Abstract
Aberrant expression of FOXN2, a member of the Forkhead box transcription factors, has been found in several types of cancer. However, the underlying mechanisms of FOXN2 deregulation in tumorigenesis remain largely unknown. Here, we find that FOXN2 binds to and is ubiquitinated by β-Trcp ubiquitin ligase and RSK2 kinase for degradation. Furthermore, we demonstrate that the Ser365 and Ser369 sites in a conserved DSGYAS motif are critical for the degradation of FOXN2 by β-Trcp and RSK2. Moreover, gain-of-function and loss-of-function studies show that FOXN2 impairs cell proliferation in vitro and in vivo and enhances the radiosensitivity of lung cancer. Importantly, β-Trcp-mediated and RSK2-mediated degradation of FOXN2 promotes tumorigenesis and radioresistance in lung cancer cells. Collectively, our study reveals a novel post-translational modification of FOXN2 and suggests that FOXN2 may be a potential therapeutic and radiosensitization target for lung cancer.
Introduction
Lung cancer has the highest morbidity and leading mortality among all cancer types worldwide, with an average 5-year survival of ~15% [1, 2]. Non-small-cell lung cancer (NSCLC) comprises ~85% of lung cancer, which mainly includes adenocarcinoma, squamous carcinoma, and large-cell carcinoma [3]. Radiotherapy is considered the main option for local advanced lung cancer patients. Unfortunately, the clinical outcomes are not satisfactory due to radioresistance. Therefore, elucidating the mechanisms and identifying novel biomarkers for radioresistance are urgently required and may provide key clues for the treatment of lung cancer.
The ubiquitin-proteasome system (UPS) is the main protein degradation system that maintains protein homeostasis through directing the proteasomal destruction [4, 5]. It encompasses ubiquitin activating enzyme E1, ubiquitin conjugating enzyme E2, and ubiquitin ligase E3. Among these, E3 ligase is the critical component of UPS because it can recognize the substrate protein for degradation [6, 7]. SCFβ-Trcp is a well-characterized E3 ubiquitin ligase and has been extensively investigated [8–10]. The F-box protein β-transducin repeat-containing protein (β-Trcp) is one of the 69 F-box proteins and often targets downstream substrates containing the classical DSGXXS phosphodegron for proteolysis [11]. It has been reported that β-Trcp has two distinct paralogs, β-Trcp1 and β-Trcp2, which share identical biological characterization [11, 12]. In addition, β-Trcp has been found to be involved in the regulation of multiple cellular processes, including cell cycle control, cell signaling, angiogenesis, lipogenesis, and tumorigenesis, by facilitating the destruction of diverse key regulatory proteins such as Wee1, Set8, VEGFR2, CHD1, and Lipin1 [13–17].
The Forkhead box (FOX) transcription factor family is classified into 19 subfamilies based on the conservation of DNA-binding domains [18–20]. FOX transcriptional factors are especially involved in mediating the expression of target genes concerning organ development, cell metabolism, immunoregulation, and ageing [21, 22]. Moreover, several lines of evidence have revealed that FOX proteins also play pivotal roles in tumorigenesis. For instance, FOXO1A serves as a tumor suppressor for HER2-overexpressing breast cancer through inhibiting the PI3K/AKT signaling pathway [23]. FOXM1 was found to be overproduced and provided advantages for tumor progression in a series of malignancies [24]. As a member of the FOX transcription factor family, the aberrant expression of FOXN2 has been observed in some types of human cancer. A recent report indicated that FOXN2 may act as a tumor suppressor in T-cell leukemia, which displays reduced FOXN2 transcript levels [25]. In addition, it has been shown that low FOXN2 expression is correlated with adverse prognosis in adult glioblastoma multiforme treated with chemotherapy or radiotherapy [26]. These studies have provided significant clues regarding FOXN2 function in cancer. Nevertheless, the biological relevance and how FOXN2 levels are regulated in tumorigenesis still remain elusive.
In this report, we identify FOXN2 as an ubiquitination substrate of β-Trcp E3 ligase and RSK2 protein kinase. We find that FOXN2 interacts with and is ubiquitinated by β-Trcp and RSK2 via a conserved DSGYAS motif. Moreover, FOXN2 suppresses cell proliferation in vitro and in vivo and enhances the radiosensitivity of lung cancer. Importantly, β-Trcp and RSK2-mediated degradation of FOXN2 promotes tumorigenesis and radioresistance in lung cancer.
Results
FOXN2 interacts with β-Trcp in cells
Recently, a large-scale protein–protein network study on the human FOX family revealed that β-Trcp1 is a potential binding partner of FOXN2 [27]. To further determine this interaction, a co-immunoprecipitation experiment was performed. As shown in Fig. 1a, exogenously expressed FOXN2 specifically interacted with β-Trcp1 but not with SKP2 and FBXW7. Moreover, exogenous FOXN2 was able to associate with endogenous β-Trcp and vice versa in H1299 cells (Fig. 1b). Importantly, the endogenous β-Trcp was capable of binding to the endogenous FOXN2 in H1299 cell (Fig. 1c). These findings support the proteomic result and demonstrate that FOXN2 is a β-Trcp-interacting protein in lung cancer cells.
