Abstract
Identification and characterization of functional molecular targets conferring stemness properties in hepatocellular carcinoma (HCC) offers crucial insights to overcome the major hurdles of tumor recurrence, metastasis and chemoresistance in clinical management. In the current study, we investigated the significance of Cripto-1 in contributing to HCC stemness. Cripto-1 was upregulated in the sorafenib-resistant clones derived from HCC cell lines and patient-derived xenograft that we previously developed, suggesting an association between Cripto-1 and stemness. By in vitro experiments, Cripto-1 fostered cell proliferation, migration, and invasion. It also enhanced self-renewal ability and conferred chemoresistance of HCC cells. Consistently, silencing of Cripto-1 suppressed in vivo tumorigenicity on serial transplantation. On the downstream signaling mechanism, expression of major components of Wnt/β-catenin pathway β-catenin, AXIN2, and C-MYC, accompanied by β-catenin activity was reduced upon Cripto-1 knockdown. The suppressive effects on stemness properties with Cripto-1 knockdown in vitro and in vivo were partially rescued by forced expression of constitutively active β-catenin. Further elucidation revealed the binding of Cripto-1 to Frizzled-7 (FZD7), low-density lipoprotein receptor-related protein 6 (LRP6) and Dishevelled-3 (DVL3) of the Wnt/β-catenin pathway and stabilized DVL3 protein. Analyses with clinical samples validated Cripto-1 overexpression in HCC tissues, as well as a positive correlation between Cripto-1 and AXIN2 expressions. High Cripto-1 level in tumor was associated with poorer disease-free survival of HCC patients. Taken together, Cripto-1 binds to FZD7/LRP6 and DVL3, stabilizes DVL3 expression and activates the Wnt/β-catenin signaling cascade to confer stemness in HCC. Our study findings substantiated the role of Cripto-1 in determining stemness phenotypes of HCC and mechanistically in modulating the Wnt/β-catenin signaling cascade, one of the most frequently deregulated pathways in liver cancer.
Introduction
Limitations of current treatment modalities for hepatocellular carcinoma (HCC) necessitate further dissection of the molecular pathogenesis of this biologically aggressive cancer. The concepts of intra-tumoral heterogeneity and cancer stem cell (CSC)/tumor-initiating cells (T-IC) have been attracting substantial attention in basic and clinical research of HCC. Stemness properties are largely responsible for tumor recurrence, metastasis, and chemoresistance [1, 2], which are the major hurdles for treatment, and from another point of view the Achilles heels of HCC. Therefore identification and characterization of molecular targets endowing stemness phenotypes in HCC carry significant clinical implications [3–5].
The canonical Wnt pathway is one of the most frequently deregulated pathways in HCC [6]. Activation of the pathway leads to stabilization and nuclear translocation of β-catenin and eventually transcriptional upregulation of target genes. β-catenin mutation is a major but not the exclusive mechanism for signaling activation [7, 8]. Other mechanisms are in place to account for the pathway activation in a proportion of HCCs. Thus the multi-level regulation and potential crosstalk related to β-catenin activation are integral parts for elucidating the molecular pathogenesis of this cancer. Notably, the Wnt/β-catenin pathway is heavily implicated in liver CSC properties [9, 10]. In this regard, our previous studies had characterized some key intrinsic molecules and modulators of this pathway in HCC including Prickle-1 [11], Dickkopf-1 (DKK1) [12], low-density lipoprotein receptor-related protein 6 (LRP6) [13], and recently Sox9 [14].
Human Cripto-1 is the founding member of the epidermal growth factor (EGF)-Cripto-1/Fibroblast Growth factor related ligand (FRL1)/Criptic (EGF-CFC) family. The family is characterized by a signal sequence, EGF-like domain, a cysteine-rich CFC motif and a short hydrophobic COOH-terminus which serves for glycosylphosphatidylinositol (GPI) cleavage and attachment [15, 16]. It has been mapped to chromosome 3p21, which comprises 6 exons spanning 4.8 kb, and encodes a 188-amino acid protein [17]. Cripto-1 is identified to exert essential biological functions during embryogenesis. It is indispensable in coordinating primitive streak formation, mesoderm and endoderm specification as well as anterior and posterior (A/P) axis orientation [18, 19]. Cripto-1 is not detected in normal adult tissues and re-expression of Cripto-1 was observed in cancers [20–25]. The association between Cripto-1 overexpression and aggressive clinicopathological features together with findings from in vitro experiments portray the oncogenic role of Cripto-1 [23, 26]. In the liver, the expression of Cripto-1 in HCC clinical samples was reported in one study [27]. High cytoplasmic expression was detected in 50% HCC tissues and correlated with larger tumor size, higher tumor stage, and poorer survival [27]. These findings suggest that Cripto-1 may as well promote carcinogenesis of HCC. Yet despite the above, the functional roles of Cripto-1 in HCC have not been fully characterized in vitro and in vivo. In particular, whether Cripto-1 regulates stemness in HCC and the molecular mechanisms remain poorly understood.
In this report, we provide evidence that Cripto-1 is a key contributor to HCC stemness through regulation of canonical Wnt signaling. By means of a series of functional assays, Cripto-1 was demonstrated to enhance self-renewal, chemoresistance, tumorigenicity and metastatic potential of HCC. Mechanistically, Cripto-1 acts as a binding partner of intrinsic components in the pathway and a modulator of DVL3 expression through post-translational mechanism.
