 95%CI: 1.07–1.28, p=0.003) and similarly for perpendicular axial size (HR: 1.17, 95%CI: 1.05–1.31, p=0.017). No other factors were significantly associated with DFS (p=0.06–0.88); however, central necrosis approached significance (HR: 1.95, 95%CI: 1.07–3.54, p=0.06). Given the strong correlation between axial size and perpendicular axial size, only axial size was included in the multivariate model. In the multivariate model, presence of a satellite nodule (HR: 2.39, 95%CI: 1.02–5.60, p=0.045) and largest axial size (HR: 1.12, 95%CI: 1.02–1.23, p=0.020) remained significantly associated with DFS.
Fig. 3.
Kaplan- Meier survival curves which illustrate that the presence of satellite nodules and vascular encasement were associated with decreased disease-free survival.
Discussion
The imaging appearance of ICC is known to be heterogeneous across patients prompting an interest in whether the phenotypic heterogeneity is predictive of biology such as tumor genetics or patient prognosis. The ability to predict tumor biology based on preoperative imaging alone could potentially benefit patient management and alter the consideration for surgery or neoadjuvant therapy. This study demonstrates an association between qualitative imaging features and survival in patients with resectable ICC. We identified that the presence of necrosis and vascular encasement were predictive of increased risk of death, while satellite nodule and largest axial dimension were predictive of DFS. It has previously been described that arterial enhancement of ICC on CT appears to correlate with DFS in surgically resected patients [11,13,14]. Our results similarly show that qualitative CT imaging features highlighting differences in enhancement patterns among resectable ICC are associated with survival. In addition to the appearance of the primary tumor itself, our results show that the presence of satellite nodules is associated with decreased disease free survival and approached significance with decreased overall survival in resected patients. This finding is concordant with those of Baheti et al in a study of 92 patients with ICC demonstrating that the presence of satellite nodules and intrahepatic metastases were associated with decreased survival [22].
Radiogenomics examines associations between tumoral imaging features and their underlying cellular genetics [23–25], with promising results emerging in many different tumors including lung, breast, brain and renal cancers [26–30]. In liver imaging, a radiogenomic biomarker of microvascular invasion has been proposed for HCC [31] and Taouli et al have recently reported a correlation between phenotypic imaging traits including infiltrative pattern, macrovascular invasion, size >5 cm, mosaic appearance, various enhancement patterns and genetic signatures of aggressive HCC [32]. For ICC, genetic signatures of aggressive tumor behavior are not yet definitively described but quantitative imaging phenotypes have previously been correlated with expression of specific markers of hypoxia [33]. Given the recent interest in defining the genomic landscape for ICC, we investigated whether CT imaging features could be associated with known ICC genetic mutations or pathways. Our study did not show an association between ICC imaging features and genetic mutations in our surgical patient population. ICC is a phenotypically heterogeneous tumor without clearly established genomic classifications at this time. Multiple genetic pathways have been implicated including IDH1, IDH2, KRAS and Chromatin remodelling complex [34,35,21]. IDH1/2 encode metabolic enzymes for the oxidative decarboxylation of isocitrate in the Kreb cycle and have been proposed as key driver mutations in multiple cancers [36,34]. KRAS is a proto-oncogene that has also been highlighted as a critical mutation in oncogenesis [37]. Chromatin remodeling genes regulate gene expression by affecting access of transcription proteins to certain portions of DNA and are among those most commonly mutated in ICC [21]. Our results show that despite a lack of associations between imaging features and genetic mutations, certain imaging features may still offer predictive information. As tumors grow larger and encase adjacent vessels, develop areas of necrosis, and spread with satellite tumors, these phenotypic differences reflect increasing biological aggressiveness.