Fig. 1.
FOXN2 interacts with β-Trcp in cells. a Immunoblot analysis of whole cell lysates (WCL) and immunoprecipitates (IP) lysed from HEK293T cells transfected with plasmids as indicated. b H1299 cells were transfected with indicated constructs. Co-IP experiments were performed using S-protein beads and blotted with indicated antibodies. c The endogenous interaction between β-Trcp and FOXN2 was detected by Co-IP. Rabbit IgG was used as a negative control
FOXN2 stability was negatively controlled by β-Trcp
Having checked a physical interaction between these two proteins, we next wondered whether FOXN2 stability is regulated by β-Trcp, since β-Trcp is a well-known E3 ligase. As shown in Fig. 2a, β-Trcp depletion using siRNAs targeting both β-Trcp1 and β-Trcp2 dramatically resulted in the accumulation of endogenous FOXN2 in different cancer cell lines. Moreover, treatment with the 26S proteasome inhibitor MG132 led to increased FOXN2 protein levels, implying the involvement of the ubiquitin–proteasome system in controlling FOXN2 stability (Fig. 2b). In line with this observation, the half-life of FOXN2 was markedly prolonged after MG132 treatment (Fig. 2c). To further explore whether FOXN2 can be degraded by β-Trcp, we detected the abundance of FOXN2 in the absence of β-Trcp after cycloheximide treatment. As seen in Fig. 2d, the half-life of FOXN2 was notably extended in β-Trcp-depleted cells. These results suggest that FOXN2 is degraded and negatively controlled by β-Trcp via the ubiquitin–proteasome pathway.
Fig. 2.
FOXN2 stability is negatively regulated by β-Trcp. a β-Trcp silencing leads to the accumulation of endogenous FOXN2. Cells were transfected with indicated siRNAs, and lysates were subjected to western blotting with the indicated antibodies. b Western blotting analysis of WCL derived from H1299 cells with or without MG132 treatment. c Left panel: H1299 cells were transfected with the indicated plasmids and harvested at the indicated points after CHX with or without MG132 treatment for immunoblotting. Right panel: quantification of the FOXN2 band intensity is shown. d Left panel: H1299 cells were transfected with the indicated siRNAs. At 48 h post transfection, cells were treated with 20 μg/mL of cycloheximide (CHX) and harvested at the indicated time points. Western blotting was conducted to detect the protein level of FOXN2. Right panel: quantification of the FOXN2 band intensity is shown
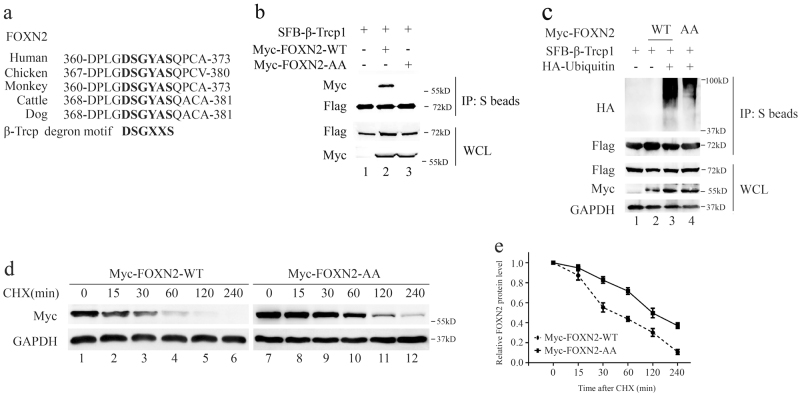
The Ser365 and Ser369 sites in a conserved DSGYAS motif are responsible for the ubiquitination of FOXN2 by β-Trcp
Most β-Trcp substrates contain a DSGXXS phosphodegron sequence that can be recognized by β-Trcp [11]. Excitingly, we noticed that FOXN2 also contains the consensus DSGXXS phosphodegron in various species (Fig. 3a). To further investigate whether this phosphodegron is associated with β-Trcp, we generated a FOXN2 mutant, termed FOXN2-AA, with Ser365 and Ser369 residues all mutated to alanine. As shown in Fig. 3b, unlike wild-type FOXN2 (FOXN2-WT), the FOXN2-AA mutant failed to associate with β-Trcp. Moreover, an in vivo ubiquitination assay demonstrated that β-Trcp substantially promoted the polyubiquitination of wild-type FOXN2 compared to the mutated form of FOXN2-AA (Fig. 3c), suggesting that the Ser365 and Ser369 sites in a conserved DSGYAS motif are responsible for the β-Trcp binding and subsequent ubiquitination. In further support of this ubiquitination site, we employed protein degradation assays. Notably, the half-life of the FOXN2-AA mutant was remarkably longer than that of wild-type FOXN2 (Fig. 3d). Taken together, these results demonstrate that Ser365 and Ser369 sites in a conserved DSGYAS motif play critical roles in the destruction of FOXN2 by β-Trcp.