Results
Cripto-1 expression is associated with stemness in HCC
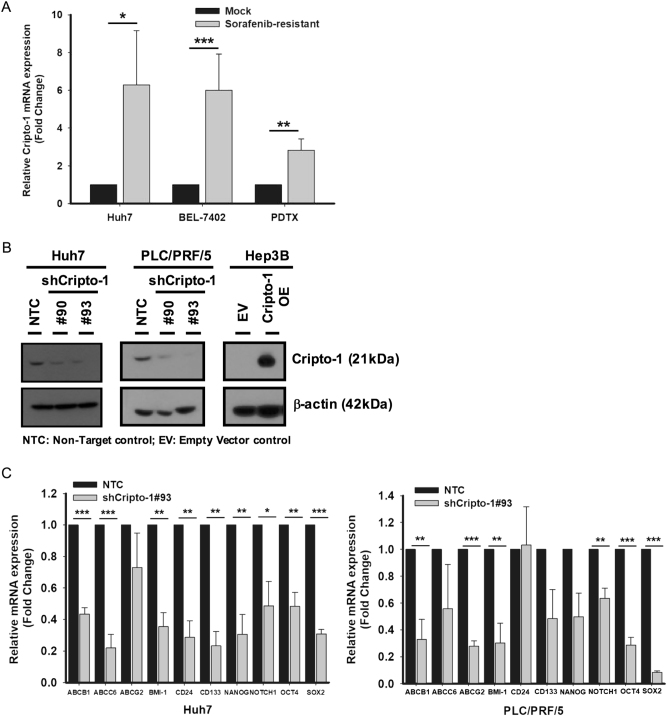
In our previous study, we developed sorafenib-resistant HCC cells both in vitro and in vivo via continuous exposure to sorafenib. The sorafenib-resistant clones demonstrated enhanced T-IC properties on functional characterization and enriched expression of CSC marker CD47 [28]. By examining the mRNA levels in the sorafenib-resistant HCC cells Huh7 and BEL-7402 as well as patient-derived tumor xenograft (PDTX) [28], Cripto-1 expression was found to be consistently upregulated as compared with control by 6.28 folds, 6.01 folds, and 2.83 folds, respectively (Fig. 1a). This prompted us to speculate that Cripto-1 could possibly be associated with stemness features in HCC. To gain further insights on this point, we employed a lentiviral-based short-hairpin (sh) knockdown approach to suppress Cripto-1 expression in Huh7 and PLC/PRF/5 HCC cells. A stable overexpression clone of Cripto-1 was also established in Hep3B cell line, which showed relatively low endogenous expression (Fig. 1b). Upon silencing of Cripto-1, we observed a downregulation of well-characterized stemness markers including BMI-1, CD24, CD133, NANOG, NOTCH1, OCT4, SOX2 and chemoresistance-related genes ABCB1, ABCG2, ABCC6 when compared with non-target control (NTC) (Fig. 1c).
Fig. 1.
Cripto-1 expression is associated with stemness in HCC. a Cripto-1 mRNA expression was upregulated in sorafenib-resistant Huh7, BEL-7402 cell lines and PDTX model (Huh7: n = 3; BEL-7402: n = 5; PDTX: n = 3). b Stable knockdown (Huh7 and PLC/PRF/5) and overexpression (Hep3B) clones of Cripto-1 were established. Alterations of Cripto-1 level were validated using western blotting. Non-target control (NTC) and empty vector control (EV) served as controls for knockdown (KD) and overexpression (OE) systems, respectively. c Knockdown of Cripto-1 (shCripto-1#93) in Huh7 and PLC/PRF/5 downregulated the expression of chemoresistance-related and stemness genes (Huh7: n = 3; PLC/PRF/5: n = 4) *p < 0.05, **p < 0.01 and ***p < 0.001, t-test. Data presented as mean ± SD
Cripto-1 determines cancer properties and self-renewal ability of HCC
We then proceeded to perform in vitro functional assays to interrogate the effects of Cripto-1 manipulation in HCC cells. Cell proliferation (Fig. 2a), migratory (Fig. 2b), and invasive (Fig. 2c) abilities of HCC cells were inhibited upon Cripto-1 knockdown. The self-renewal ability was tested by tumorsphere formation assay. Silencing of Cripto-1 in Huh7 and PLC/PRF/5 cell lines decreased the number and size of tumorspheres in both primary and secondary generations, whereas forced expression of Cripto-1 in Hep3B enhanced self-renewal of HCC (Fig. 2d).
Fig. 2.
Cripto-1 determines cancer properties and self-renewal ability of HCC. a Silencing of Cripto-1 inhibited cell proliferation in both Huh7 and PLC/PRF/5 cells (n = 3). Knockdown of Cripto-1 suppressed b migratory ability and c invasiveness of HCC cells (migration: n = 3; invasion: Huh7 n = 3; PLC/PRF/5 n = 4). d Silencing of Cripto-1 reduced the number and size of the tumorspheres in primary and secondary generations when comparing with non-target control (NTC) group in both Huh7 and PLC/PRF/5, whereas Cripto-1-overexpressing Hep3B cells enhanced the self-renewal ability (Huh7: n = 3; PLC/PRF/5: n = 4; Hep3B: n = 5; Scale bar: 50 μm). */#p < 0.05, **/##p < 0.01, and ***/###p < 0.001, t-test. Data presented as mean ± SD
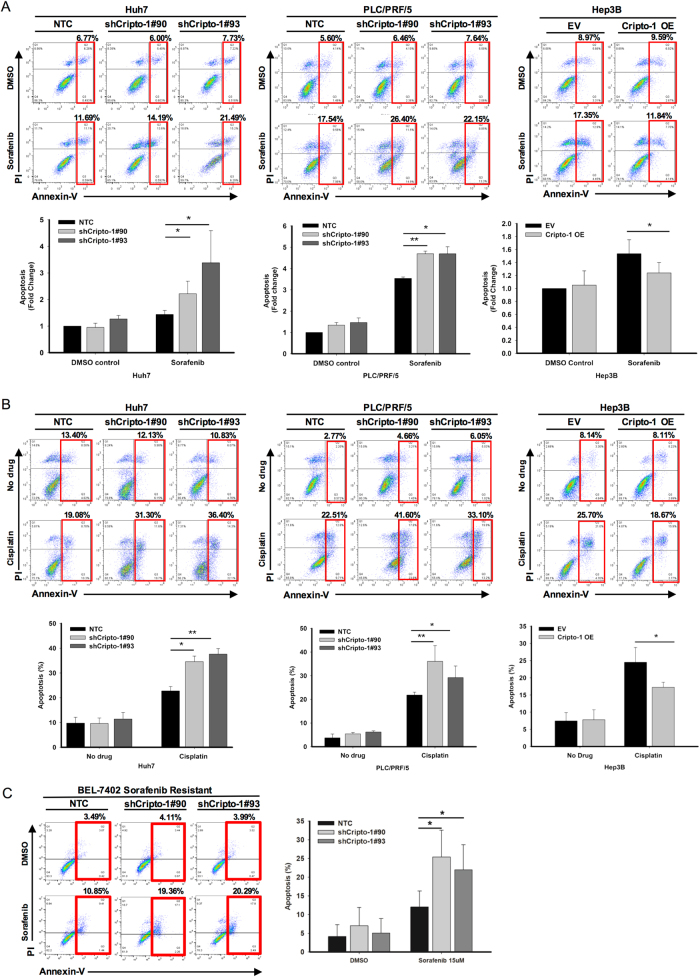
Cripto-1 confers resistance to sorafenib and cisplatin in HCC cells
Chemoresistance is a key feature of cancer stemness. Hence, we examined the effect on chemosensitivity of HCC cells upon altered expression of Cripto-1. By means of Annexin V apoptosis assay, silencing of Cripto-1 increased the proportion of HCC cells in apoptotic phase upon treatment with sorafenib (Fig. 3a) or cisplatin (Fig. 3b), in other words sensitized HCC cells to these chemotherapeutic agents. Force-expression of Cripto-1, in contrary, increased the chemoresistance of HCC cells. Intriguingly, knockdown of Cripto-1 in sorafenib-resistant BEL-7402 cells enhanced cell apoptosis and sensitivity towards sorafenib (Fig. 3c).