This study is a hypothesis generating study with several limitations. It was retrospective, involving a single institution and had a moderate sample size of 66 patients. However, ICC is a rare cancer limiting our ability to generate larger patient population without performing multi-center studies, which will be required to validate our results. Second, the exclusion of unresectable tumors limited the sample size, but increased the homogeneity of the study population tumors. Another limitation was the inclusion of CT studies performed at multiple institutions, using different CT scanners with varying imaging protocols, including both single phase and multi-phase studies. This inclusion criteria kept our population as large as possible, but certain imaging features were not evaluable, such as quantitative measures of tumor enhancement that have been previously described for ICC [11]. We also excluded the evaluation of arterial phase enhancement pattern in our patients for this reason. Certain features such as fibrosis may not have been optimally evaluated in the absence of delayed imaging. Inter-reader variability in the assessment of ICC imaging features described would also need to be performed in future validation studies, preferably with uniform multiphasic imaging protocols. For our genetic analysis, our approach provides extensive analysis for over 400 cancer-associated genes, but it does not fully replace a whole genome sequencing platform. Unknown, potentially more relevant genetic pathways may not have been included; for example, Jusakul et al have recently described new distinct subtypes of cholangiocarcinomas based on 489 cases from 10 countries [38]. Use of single tissue samples may also be a limitation given that intratumoral heterogeneity may be significant for ICC. Finally, given the moderate sample size of our patient population, limited genome sequencing performed, and the rapidly evolving field of ICC genomics, we may need to revisit the associations between CT imaging features and ICC genetic pathways in future, larger multi-center studies.
In conclusion, in this retrospective analysis, our results are consistent with prior studies and highlight the potential prognostic significance of visually assessed imaging features in resectable ICC. No association was identified between these imaging features and currently known genetic pathways in ICC. The lack of associations between imaging features and tumor genetics may reflect limitations of the genetic profiling methodology used, but may also highlight the non-redundant information within observable radiographic phenotypic variations in resectable ICC patients.
Acknowledgments
Linda M. Pak, MD, was supported by the Clinical and Translational Science Center at Weill Cornell Medical Center and MSKCC Award Number UL1TR00457. This work was also supported in part by NIH/NCI P30 CA008748 Cancer Center Support Grant.
Footnotes
Compliance with Ethical Standards
Conflict of interest: The authors declare that they have no conflict of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent: For this type of study formal consent is not required.
References
- 1.Blechacz B, Komuta M, Roskams T, Gores GJ. Clinical diagnosis and staging of cholangiocarcinoma. Nature reviews Gastroenterology & hepatology. 2011;8(9):512–522. doi: 10.1038/nrgastro.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mar WA, Shon AM, Lu Y, Yu JH, Berggruen SM, Guzman G, Ray CE, Jr, Miller F. Imaging spectrum of cholangiocarcinoma: role in diagnosis, staging, and posttreatment evaluation. Abdominal radiology (New York) 2016;41(3):553–567. doi: 10.1007/s00261-015-0583-9. [DOI] [PubMed] [Google Scholar]
- 3.Yamasaki S. Intrahepatic cholangiocarcinoma: macroscopic type and stage classification. Journal of hepato-biliary-pancreatic surgery. 2003;10(4):288–291. doi: 10.1007/s00534-002-0732-8. [DOI] [PubMed] [Google Scholar]
- 4.Khan SA, Toledano MB, Taylor-Robinson SD. Epidemiology, risk factors, and pathogenesis of cholangiocarcinoma. HPB : the official journal of the International Hepato Pancreato Biliary Association. 2008;10(2):77–82. doi: 10.1080/13651820801992641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park J, Kim MH, Kim KP, Park DH, Moon SH, Song TJ, Eum J, Lee SS, Seo DW, Lee SK. Natural History and Prognostic Factors of Advanced Cholangiocarcinoma without Surgery, Chemotherapy, or Radiotherapy: A Large-Scale Observational Study. Gut and liver. 2009;3(4):298–305. doi: 10.5009/gnl.2009.3.4.