Fig. 3.
The Ser365 and Ser369 sites in a conserved DSGYAS motif are responsible for the ubiquitination of FOXN2 by β-Trcp. a Alignment of the candidate phosphodegron sequence in FOXN2 in different species. b HEK293T cells were transfected with the indicated plasmids and harvested 24 h after transfection. Co-IP experiments were performed using S-protein beads and blotted with the indicated antibodies. c HeLa cells transfected with plasmids as indicated. At 24 h post transfection, cells were treated with MG132 (10 μM) for 4 h before harvesting. The lysates were incubated with S beads and then subjected to immunoblot. d HeLa cells were transfected with Myc-FOXN2-WT or Myc-FOXN2 AA plasmid. At 24 h post transfection, cells were treated with 20 μg/mL of cycloheximide (CHX) and collected at the indicated time points. Western blotting was performed to examine the protein level of FOXN2. e Quantification of the FOXN2-WT and FOXN2-AA band intensity in d
RSK2 binds to and promotes the polyubiquitination and degradation of FOXN2
It has been well established that a protein kinase is required to phosphorylate downstream substrates in the phosphodegron motif DSGXXS before the substrate is recognized and degraded by β-Trcp ligase [11]. To this end, we utilized NetPhos 3.1 Server, a kinase prediction program [28], to search for the candidate kinases responsible for the phosphorylation of FOXN2 at Ser365 and Ser369 sites. As shown in Supplementary Table 1, several potential kinases may simultaneously phosphorylate these two sites. To further explore which kinase was involved in the β-Trcp-mediated degradation of FOXN2, we first employed an immunoprecipitation assay and found that only RSK2 specifically interacted with FOXN2 at the exogenous and endogenous level (Fig. 4a–c). Moreover, knockdown of endogenous RSK2 with two validated siRNAs led to an accumulation of FOXN2 in two different lung cancer cell lines (Fig. 4d). To further investigate whether RSK2 affected the ubiquitination of FOXN2, we performed an in vivo ubiquitination assay and found that the polyubiquitination of FOXN2 was dramatically reduced when RSK2 was depleted (Fig. 4e). Consistently, we demonstrated that depletion of endogenous RSK2 extended the half-life of FOXN2 (Fig. 4f). Collectively, these results suggest that RSK2 is the upstream kinase that phosphorylates FOXN2 at the Ser365 and Ser369 sites and subsequently promotes its ubiquitination and degradation by β-Trcp ligase.
Fig. 4.
RSK2 binds to FOXN2 and promotes its ubiquitination and degradation. a H1299 cells were transfected with the indicated constructs, followed by IP with S-protein beads and immunoblotting with the indicated antibodies. b H1299 cells were transfected with SFB-tagged RSK2 plasmid. After 24 h, cells were collected and then incubated with S-protein beads. The samples were analyzed by western blotting with the indicated antibodies. c H1299 cells were harvested and subjected to immunoprecipitation using anti-IgG or anti-RSK2 antibody and then analyzed by western blotting. d Immunoblot analysis of H1299 and A549 cells transfected with the indicated siRNAs. e RSK2 depletion inhibits FOXN2 ubiquitination in vivo. HeLa cells were transfected with the indicated siRNAs and plasmids, followed by the treatment with MG132 (10 μM) for 4 h prior to collection. The lysates were incubated with S beads overnight and then subjected to immunoblotting to examine the polyubiquitylation of exogenous FOXN2. f Upper panel: H1299 cells were transfected with the indicated siRNAs. At 48 h post transfection, cells were treated with 20 μg/mL of cycloheximide (CHX) and harvested at the indicated time points. Western blotting was conducted to detect the protein level of FOXN2. Lower panel: quantification of the FOXN2 band intensity is shown
FOXN2 silencing promotes cell cycle redistribution and tumorigenesis
To explore the potential downstream signaling pathways that are regulated by FOXN2, we performed gene set enrichment analysis (GSEA) and found that FOXN2 may participate in the cell cycle regulation, mismatch repair, nucleotide excision repair and homologous recombination (Fig. 5a). Interestingly, FOXN2 was shown to be strongly associated with ubiquitin-mediated proteolysis (Fig. 5a), which is consistent with our results above. To further validate these predictions, we examined the cell cycle distribution after FOXN2 depletion. FOXN2 was effectively knocked down by two different siRNAs in two lung cancer cell lines (Fig. 5b). As shown in Fig. 5c, cell cycle analysis by flow cytometry exhibited an increase in the percentage of cells in the S phase, further supporting the notion that FOXN2 is involved in cell cycle control.
Fig. 5.