Fig. 3.
Cripto-1 confers chemoresistance to sorafenib and cisplatin in HCC cells. Knockdown of Cripto-1 increased the proportion of HCC cells in the apoptotic phase when treated with a sorafenib and b cisplatin. Cripto-1-overexpressing cells were more resistant to both chemotherapeutic agents when compared with empty vector (EV) control. c Suppression of Cripto-1 in sorafenib-resistant BEL-7402 cells enhanced chemosensitivity to sorafenib. Representative Annexin V-PI flow analysis plots of each treatment were shown (sorafenib treatment: n = 3–5; cisplatin treatment: n = 3–4) *p < 0.05, **p < 0.01, t-test). Data presented as mean ± SD
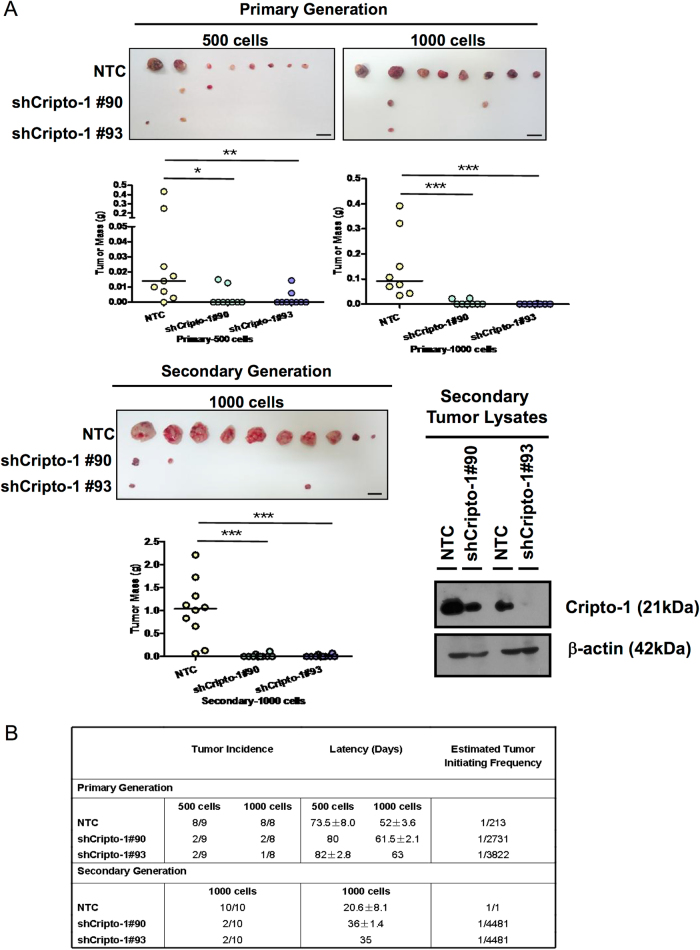
Cripto-1 enhances in vivo tumorigenicity
The regulatory role of Cripto-1 in tumor growth and stemness properties of HCC was further investigated in a biological environment using in vivo mouse model. By means of subcutaneous inoculation in NOD/SCID mice and a limiting dilution approach with 500 and 1000 cells, knockdown of Cripto-1 in Huh7 suppressed in vivo tumorigenicity, as evidenced by reduced tumor mass (Fig. 4a), decreased tumor incidence (500 cells—NTC: 8/9 versus shCripto-1: 2/9; 1000 cells—NTC: 8/8 versus shCripto-1: 2/8 and 1/8) and prolonged latency period (500 cells—NTC: 73.5 days versus shCripto-1: 80 days and 82 days; 1000 cells—NTC: 52 days versus shCripto-1: 61.5 days and 63 days). The effects on tumorigenicity in terms of these parameters were propagated in the secondary inoculation upon serial transplantation (tumor incidence—NTC: 10/10 versus shCripto-1: 2/10; latency period—NTC: 20.6 days versus shCripto-1: 36 days and 35 days). Estimated tumor initiating frequency was reduced from 1/213 to 1/2731 and 1/3822 after silencing of Cripto-1 in primary generation, whereas it was further diminished from 1/1 to 1/4481 in secondary generation (Fig. 4b).
Fig. 4.