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doussot A, Gonen M, Wiggers JK, Groot-Koerkamp B, DeMatteo RP, Fuks D, Allen PJ, Farges O, Kingham TP, Regimbeau JM, D'Angelica MI, Azoulay D, Jarnagin WR. Recurrence Patterns and Disease-Free Survival after Resection of Intrahepatic Cholangiocarcinoma: Preoperative and Postoperative Prognostic Models. Journal of the American College of Surgeons. 2016;223(3):493–505. e492. doi: 10.1016/j.jamcollsurg.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie B, Tomaszewski MR, Neves A, Ros S, Hu DE, McGuire S, Mullins SR, Tice DA, Sainson RCA, Bohndiek SE, Wilkinson RW, Brindle KM. Optoacoustic detection of early therapy-induced tumor cell death using a targeted imaging agent. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017 doi: 10.1158/1078-0432.ccr-17-1029. [DOI] [PubMed] [Google Scholar]
- 8.Kuo MD, Yamamoto S. Next generation radiologic-pathologic correlation in oncology: Rad-Path 2.0. AJR American journal of roentgenology. 2011;197(4):990–997. doi: 10.2214/ajr.11.7163. [DOI] [PubMed] [Google Scholar]
- 9.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsayes KM, Kielar AZ, Agrons MM, Szklaruk J, Tang A, Bashir MR, Mitchell DG, Do RK, Fowler KJ, Chernyak V, Sirlin CB. Liver Imaging Reporting and Data System: an expert consensus statement. Journal of hepatocellular carcinoma. 2017;4:29–39. doi: 10.2147/jhc.s125396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SA, Lee JM, Lee KB, Kim SH, Yoon SH, Han JK, Choi BI. Intrahepatic mass-forming cholangiocarcinomas: enhancement patterns at multiphasic CT, with special emphasis on arterial enhancement pattern--correlation with clinicopathologic findings. Radiology. 2011;260(1):148–157. doi: 10.1148/radiol.11101777. [DOI] [PubMed] [Google Scholar]
- 12.Konstantinidis IT, Do RK, Gultekin DH, Gonen M, Schwartz LH, Fong Y, Allen PJ, D'Angelica MI, DeMatteo RP, Klimstra DS, Kemeny NE, Jarnagin WR. Regional chemotherapy for unresectable intrahepatic cholangiocarcinoma: a potential role for dynamic magnetic resonance imaging as an imaging biomarker and a survival update from two prospective clinical trials. Annals of surgical oncology. 2014;21(8):2675–2683. doi: 10.1245/s10434-014-3649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turkoglu MA, Yamamoto Y, Sugiura T, Okamura Y, Ito T, Ashida R, Uemura S, Miyata T, Kakuda Y, Nakanuma Y, Uesaka K. The favorable prognosis after operative resection of hypervascular intrahepatic cholangiocarcinoma: A clinicopathologic and immunohistochemical study. Surgery. 2016;160(3):683–690. doi: 10.1016/j.surg.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Fujita N, Asayama Y, Nishie A, Ishigami K, Ushijima Y, Takayama Y, Okamoto D, Moirta K, Shirabe K, Aishima S, Wang H, Oda Y, Honda H. Mass-forming intrahepatic cholangiocarcinoma: Enhancement patterns in the arterial phase of dynamic hepatic CT - Correlation with clinicopathological findings. European radiology. 2017;27(2):498–506. doi: 10.1007/s00330-016-4386-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhu AX, Borger DR, Kim Y, Cosgrove D, Ejaz A, Alexandrescu S, Groeschl RT, Deshpande V, Lindberg JM, Ferrone C, Sempoux C, Yau T, Poon R, Popescu I, Bauer TW, Gamblin TC, Gigot JF, Anders RA, Pawlik TM. Genomic profiling of intrahepatic cholangiocarcinoma: refining prognosis and identifying therapeutic targets. Annals of surgical oncology. 2014;21(12):3827–3834. doi: 10.1245/s10434-014-3828-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, Hiraoka N, Ojima H, Shimada K, Okusaka T, Kosuge T, Miyagawa S, Shibata T. Genomic spectra of biliary tract cancer. Nature genetics. 2015;47(9):1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 17.Moeini A, Sia D, Bardeesy N, Mazzaferro V, Llovet JM. Molecular Pathogenesis and Targeted Therapies for Intrahepatic Cholangiocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22(2):291–300. doi: 10.1158/1078-0432.ccr-14-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, Cornella H, Klotzle B, Fan JB, Cotsoglou C, Thung SN, Fuster J, Waxman S, Garcia-Valdecasas JC, Bruix J, Schwartz ME, Beroukhim R, Mazzaferro V, Llovet JM. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144(4):829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, Offerhaus GJ, Roa JC, Roberts LR, Gores GJ, Popescu I, Alexandrescu ST, Dima S, Fassan M, Simbolo M, Mafficini A, Capelli P, Lawlor RT, Ruzzenente A, Guglielmi A, Tortora G, de Braud F, Scarpa A, Jarnagin W, Klimstra D, Karchin R, Velculescu VE, Hruban RH, Vogelstein B, Kinzler KW, Papadopoulos N, Wood LD. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nature genetics. 2013;45(12):1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baheti AD, Tirumani SH, Shinagare AB, Rosenthal MH, Hornick JL, Ramaiya NH, Wolpin BM. Correlation of CT patterns of primary intrahepatic cholangiocarcinoma at the time of presentation with the metastatic spread and clinical outcomes: retrospective study of 92 patients. Abdominal imaging. 2014;39(6):1193–1201. doi: 10.1007/s00261-014-0167-0. [DOI] [PubMed] [Google Scholar]