FOXN2 knockdown results in cell cycle redistribution in two lung cancer cells. a Gene set enrichment analysis (GSEA) of the primary lung cancer database from the Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/). The enriched pathway analysis shows that FOXN2 is associated with ubiquitin-mediated proteolysis, cell cycle, mismatch repair, nucleotide excision repair, and homologous recombination. b Western blotting shows the knockdown efficiency of FOXN2 by two different siRNAs in H1299 and A549 cells. c Cell cycle analysis of H1299 and A549 cells after transfection with the indicated siRNAs for 48 h (left panel). Histograms show the percentage of cells in different phases (right panel). **P < 0.01 compared to control cells
A recent study reported that β-Trcp1 was overexpressed in lung cancer tissues [29]. Our results indicate that the protein level of FOXN2 was negatively regulated by β-Trcp; thus, we speculated that FOXN2 may function as a tumor suppressor in the tumorigenesis of lung cancer. First, we detected the cell growth and proliferation capability in FOXN2-depleted lung cancer cells. We showed that inhibition of FOXN2 in H1299 and A549 cells led to cell growth promotion (Fig. 6a). Moreover, compared to control cells, FOXN2 silencing resulted in marked augmentation in colony numbers (Fig. 6b). These results suggest that FOXN2 impairs cell growth and proliferation in vitro. To further investigate whether FOXN2 suppresses tumorigenesis in vivo, we performed subcutaneous xenograft experiments in T-cell-deficient athymic nude mice and found that mice injected with FOXN2 knockdown H1299 cells showed significantly increased tumor growth compared to those injected with shControl cells (Fig. 6c–e). Moreover, the tumor weight was also substantially increased in mice injected with shFOXN2 lung cancer cells (Fig. 6f). Together, these results demonstrate that loss of FOXN2 promotes tumorigenesis, indicating that FOXN2 has potential tumor suppressor activity in lung cancer.
Fig. 6.
Loss of FOXN2 promotes lung cancer cell growth in vitro and in vivo. a FOXN2 depletion promoted cell growth. Cells were transfected with the indicated siRNAs for 48 h and then seeded at low density in plates. The cell numbers were counted every other day. b Colony formation was significantly increased in FOXN2-depleted cells. Cells transfected with the indicated siRNAs were grown for 2 weeks. Representative violet staining pictures are shown (upper panel). The numbers of colonies were counted and shown with histograms (lower panel). ***P < 0.001 compared to control cells. c Western blotting shows the knockdown efficiency of FOXN2 by two different shRNAs in H1299 cells. d FOXN2 inhibition promotes tumor growth in vivo. Representative images of the xenograft tumors in three groups are shown. e The growth curves of tumors in the three groups are presented (five mice/group). f Tumor weights in the three groups are shown (five mice/group)
FOXN2 depletion results in radioresistance in lung cancer cells
A previous study has shown that low FOXN2 expression is correlated with adverse outcomes in glioblastoma treated with radiotherapy [26]. In addition, the GSEA report displayed that FOXN2 is associated with the DNA damage and repair pathway (Fig. 5a), suggesting that FOXN2 may participate in the regulation of cell radiosensitivity. To confirm this hypothesis, γ-H2AX foci experiments and cell survival assays were performed. It has been well documented that γ-H2AX foci are considered to be an indicator for DNA double-strand breaks and closely associated with DNA damage repair. As shown in Fig. 7a, b, the number of γ-H2AX foci was significantly decreased at 4 and 24 h post irradiation in FOXN2-depleted lung cancer cells. In addition, FOXN2-depleted cells were more resistant to radiation than control cells (Fig. 7c). Therefore, these results support our hypothesis and demonstrate that FOXN2 enhances the radiosensitivity of lung cancer cells, indicating that FOXN2 plays a key role in the DNA damage response.
Fig. 7.
FOXN2 enhances the radiosensitivity of lung cancer cells. a FOXN2-depleted H1299 cells (upper panel) and A549 cells (lower panel) were irradiated with 2 Gy and collected at the indicated time points. An immunostaining experiment was performed to examine γ-H2AX foci formation. DAPI was applied to counterstain the nuclei. b Quantification of the γ-H2AX foci result is shown in a. More than 100 cells were counted in each group, and data are presented as means ± SD. ***P < 0.001 compared to control cells. c Radioresistance of cells lacking FOXN2. H1299 cells (left panel) and A549 cells (right panel) were transfected with control or FOXN2 siRNAs and irradiated with doses as indicated. The colonies were counted 14 days later. Experiments were performed in triplicate, and the results are presented as the means ± SD
β-Trcp and RSK2-mediated ubiquitination of FOXN2 promotes tumorigenesis and radioresistance in lung cancer
Functional studies have clearly shown that FOXN2 knockdown promoted tumorigenesis and radioresistance in lung cancer. Thus, to further determine the biological functions of FOXN2 and explore the significance of ubiquitination modification, we reconstituted FOXN2-deficient cells with vectors expressing FOXN2-WT (wild-type) or FOXN2-AA mutant (Fig. 8a). As shown in Fig. 8b–f, the reconstitution with siRNA-resistant wild-type FOXN2 reversed the cell proliferation and radioresistance in FOXN2-knockdown cells. These findings support the role of FOXN2 in tumorigenesis and DNA damage response. More importantly, restoring the siRNA-resistant FOXN2 mutant, which is much more resistant to degradation by β-Trcp, led to a greater inhibition of cell proliferation and clonogenic survival in response to DNA damage compared to the expression of wild-type FOXN2 (Fig. 8b, c, f). Moreover, FOXN2-AA mutant restoration caused a larger γ-H2AX foci number than did wild-type FOXN2 at 4 and 24 h post irradiation (Fig. 8d, e). These results not only reveal that FOXN2 may function as a tumor suppressor protein but also suggest that β-Trcp and RSK2-mediated ubiquitination and degradation of FOXN2 promote tumorigenesis and radioresistance in lung cancer cells.