Cripto-1 regulates in vivo tumorigenicity. Silencing of Cripto-1 (shCripto-1#90 and shCripto-1#93) suppressed in vivo tumorigenicity in limited dilution manner upon serial transplantation in NOD/SCID mice. a Knockdown of Cripto-1 decreased the tumor mass in both primary and secondary generations. Knockdown of Cripto-1 in shCripto-1 was validated in tumors from secondary generation. (Primary n = 8–9, Secondary n = 10, *p < 0.05, **p < 0.01, and ***p < 0.001, Mann–Whitney’s U-test; Scale bar: 1 cm). b Silencing of Cripto-1 reduced tumor incidence and prolonged tumor latency period. Latency was presented as mean ± SD. The suppression of Cripto-1 diminished the tumor initiating frequency in both primary and secondary generations when compared with the respective NTC groups
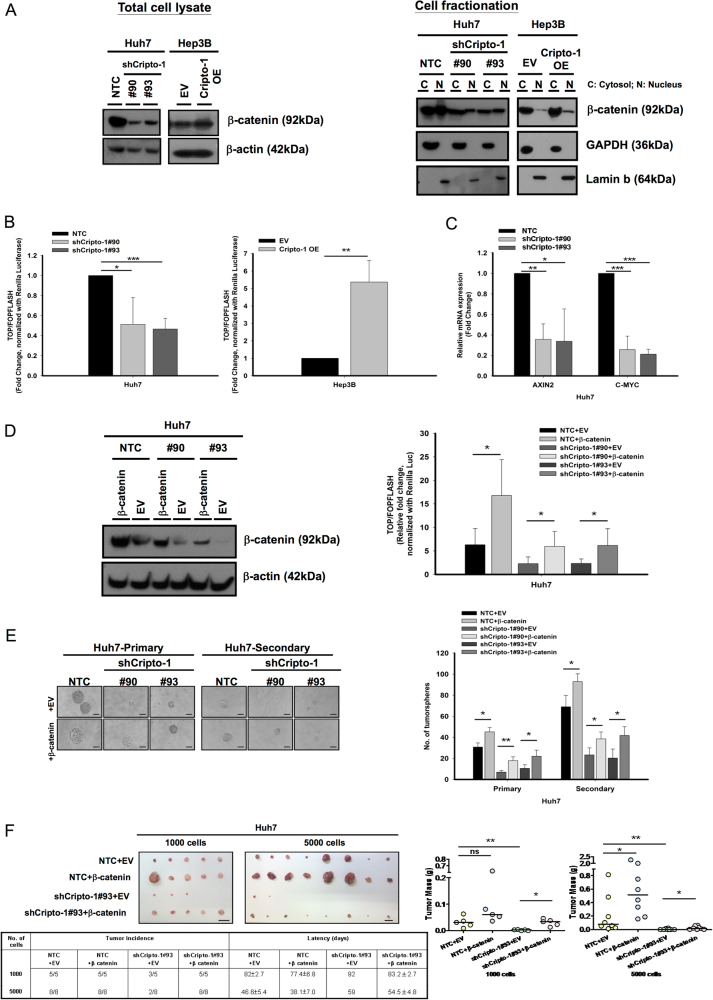
Cripto-1 regulates stemness properties in HCC via Wnt/β-catenin signaling
To delineate the molecular mechanism mediating the effect of Cripto-1 in HCC, we examined the expression of key molecular targets of the Wnt/β-catenin pathway upon manipulated Cripto-1 expression. Consistently in both Huh7 and PLC/PRF/5 cell lines, knockdown of Cripto-1 downregulated β-catenin expression in total cell lysate and the nuclear fraction. On the contrary, forced expression of Cripto-1 enhanced β-catenin expression (Fig. 5a and Supplementary Figure S1A). These were accompanied by corresponding alterations in Wnt/β-catenin activity measured by TOP/FOPFlash reporter assay (Fig. 5b and Supplementary Figure S1B) and expression levels of β-catenin downstream targets AXIN2 and C-MYC (Fig. 5c and Supplementary Figure S1C). To ascertain that the stemness properties endowed by Cripto-1 were mediated through Wnt/β-catenin signaling, we carried out rescue experiments through stable expression of constitutively active β-catenin (Plasmid 16520, Addgene, Cambridge, MA, USA) in Cripto-1 silenced HCC cells. Stable expression of β-catenin was confirmed by western blotting. The suppression in signaling activation with Cripto-1 knockdown was abrogated by enforcement of β-catenin (Fig. 5d and Supplementary Figure S1D). In addition, the functional effects on in vitro self-renewal (Fig. 5e and Supplementary Figure S1E) and in vivo tumorigenicity upon silencing of Cripto-1 were partially rescued by forced expression of β-catenin (Fig. 5f).
Fig. 5.
Cripto-1 modulates stemness properties in HCC via regulation of Wnt/β-catenin signaling. a Silencing of Cripto-1 suppressed, whereas forced expression of Cripto-1 in Hep3B enhanced, the protein expression of total and nuclear β-catenin (C cytosol and N nucleus). b Knockdown of Cripto-1 suppressed the activity of Wnt/β-catenin signaling in Huh7. Forced expression of Cripto-1 enhanced the signal activation as measured by TOP/FOPFLASH luciferase assay (n = 3, *p < 0.05, **p < 0.01, and ***p < 0.001, t-test). c Silencing of Cripto-1 downregulated the transcript level of β-catenin downstream targets AXIN2 and C-MYC in Huh7 (n = 3, *p < 0.05, **p < 0.01, and ***p < 0.001, t-test). d Stable expression of β-catenin was enforced in shCripto-1 clones in Huh7 and the expression of β-catenin was confirmed by Western blotting. The suppression of Wnt/β-catenin signaling activity in shCripto-1 cells was partially nullified upon enforcement of active β-catenin when compared with empty vector (EV) group (n = 5, *p < 0.05, t-test). e The suppression of tumorsphere formation ability upon Cripto-1 knockdown in Huh7 was abrogated by enforcement of active β-catenin (primary and secondary generations n = 3, *p < 0.05, and **p < 0.01, t-test; Scale bar: 50 μm). Data presented as mean ± SD. f β-catenin stable expression or empty vector (EV) were enforced in both shCripto-1#93 and non-target control (NTC) Huh7 cells and subcutaneously inoculated with 1000 and 5000 cells in NOD/SCID mice. The effects on tumor growth, tumor incidence, and latency period conferred by Cripto-1 knockdown were partially rescued with expression of β-catenin. Latency was presented as mean ± SD. (n = 5–8, ns not significant, *p < 0.05 and **p < 0.01, Mann–Whitney’s U-test; Scale bar: 1 cm)
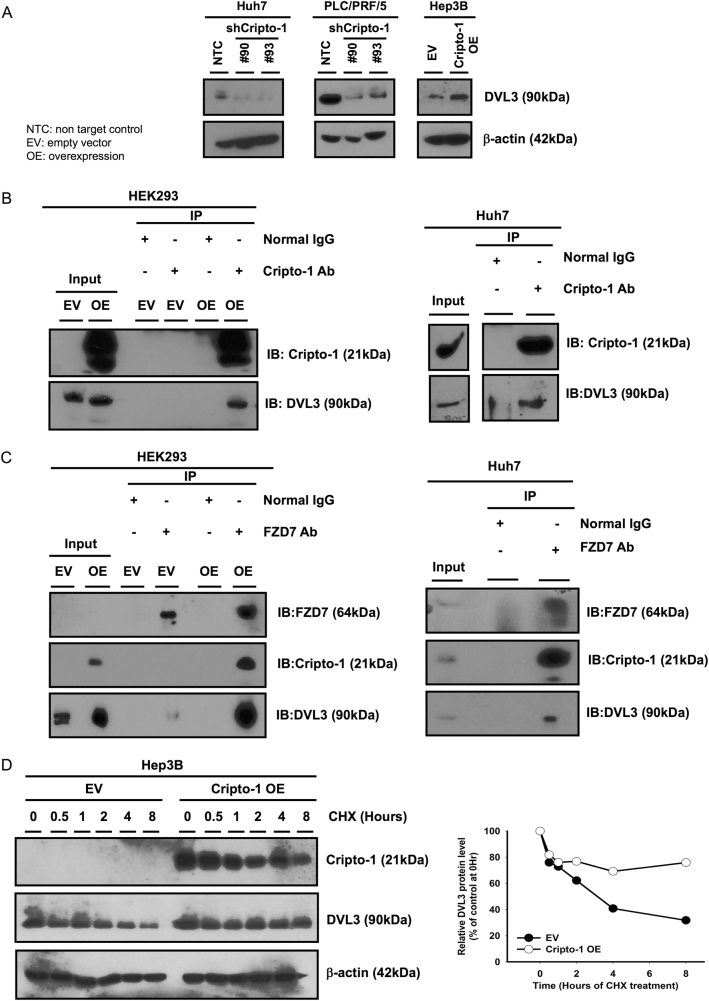
Cripto-1 binds to FZD7/LRP6 and DVL3 and attenuates DVL3 protein degradation