- 23.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nature biotechnology. 2007;25(6):675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 24.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, Aldape K, Cha S, Kuo MD. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(13):5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair VS, Gevaert O, Davidzon G, Napel S, Graves EE, Hoang CD, Shrager JB, Quon A, Rubin DL, Plevritis SK. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer research. 2012;72(15):3725–3734. doi: 10.1158/0008-5472.can-11-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269(1):8–15. doi: 10.1148/radiol.13122697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gevaert O, Xu J, Hoang CD, Leung AN, Xu Y, Quon A, Rubin DL, Napel S, Plevritis SK. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012;264(2):387–396. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR American journal of roentgenology. 2012;199(3):654–663. doi: 10.2214/ajr.11.7824. [DOI] [PubMed] [Google Scholar]
- 29.Zinn PO, Mahajan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, Colen RR. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PloS one. 2011;6(10):e25451. doi: 10.1371/journal.pone.0025451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlo CA, Di Paolo PL, Chaim J, Hakimi AA, Ostrovnaya I, Russo P, Hricak H, Motzer R, Hsieh JJ, Akin O. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology. 2014;270(2):464–471. doi: 10.1148/radiol.13130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, Rutman AM, Siripongsakun S, Lu D, Imanbayev G, Kuo MD. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology (Baltimore, Md) 2015;62(3):792–800. doi: 10.1002/hep.27877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taouli B, Hoshida Y, Kakite S, Chen X, Tan PS, Sun X, Kihira S, Kojima K, Toffanin S, Fiel MI, Hirschfield H, Wagner M, Llovet JM. Imaging-based surrogate markers of transcriptome subclasses and signatures in hepatocellular carcinoma: preliminary results. European radiology. 2017 doi: 10.1007/s00330-017-4844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadot E, Simpson AL, Do RK, Gonen M, Shia J, Allen PJ, D'Angelica MI, DeMatteo RP, Kingham TP, Jarnagin WR. Cholangiocarcinoma: Correlation between Molecular Profiling and Imaging Phenotypes. PloS one. 2015;10(7):e0132953. doi: 10.1371/journal.pone.0132953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, Kwak EL, Clark JW, Ryan DP, Deshpande V, Dias-Santagata D, Ellisen LW, Zhu AX, Iafrate AJ. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. The oncologist. 2012;17(1):72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson S, Hyder O, Dodson R, Nayar SK, Poling J, Beierl K, Eshleman JR, Lin MT, Pawlik TM, Anders RA. The frequency of KRAS and BRAF mutations in intrahepatic cholangiocarcinomas and their correlation with clinical outcome. Human pathology. 2013;44(12):2768–2773. doi: 10.1016/j.humpath.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, Andersen JB, Jiang W, Savich GL, Tan TX, Auman JT, Hoskins JM, Misher AD, Moser CD, Yourstone SM, Kim JW, Cibulskis K, Getz G, Hunt HV, Thorgeirsson SS, Roberts LR, Ye D, Guan KL, Xiong Y, Qin LX, Chiang DY. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32(25):3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Churi CR, Shroff R, Wang Y, Rashid A, Kang HC, Weatherly J, Zuo M, Zinner R, Hong D, Meric-Bernstam F, Janku F, Crane CH, Mishra L, Vauthey JN, Wolff RA, Mills G, Javle M. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PloS one. 2014;9(12):e115383. doi: 10.1371/journal.pone.0115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, Choo SP, Myint SS, Thanan R, Nagarajan S, Lim WK, Ng CCY, Boot A, Liu M, Ong CK, Rajasegaran V, Lie S, Lim AST, Lim TH, Tan J, Loh JL, McPherson JR, Khuntikeo N, Bhudhisawasdi V, Yongvanit P, Wongkham S, Totoki Y, Nakamura H, Arai Y, Yamasaki S, Chow PK, Chung AYF, Ooi L, Lim KH, Dima S, Duda DG, Popescu I, Broet P, Hsieh SY, Yu MC, Scarpa A, Lai J, Luo DX, Carvalho AL, Vettore AL, Rhee H, Park YN, Alexandrov LB, Gordan R, Rozen SG, Shibata T, Pairojkul C, Teh BT, Tan P. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer discovery. 2017;7(10):1116–1135. doi: 10.1158/2159-8290.cd-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]