Fig. 8.
β-Trcp and RSK2-mediated degradation of FOXN2 promotes cell proliferation and radioresistance in lung cancer cells. a FOXN2 knockdown cells were reconstituted with either FOXN2-WT (wild type) or FOXN2-AA mutant as indicated. b The growth curve of H1299 cells is shown. Cells were transfected with the indicated siRNAs and plasmids and then seeded at low density in plates. The cell numbers were calculated every other day. c H1299 cells were transfected with the indicated siRNAs, and plasmids were allowed to grow 2 weeks to form colonies. Representative violet staining results are shown (upper panel). The numbers of colonies were counted and shown with histograms (lower panel); **P < 0.01. d H1299 cells transfected with the indicated siRNAs and plasmids were irradiated with 2 Gy and harvested at the indicated time points. An immunostaining experiment was carried out to detect the γ-H2AX foci formation. e Quantification of γ-H2AX foci is shown in d; **P < 0.01, ***P < 0.001. f Cell survival curves derived from H1299 cells. Cells were transfected with the indicated siRNAs and plasmids and treated with the radiation dose as indicated. Percentages of surviving colonies were measured 2 weeks later. The experiments were conducted in triplicate, and the results are presented as the means ± SD
Discussion
In the present study, we identify FOXN2 as a new substrate of β-Trcp ubiquitin ligase and RSK2 protein kinase and uncover a novel post-translational modification of FOXN2 via the ubiquitination pathway. Furthermore, we illustrate the roles of FOXN2 in tumorigenesis and DNA damage response. Our results provide new mechanistic insight into the regulation of FOXN2 and suggest that FOXN2 may be a tumor suppressor protein and radiosensitization target in lung cancer.
Post-translational modifications including ubiquitination, phosphorylation, and acetylation play crucial roles in the control of the activity, localization and/or stability of several key proteins involved in a wide spectrum of cellular processes [30]. Accumulating evidence have revealed that several Forkhead factors can be regulated by many post-translational modifications. For instance, the phosphorylation of FOXO1 by CDK2 led to cytoplasmic localization and diminished DNA damage-induced apoptotic cell death [31]. The ubiquitin ligase Stub1 modulated the immune responses by promoting polyubiquitination and degradation of FOXP3 [32]. Our study revealed that FOXN2 is ubiquitinated and destructed by β-Trcp E3 ubiquitin ligase. Moreover, FOXN2 contains a canonical DSGXXS phosphodegron, which is recognized by β-Trcp. It is well established that prior phosphorylation of the phosphodegron motif is critical for the β-Trcp recognition. Our results demonstrate that the Ser365 and Ser369 phosphorylation sites in the DSGXXS motif are responsible for β-Trcp binding and subsequent ubiquitination. Importantly, we identified RSK2 kinase as the upstream kinase for the FOXN2 phosphodegron. RSK2 has been reported to promote cell proliferation and tumor metastasis through modulating microtubule dynamics and the proapoptotic signaling pathway [33, 34]. In addition, RSK2 has also been shown to function as a crucial kinase that is involved in β-Trcp-mediated degradation of BimEL in lung cancer cells [35]. Our data clearly showed that RSK2 binds to and triggers the destruction of FOXN2, indicating that RSK2 acts cooperatively with β-Trcp to regulate the stability of FOXN2. Taken together, our findings strongly suggest that FOXN2 is a novel physiological substrate of the β-Trcp ubiquitin ligase and RSK2 kinase.