The above findings led us to further investigate how exactly Cripto-1 participates in the Wnt/β-catenin signaling cascade. To this end, we explored the potential direct target or interacting partner of Cripto-1. With western blotting, we found that Cripto-1 positively regulated the expression of DVL3, a modulator of the β-catenin degradation complex (Fig. 6a); while it did not significantly alter the DVL3 mRNA level (Supplementary Figure S2). In view of this we first examined whether there is any binding relationship between the two. Results from co-immunoprecipitation (Co-IP) experiments showed that Cripto-1 bound to DVL3 in both HEK293 cells with Cripto-1 transient expression and parental Huh7 cells (Fig. 6b), which showed a relatively high endogenous Cripto-1 level among HCC cell lines. Next, we studied the possibility of other binding partners for Cripto-1. Previous studies have shown that DVL3 physically associates with FZD receptors [29]. Thus, we speculated that Cripto-1 might also interact with the FZD receptors. FZD7 is a well-characterized member among the FZD receptor family in hepatocarcinogenesis [30–32]. We then proceeded to Co-IP assays similarly and interestingly, a binding interaction between Cripto-1 and FZD7 was also observed (Fig. 6c). Similarly a protein–protein physical interaction was also detected between Cripto-1 and LRP6, the latter of which is known to physically associates with FZD receptors (Supplementary Figure S3). Taking a step further, we interrogated the mechanism through which Cripto-1 increase the protein expression of DVL3. By cycloheximide chase assay, we observed a slower rate of DVL3 protein degradation upon Cripto-1 overexpression when compared with the EV control. Overexpression of Cripto-1 enhanced the half-life of DVL3 from 3 hours to >8 hours (Fig. 6d). The above findings suggest that Cripto-1 binds to DVL3 and FZD7/LRP6, and such interaction upregulates DVL3 expression through protein stabilization and activates Wnt/β-catenin signaling.
Fig. 6.
Cripto-1 binds to DVL3 and FZD7 in the Wnt/β-catenin signaling cascade and modulates DVL3 expression. a Protein expression of DVL3 was downregulated upon Cripto-1 knockdown compared with the non-target control (NTC) in Huh7 and PLC/PRF/5. DVL3 expression was enhanced under forced expression of Cripto-1. b Cripto-1 was immunoprecipitated using anti-Cripto-1 antibody and the immunoprecipitated proteins were analyzed by western blotting (IB) with anti-DVL3 antibody. DVL3 was detected from the immunoprecipitated protein when compared with normal IgG control in both HEK293 with Cripto-1 transient expression and parental Huh7 cells, suggesting that Cripto-1 bound to DVL3. c FZD7 was immunoprecipitated using anti-FZD7 antibody. The immunoprecipitated proteins were analyzed with anti-DVL3 and Cripto-1 antibodies. Cripto-1 and DVL3 were detected when compared with normal IgG control in both HEK293 with Cripto-1 transient expression and parental Huh7 cells, showing a binding interaction between Cripto-1, FZD7, and DVL3. d With cyclohexamide treatment, the stability of DVL3 protein was enhanced upon overexpression of Cripto-1. The expression levels showed a time-dependent decrease in the EV control groups. The DVL3 protein level at each time point was quantified using ImageJ, and normalized with respective β-actin. The degradation curve was plotted as percentage relative to the expression level at 0 h of treatment. Representative dataset from three independent experiments was shown
Cripto-1 level in clinical samples is correlated with prognosis of HCC patients
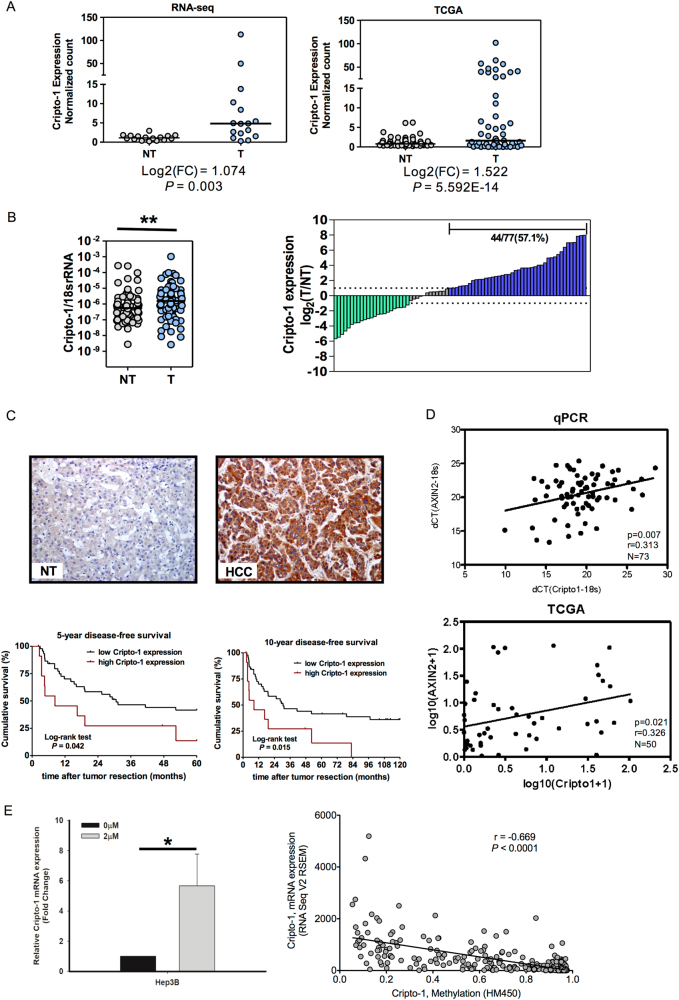
Lastly, we performed some expression analyses with HCC clinical samples. From both our in-house RNA sequencing dataset (available in Sequence Read Archive of National Center for Biotechnology Information with the accession number SRP062885) and the TCGA database, Cripto-1 was significantly upregulated in HCC tumor tissues when compared with the corresponding non-tumoral liver tissues (Fig. 7a). We then carried out qRT-PCR analysis in an independent cohort of 77 HCC clinical samples and confirmed the overexpression of Cripto-1 in HCC. 44 (57.1%) cases showed Cripto-1 upregulation (tumor/non-tumor ≥ 2 folds) (Fig. 7b). Cripto-1 overexpression in HCC was also detected by immunohistochemical staining (Fig. 7c). Higher Cripto-1 expression was associated with poorer disease-free survival of HCC patients at 5 years (p = 0.042) and 10 years (p = 0.015). (Fig. 7c). Moreover, the expression levels of Cripto-1 and AXIN2 were positively correlated in both our clinical cohort (p = 0.007) and the TCGA database (p = 0.021) (Fig. 7d). Immunohistochemical expression