FOXN2 has been less studied in recent years even though it has been considered a critical transactional factor. Therefore, the biological function of FOXN2, especially in human lung cancer, is totally unknown. Our work showed for the first time that FOXN2 impairs cell proliferation and enhances the radiosensitivity of lung cancer. The opposite effects were observed when FOXN2 was knocked down in lung cancer cells. Accumulating evidence have demonstrated that there are many molecular mechanisms determining cell radiosensitivity, including cell proliferation and apoptosis, cell cycle regulation, DNA damage sensing and repair, and tumor immune microenvironment [36–38]. Our data indicated that FOXN2 depletion promotes cell growth, leads to increased cell numbers in the S phase and inhibits DNA damage-induced γ-H2AX foci formation. Additionally, using the GSEA database, we discovered that FOXN2 may be associated with cell cycle regulation, mismatch repair, nucleotide excision repair, and homologous recombination, suggesting that FOXN2 may regulate the expression of genes involved in these processes; this issue should be further investigated in the future. However, our findings strongly indicate that loss of FOXN2 enhances radioresistance of lung cancer cells by promoting cell proliferation and cell cycle redistribution, as well as inhibiting DNA damage response. More importantly, we found that the FOXN2 mutant was more potent than wild-type FOXN2 in suppressing cell proliferation and enhancing radiosensitivity, suggesting that β-Trcp and RSK2-mediated FOXN2 degradation is required for these biological functions, further supporting the roles of FOXN2 in tumorigenesis and DNA damage response. Using the online Oncomine database and Kaplan–Meier plot analysis, we showed that FOXN2 is downregulated and that low levels of FOXN2 expression correlate with poor overall survival in lung cancer patients (Supplementary Figure 1). We were unable to examine the protein levels of FOXN2 in lung cancer tissues due to the lack of antibodies suitable for immunohistochemistry. However, all of the data presented indicate that FOXN2 may act as a novel tumor suppressor in lung cancer. Notably, high protein levels of β-Trcp were observed in multiple types of human cancers [15, 39, 40], especially in lung cancer [29]. Thus, our results imply that the accumulation of β-Trcp may account for FOXN2 downregulation in human lung cancer.
In summary, our work uncovered that FOXN2 is a bona fide substrate of β-Trcp ubiquitin ligase, providing insight into the molecular mechanisms by which FOXN2 is regulated and the biological significance of β-Trcp and RSK2-mediated FOXN2 degradation in lung cancer. Importantly, we also provided evidence that FOXN2 may be downregulated and function as a tumor suppressor protein by inhibiting tumorigenesis and radioresistance. Given that β-Trcp is frequently overproduced and results in the degradation of FOXN2 in lung cancer, targeting β-Trcp could be a potential new therapeutic strategy for lung cancer, displaying elevated levels of β-Trcp and low levels of FOXN2.
Materials and methods
Cell culture
HEK293T, HeLa, H1299, and A549 cells were purchased from the American Type Culture Collection (ATCC) cultured in Dulbecco’s modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 μg/mL of penicillin and streptomycin at 37 °C with 5% CO2.
Plasmids
FOXN2, β-Trcp1, and RSK2 plasmids were purchased from DF/HCC DNA Resource Core. Myc-β-Trcp1 plasmid was kindly provided by Dr. Wenyi Wei (Harvard Medical School, USA). All constructs were subcloned into the pDOR201 vector and then transferred to a Gateway-compatible destination vector with indicated SFB and Myc tag using Gateway Technology (Invitrogen). The KOD hot start kit (Novagen) was used to generate mutations, and all mutations in this study were confirmed by DNA sequencing.
Antibodies and reagents
Anti-FOXN2 antibody was obtained from Mybiosource. Anti-β-Trcp antibody, anti-ATM antibody, anti-GSK3β antibody, and anti-HA antibodies were obtained from Cell Signaling Technology. Anti-GAPDH antibody, anti-Myc antibody, anti-CDK1 antibody, anti-CK1α antibody, and anti-CK2α antibodies were purchased from Santa Cruz Biotechnology. Anti-RSK1 antibody, anti-RSK2 antibody, and anti-PKCα antibodies were obtained from Proteintech. Anti-Flag antibody was purchased from Sigma-Aldrich. Cycloheximide and MG132 were purchased from Sigma-Aldrich and Millipore, respectively.
RNA interference
The sequences of siRNAs were as follows: β-Trcp siRNA: 5′-AAGUGGAAUUUGUGGAACAUC-3′, which has been described previously [14–16]. RSK2 siRNA-1: 5′-GGGAGGAGAUUUGUUUACACGCUUA-3′; RSK2 siRNA-2: 5′- AGCGCTGAGAATGGACAGCAA-3′, which has been described previously [35, 41]. FOXN2 siRNA-1: 5′-GAGGAGUUACGGCAAUGCA-3′; FOXN2 siRNA-2: 5′-CCUUGCUGGAAUUCGUACA-3′. All siRNAs were transfected into cells at 100 nM using LipofectamineRNAimax (Invitrogen). The cells were harvested 48 h after transfection. To construct FOXN2 siRNA-resistant plasmids, six nucleotide substitutions were introduced into the FOXN2 siRNA target sequence.
Western blotting and immunoprecipitation
Cells were lysed in NETN buffer (20 mM of Tris HCl, pH 8.0, 100 mM of NaCl, 1 mM of EDTA, and 0.5% Nonidet P-40) on ice for 30 min. Proteins were resolved on SDS-PAGE and then transferred to PVDF membranes for western blotting. For exogenous immunoprecipitation, samples were incubated with S-protein agarose beads (Novagen), shaking overnight at 4 °C. For the endogenous binding, the supernatants were first incubated with anti-β-Trcp for 2 h and then protein A/G-agarose overnight. The precipitates were washed five times with NETN buffer and analyzed by western blotting.
Establishment of stable cell lines
This assay was performed as described previously [42]. Briefly, HEK293T cells were transiently transfected with FOXN2 shRNAs and packaging plasmids pSPAX2 and pMD2G. After 48 h, lentiviral supernatants were collected and used to infect H1299 cells with the addition of 8 μg/mL of Polybrene (Sigma). Stable cell lines were selected with media containing 2 μg/mL of puromycin and validated by Western blotting. The shRNA sequences were as follows:
Control shRNA: 5′-TCTCGCTTGGGCGAGAGTAAG-3′;
FOXN2 shRNA-1: 5′-CCTTGCTGGAATTCGTACA-3′; and
FOXN2 shRNA-2: 5′-GAGGAGTTACGGCAATGCA-3′.
In vivo ubiquitination assay
Cells were transfected with the indicated plasmids for 24 h. Proteasome inhibitor MG132 was added 4 h before cells were lysed. Samples were subjected to western blotting using the indicated antibodies.
Protein degradation assay
Cells were transfected with indicated siRNAs for 48 h. Alternatively, cells were directly transfected with Myc-tagged FOXN2-WT or FOXN2-AA mutant plasmids. After transfection, 20 μg/mL of cycloheximide (CHX) was added to the medium at the indicated time. Cells were lysed, and proteins were subjected to immunoblot using the indicated antibodies.
Cell growth and colony formation analysis
This assay was performed as described previously [43]. Cells transfected with the indicated siRNAs or plasmids were seeded into six-well plates at 1 × 104/well. Cell numbers were counted every other day by digesting into suspension with trypsin/EDTA. For colony formation assays, cells were seeded in a six-well plate at a density of 500/well and then cultured for 2 weeks. The numbers of colonies containing more than 50 cells were counted.
Flow cytometry
This assay was performed as described previously [43]. A549 and H1299 cells transfected with the indicated siRNAs were collected and stained with propidium iodide (PI) and RNase A. The samples were analyzed by flow cytometry.
Immunofluorescence staining
Cells transfected with the indicated siRNAs or plasmids were cultured on coverslips and then were exposed to 2 Gy IR. Cells were then fixed with 4% paraformaldehyde solution for 15 min at various time points after radiation (30 min, 4 h and 24 h) and extracted with buffer containing 0.2% Triton X-100 for 5 min. The samples were blocked with 5% bovine serum albumin (BSA) and then incubated with γ-H2AX antibody overnight. Cells were washed and incubated with secondary antibody for 1 h. Finally, cells were counterstained with DAPI for 10 min to visualize the morphology.
Clonogenic cell survival assays
This assay was performed as described previously [43]. A549 and H1299 cells transfected with the indicated siRNAs or plasmids were seeded in triplicate into six-well plates and irradiated with indicated doses. Cells were further grown for 2 weeks to form distinct cell clusters. The colonies comprising more than 50 cells were evaluated.
Xenograft study
All animal experiments were approved by the Medical Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology. BALB/c nude mice (5–6 weeks old) were used for animal studies. Mice were randomly divided into three groups consisting of five mice and injected with 5 × 106 H1299 stable cells (shControl, shFOXN2–1, and shFOXN2–2). Tumor growth was measured with calipers every 3 days, and tumor volumes were calculated according to the formula V = (L × W2)/2, where V = volume (mm3), L = length (mm), and W = width (mm). Mice were euthanized after 3 weeks. The tumors were removed, photographed and weighed.
Statistical analysis
In our study, three (or at least three) independent experiments were performed in each assay. Data are shown as the means ± SD. The significance of the data between different groups was analyzed by Student’s t-test. P < 0.05 was considered significant.
Electronic supplementary material
Acknowledgements
This work was supported by National Key R&D Program of China (2016YFC0106700 to GW and SZ), National Key R&D Program of China (2016YFC1303800 to LL), the National Natural Science Foundation of China (81672289 to SX), a grant from the Wuhan Science and Technology Bureau (2015060101010041 to SX), and the National Natural Science Foundation of China (81372435 to KY).