in tissues from the normal livers (n = 4), livers with chronic hepatitis (n = 32), cirrhotic livers (n = 41), and liver dysplastic nodules (n = 14) were also analyzed. There was minimal Cripto-1 expression in normal livers, chronic hepatitis or cirrhotic livers, while 7 (50%) dysplastic nodules showed focal Cripto-1 expression (Supplementary Figure S4). To interrogate the regulatory mechanism for Cripto-1 expression in HCC, we first treated Hep3B cells, which have low endogenous Cripto-1 expression, with demethylating agent 5-aza-2′-deoxycytidine (5-Aza) and examined the change in Cripto-1 expression. Treatment with 5-Aza significantly restored Cripto-1 mRNA level. Further analysis from the TCGA database revealed a negative correlation between the DNA methylation status of Cripto-1 and Cripto-1 expression in HCC (Fig. 7e).
Fig. 7.
Cripto-1 levels in HCC clinical samples carry prognostic significance and are correlated with AXIN2 expressions. a Cripto-1 was significantly upregulated in HCC tumor tissues (T) versus the corresponding non-tumorous liver tissues (NT) from both in-house RNA sequencing dataset (n = 16) and the TCGA database (n = 50). b Cripto-1 level was significantly upregulated in HCC tumor (T) versus non-tumorous liver (NT) tissues by qRT-PCR (n = 77, **p < 0.01, Wilcoxon signed-rank test). Cripto-1 upregulation was identified in 57.1% (44/77) of the primary HCC cohort. Data presented as log2 ratio of Cripto-1 mRNA level in HCC tissues as compared to NT. Upregulation was defined as log2 (T/NT) ≥1. c Cripto-1 protein level was overexpressed in HCC by immunohistochemical staining (IHC). Representative images were shown. High Cripto-1 immunoexpression was associated with poorer 5-year (p = 0.042) and 10-year (p = 0.015) disease-free survival of HCC patients (log-rank test). d Cripto-1 and AXIN2 expressions were positively correlated with both in-house clinical HCC samples (n = 73, r = 0.313, and p = 0.007) and TCGA dataset (n = 50, r = 0.326, p = 0.021, and Pearson correlation). e Treatment of Hep3B cells with 5-Aza-2′-deoxycytidine significantly upregulated Cripto-1 mRNA expression (n = 3, *p < 0.05). Analysis of the TCGA database revealed a negative correlation between DNA methylation status and expression of Cripto-1 in HCC (r = −0.669, p < 0.0001, and Pearson correlation)
Discussion
In the current study, by means of functional assays and expression analyses we unraveled the regulatory role of Cripto-1 on stemness phenotypes in HCC. We delineated that Cripto-1 mediates HCC stemness through canonical Wnt signaling via binding with FZD7/LRP6 and DVL3 and modulation of DVL3 level (Supplementary Figure S5). Our findings add a new layer of knowledge on key molecular targets driving HCC stemness and the mechanistic events leading to activation of Wnt/β-catenin pathway in HCC. A recent report by Florenzano et al. described Cripto-1 as an epiblast marker which is deterministic for maintaining pluripotent states. The authors showed that Cripto-1 sustains self-renewal ability of mouse embryonic stem cell (ESC) through Wnt/β-catenin signaling [33]. The findings of our current study probably indicate that re-expression of Cripto-1 in liver cancer recapitulates the functions and signaling mechanism of embryonic processes.
For cancers, studies using murine cell lines also provided a clue on the link between Cripto-1 and canonical Wnt pathway [34], and a recent report illustrated this association in human prostate cancer by expression correlations [35]. Interestingly, in other reports Cripto-1 was shown to take part in stem cell maintenance and pluripotency regulation in embryonal carcinoma, colorectal and esophageal cancers [36–38]. In these cancer models, the stemness properties were demonstrated by functional characterization in vitro and in vivo. Yet, only the study on colorectal cancer delineated the downstream molecular mechanism, which focused on the Src/Akt pathway [37]. Our experimental results provide solid comprehensive evidence on the characterization of Cripto-1 in HCC stemness including its critical role in the modulation of Wnt/β-catenin signaling in human cancer models. Although Cripto-1 was found to be a direct downstream target of β-catenin in one report in which Cripto-1 expression was examined at transcript level [39], upon forced expression of β-catenin we did not observe increased protein expression of Cripto-1 in our system using Huh7 cell line (Supplementary Figure S6).
FZD/LRP receptors together with DVL are intrinsic key players in the Wnt/β-catenin pathway. Studies have shown that alteration of these targets regulates the pathway activity in cancers including HCC [13, 31, 40]. Overexpression of these targets serves to activate Wnt/β-catenin signaling. Previously we identified Prickle-1 and Sox9 as upstream regulators of DVL3 and FZD7 respectively in HCC [11, 14]. In the current study, we found that Cripto-1 acts as a binding partner for both DVL3 and FZD7/LRP6 and a modulator of DVL3 expression through altering its protein stability. Given that DVL and FZD are known interacting partners in the pathway [29], we postulate that Cripto-1 forms a functional complex with FZD7, LRP6, and DVL3 in HCC and potentially serves to facilitate the interaction among these targets. This finding provides novel insights to the molecular events in the activation of the canonical Wnt signaling cascade in liver cancer. Furthermore, it substantiates the significance of Cripto-1 in governing Wnt/β-catenin signal transduction through simultaneous interaction with multiple key molecules in the signaling cascade.
Apart from functioning in the Wnt/β-catenin signal transduction axis, Cripto-1 potentially serves as a link with other signaling pathways in HCC. An earlier study showed that NANOG regulated Cripto-1 expression and induced cell migration and invasion through epithelial–mesenchymal transition in HCC cell lines [41]. In this regard, several studies demonstrated the potential crosstalk between NANOG and the canonical Wnt pathway in different manners [42–44]. From our analysis of both in-house cohort and the TCGA database, the expression levels of Cripto-1 and NANOG in HCC clinical samples were not significantly correlated (Supplementary Figure S7). Whether Cripto-1 and NANOG work together to regulate HCC stemness features and the detailed molecular mechanism is yet to be clarified.
With our clinical samples Cripto-1 level was associated with prognostic significance, a finding consistent with that from a previous study [27]. The focal expression in dysplastic nodules, a precursor lesion of HCC, suggested that Cripto-1 upregulation is involved in the multi-step hepatocarcinogenesis. Cripto-1 may also be a potential strategic target for sorafenib-resistant HCC. In addition, the significant role of Cripto-1 in the signal transduction of canonical Wnt pathway renders it an appealing therapeutic target. Exploration of the regulatory mechanism for Cripto-1 expression in HCC would be one of the future directions to enhance the translational value of our findings. In this regard, epigenetics is gaining importance as a common and potent regulatory mechanism of gene expression in HCC. Notably, the epigenetic makeup is an efficient machinery in governing transcription patterns to maintain cell state, manipulate cell plasticity and to orchestrate intratumoral heterogeneity [45, 46]. We observed a negative correlation between promoter DNA methylation level and Cripto-1 transcript level. DNA hypomethylation coupling with overexpression of gene targets has been observed in human cancers [47–49]. Our findings suggest that DNA hypomethylation is a possible mechanism contributing to Cripto-1 overexpression in HCC. As far as other epigenetic mechanisms are concerned, microRNAs (miR-15a-16 and miR-15b) were found to regulate Cripto-1 expression in brain tumor and lung cancer [26, 50]. In HCC, by means of two computational analysis programs TargetScan and miRANDA, we identified a list of miRNAs potentially regulating Cripto-1 expression. We further shortlisted the candidates to those at the same time being downregulated in HCC according to our database, and further to those being proven tumor suppressors in HCC [51–55]. By means of this algorithm, we identified Let-7, miR-26, miR-195, miR-424, and miR-422a as potential targets in HCC. Validation of these findings could possibly help to pave the knowledge platform for targeting Cripto-1 in HCC by various epigenetic approaches.
Materials and methods
Clinical samples
Human liver tissue samples were obtained from patients with liver resection at Queen Mary Hospital, Hong Kong. All specimens collected were either snap-frozen in liquid nitrogen and stored at −80 °C, or fixed in 10% formalin for paraffin embedding. Frozen sections from tumorous and non-tumorous liver samples were cut and stained for histological examination to ensure homogenous cell population of tissues. Use of human specimens was approved by the institutional review board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster.
Cell line models
PLC/PRF/5 and Hep3B cell lines were purchased from American Type Culture Collection (Manassas, VA). Huh7 was obtained from Dr. H. Nakabayashi at Hokkaido University School of Medicine (Sapporo, Japan). HEK293 was purchased from Invitrogen. All cell lines were cultured at 37 °C in a humidified incubator with 5% carbon dioxide (CO2). The cell lines were authenticated by morphological observation and short tandem repeat-based assay with AuthentiFiler PCR amplification kit (ABI).
Immunohistochemistry (IHC) and assessment
Immunohistochemical staining for Cripto-1 was performed on formalin-fixed, paraffin-embedded sections of 4-μm thick using labeled horseradish peroxidase (HRP) method. After 15 min-heat antigen retrieval with Target Retrieval Solution (Dako, Glostrup, Denmark), endogenous peroxidase activities were quenched by 3% H2O2. The sections were immersed in serum free-protein block solution (Dako) and incubated with antibody against Cripto-1 (1:100, ab19917, Abcam, Cambridge) diluted in antibody diluent at 4 °C overnight. The sections were then washed thoroughly and incubated with EnvisionTM HRP-conjugated secondary antibody (Dako). Positive signals were visualized using 3,3′-diaminobenzidine (Dako). Nuclei were counterstained with hematoxylin. Immunohistochemical staining was assessed by a histopathologist (RCL) using a semi-quantitative method. Cytoplasmic expression in tumor tissue was graded as low for 0–33% staining and high for >33% staining, respectively.
Preparation of protein extracts and western blot analyses
Whole cell lysates were extracted using RIPA buffer supplemented with protease inhibitor cocktail. Protein lysate were separated by SDS-polyacrylaminde gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membrane (Millipore, MA, USA) for western blotting analyses. Primary antibodies against Cripto-1 (1:1000, ab108391, Abcam), total β-catenin (1:2500, 610153, BD Transduction Laboratories, San Jose, CA), Dishevelled-3 (DVL3) (1:1000, sc-8027, Santa Cruz Biotechnology), Frizzled-7 (FZD7) (1:1000, ab51049, Abcam), LRP6 (1:1000, #2560, Cell Signaling) and β-actin (1:80,000, A5316, Sigma-Aldrich, St. Louis, MO, USA) were incubated at 4 °C overnight. HRP-conjugated antirabbit or antimouse IgG were used as secondary antibodies at 1:5000 where appropriate (Santa Cruz Biotechnology, Texas, USA). The signals were visualized using the enhanced chemiluminescence method.