Author contributions’
SX designed and conceived the study. JM, YL, and SZ performed experiments and analyzed the data. YL, JH, ZY, JR, KH, LL, KY, and GW provided advice and technical assistance, SX wrote the manuscript. All authors have contributed to and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41418-017-0055-6) contains supplementary material, which is available to authorized users.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 5.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–81. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 7.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 8.Skaar JR, D’Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009;137:1358. doi: 10.1016/j.cell.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 9.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–81. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng N, Zhou Q, Wang Z, Wei W. Recent advances in SCF ubiquitin ligase complex: clinical implications. Biochim Biophys Acta. 2016;1866:12–22. doi: 10.1016/j.bbcan.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–49. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa T, Araki T, Nakagawa M, Hirao A, Unno M, Nakayama K. S6 kinase- and beta-TrCP2-dependent degradation of p19Arf is required for cell proliferation. Mol Cell Biol. 2015;35:3517–27. doi: 10.1128/MCB.00343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, et al. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci USA. 2004;101:4419–24. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Dai X, Zhong J, Inuzuka H, Wan L, Li X, et al. SCF(beta-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat Commun. 2015;6:10185. doi: 10.1038/ncomms10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaik S, Nucera C, Inuzuka H, Gao D, Garnaas M, Frechette G, et al. SCF(beta-TRCP) suppresses angiogenesis and thyroid cancer cell migration by promoting ubiquitination and destruction of VEGF receptor 2. J Exp Med. 2012;209:1289–307. doi: 10.1084/jem.20112446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu K, Fukushima H, Ogura K, Lien EC, Nihira NT, Zhang J, et al. The SCFbeta-TRCP E3 ubiquitin ligase complex targets Lipin1 for ubiquitination and degradation to promote hepatic lipogenesis. Sci Signal. 2017;10:eaah4117. [DOI] [PMC free article] [PubMed]
- 17.Zhao D, Lu X, Wang G, Lan Z, Liao W, Li J, et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 2017;542:484–8. doi: 10.1038/nature21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh M, Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25:1495–500. [PubMed] [Google Scholar]
- 19.Tuteja G, Kaestner KH. SnapShot: forkhead transcription factors I. Cell. 2007;130:1160. doi: 10.1016/j.cell.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Tuteja G, Kaestner KH. Forkhead transcription factors II. Cell. 2007;131:192. doi: 10.1016/j.cell.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 22.Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Development. 2016;143:4558–70. doi: 10.1242/dev.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Shang X, Sarkissyan M, Slamon D, Vadgama JV. FOXO1A is a target for HER2-overexpressing breast tumors. Cancer Res. 2010;70:5475–85. doi: 10.1158/0008-5472.CAN-10-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–9. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 25.Nagel S, Pommerenke C, Meyer C, Kaufmann M, MacLeod RA, Drexler HG. Identification of a tumor suppressor network in T-cell leukemia. Leuk Lymphoma. 2017;58:1–15. doi: 10.1080/10428194.2017.1283029. [DOI] [PubMed] [Google Scholar]
- 26.Robertson E, Perry C, Doherty R, Madhusudan S. Transcriptomic profiling of Forkhead box transcription factors in adult glioblastoma multiforme. Cancer Genom Proteom. 2015;12:103–12. [PubMed] [Google Scholar]
- 27.Li X, Wang W, Wang J, Malovannaya A, Xi Y, Li W, et al. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol Syst Biol. 2015;11:775. doi: 10.15252/msb.20145504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–49. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Zhou W, Yang F, Chen G, Li H, Zhao Y, et al. The beta-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat Commun. 2017;8:14002. doi: 10.1038/ncomms14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benayoun BA, Caburet S, Veitia RA. Forkhead transcription factors: key players in health and disease. Trends Genet. 2011;27:224–32. doi: 10.1016/j.tig.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–7. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Barbi J, Bu S, Yang HY, Li Z, Gao Y, et al. The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity. 2013;39:272–85. doi: 10.1016/j.immuni.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lara R, Seckl MJ, Pardo OE. The p90 RSK family members: common functions and isoform specificity. Cancer Res. 2013;73:5301–8. doi: 10.1158/0008-5472.CAN-12-4448. [DOI] [PubMed] [Google Scholar]
- 34.Alesi GN, Jin L, Li D, Magliocca KR, Kang Y, et al. RSK2 signals through stathmin to promote microtubule dynamics and tumor metastasis. Oncogene. 2016;35:5412–21. doi: 10.1038/onc.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, et al. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–16. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59:928–42. doi: 10.1016/j.ijrobp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer. 2015;15:409–25. doi: 10.1038/nrc3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seshacharyulu P, Baine MJ, Souchek JJ, Menning M, Kaur S, Yan Y, et al. Biological determinants of radioresistance and their remediation in pancreatic cancer. Biochim Biophys Acta. 2017;1868:69–92. doi: 10.1016/j.bbcan.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ougolkov A, Zhang B, Yamashita K, Bilim V, Mai M, Fuchs SY, et al. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J Natl Cancer Inst. 2004;96:1161–70. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- 40.Koch A, Waha A, Hartmann W, Hrychyk A, Schuller U, Waha A, et al. Elevated expression of Wnt antagonists is a common event in hepatoblastomas. Clin Cancer Res. 2005;11:4295–304. doi: 10.1158/1078-0432.CCR-04-1162. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y, Yamada N, Tanaka T, Hori T, Yokoyama S, Hayakawa Y, et al. Crucial roles of RSK in cell motility by catalyzing serine phosphorylation of EphA2. Nat Commun. 2015;6:7679. doi: 10.1038/ncomms8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Y, Ma J, Li Y, Huang J, Zhang S, Yin Z, et al. CDP138 silencing inhibits TGF-β/Smad signaling to impair radioresistance and metastasis via GDF15 in lung cancer. Cell Death Dis. 2017;8:e3036. doi: 10.1038/cddis.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Ma J, Lu Y, Zhang S, Huang J, Chen J, et al. CDK20 interacts with KEAP1 to activate NRF2 and promotes radiochemoresistance in lung cancer cells. Oncogene. 2017;36:5321–30. doi: 10.1038/onc.2017.161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.