Cripto-1 knockdown and overexpressing HCC cells
Short-hairpin RNAs (shRNAs) targeting Critpo-1 (shCripto-1) were ordered from Sigma-Aldrich (Mission® shRNA bacterial glycerol stock). The clone IDs were TRCN0000004890 (shCripto-1#90) and TRCN0000004893 (shCripto-1#93). To package the virus, 293FT cells were seeded 24 h prior to transfection. Either shCripto-1 or non-target control (NTC) plasmid were co-transfected with packaging plasmid mix (System Biosciences, Mountain View, CA) using Lipofectamine® 2000 (Invitrogen, Thermo Fisher Scientific, MA, USA). The viral supernatant was collected and used to infect Huh7 and PLC/PRF/5 in polybrene (Sigma-Aldrich) supplemented media. The infected cells were selected with gradient concentration of puromycin (Sigma-Aldrich). The success of silencing of Cripto-1 in HCC cells was confirmed using western blotting. For overexpression study, Cripto-1 coding region was amplified with primers indicated at Supplementary Table S1 and was cloned into pcDNA3.1 (Cripto-1-OE) (Invitrogen). Either Cripto-1-OE or empty vector (EV) was transfected into Hep3B using Lipofectamine® 2000. Empty vector and stable Cripto-1 expressing clones were established with G418 selection.
Cell proliferation assay
Proliferation of Cripto-1 knockdown cells was determined using cell counting method. Total number of cells was counted in triplicates each day for six days using trypan blue exclusion method.
Cell migration and matrigel invasion transwell assays
Migration assays were performed using polycarbonate membrane transwell inserts with pore size of 8.0 μm (Millpore). Invasion assays were performed using Matrigel (BD Biosciences, San Jose, CA) coated transwell inserts. Cells in serum-free medium were seeded in upper chamber with the lower chamber supplemented with medium supplemented with 10% FBS as chemoattractant. Cells were incubated in humidified incubator at 37 °C for 24 h. The transwell membranes were fixed with methanol and stained with 1% crystal violet. The membranes were cleaned, air-dried, mounted on slides, and documented. The number of cells migrated or invaded was counted in five random fields.
Tumorsphere formation assay
Five hundred Huh7 or 300 PLC/PRF/5 cells were seeded to 24-well plates coated with polyHEMA (Sigma Aldrich). Cells were grown in DMEM/F12 medium (Invitrogen) for 10 day supplemented with 4 μg/mL insulin (Sigma-Aldrich), B27 (Invitrogen), 20 ng/mL EGF (Sigma-Aldrich), and 20 ng/mL basic FGF (Invitrogen). The medium was supplemented with 0.25% methylcellulose (Sigma-Aldrich) for PLC/PRF/5. The number of tumorspheres was counted under microscope.
RNA extraction and quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen) according to manufacturer’s protocol. One microgram of RNA was used for cDNA synthesis using GeneAmp RNA PCR Kit (Applied Biosystems, Thermo Fisher Scientific). Expression of Cripto-1 of clinical samples was examined using TaqMan® Assay target probes (Assay ID: Hs02339499_g1) and TaqMan® Universal PCR Master Mix, normalized using 18s rRNA. Expression of chemoresistance-associated genes, stemness genes and β-catenin downstream targets were examined by Power SYBR® Green PCR Master Mix (Applied Biosystems) with Applied Biosystems 7900HT fast real-time PCR System (Supplementary Table S2). Relative expression differences were calculated using 2−ΔΔCT method with reference to 18s rRNA.
Annexin V-apoptosis assay
Annexin V apoptosis assays were performed using Annexin V-FITC reagent (BD Pharmingen™) and propidium iodide (PI) according to manufacturer’s protocol. The stained cells were resuspended in binding buffer and subjected to analyses using FACSCalibur flow cytometer and CellQuest software (BD BioSciences).
β-catenin TCF binding assay
β-catenin activity was examined using luciferase reporter assay of TCF/LEF-dependent transcription (TOP/FOPFLASH reporter assay). Either Firefly luciferase pSuper8XTOPflash or pSuper8XFOPflash constructs (gifts from Dr. Moon R, University of Washington, USA), together with Renilla luciferase construct pRL-CMV (Promega, Madison, WI, USA) for normalization of transfection efficiency, were transfected using Lipofectamine® 2000. Luciferase activities were assayed using Dual-Luciferase® Reporter Assay System (Promega) according to manufacturer’s protocol.
In vivo subcutaneous xenograft inoculation
The study protocol was approved by the Committee of the Use of Live Animals in Teaching and Research at the University of Hong Kong. Limited dilution assays were performed with Huh7 cells. They were resuspended in 100 μL PBS/Matrigel (1:1) and injected subcutaneously at both sides of posterior flanks of 4 to 6-week-old male non-obese diabetic (NOD)/SCID mice (NOD.CB17-Prkdcscid/J). The animals were sacrificed and tumor bulks were harvested. Tumor mass, tumor incidence and latency period were recorded. Primary tumors were mechanically dissociated and subjected to Liberase (Roche Diagnostics, Basel, Switzerland) digestion supplemented with DNase I (Roche Diagnostics). The enzyme suspensions were inactivated with DMEM/F12 medium containing 10% FBS. The dispersed cells were filtered with 100 μm followed by 70 μm cell strainers and pelleted. One thousand cells of each group were subcutaneously injected to secondary recipient mice as indicated previously. Tumors harvested from the secondary recipients were snapped frozen, lyophilized, and lysed in RIPA buffer for western blot analyses. Tumor initiating frequency was calculated using extreme limiting dilution analysis (http://bioinf.wehi.edu.au/software/elda/).
Co-immunoprecipitation (Co-IP) assays
Cripto-1 was transiently overexpressed in HEK293 cells using the Cripto-1-OE vector established. Empty vector (pcDNA3.1) was transfected as control. Cells were harvested at 48 h-post-transfection and lysed using RIPA buffer supplemented with protease inhibitor cocktail. Either Cripto-1 or FZD7 (16974-1AP, Proteintech Group Inc., Chicago, USA) antibodies or normal rabbit IgG (Santa Cruz Biotechnology) were incubated with cell lysates overnight with rotation at 4 °C. The protein-antibody complexes were captured using Dynabeads® Protein G (Invitrogen). The immunoprecipitated protein complexes were washed, eluted and subjected to western blot analyses. The experiment was also performed using parental Huh7 cells.
Cycloheximide chase experiment
Either Hep3B empty-vector (EV) or Cripto-1 stable-expressing clone was treated with translational inhibitor cycloheximide (CHX, Sigma) at concentration of 50 μg/mL for indicated time. Target protein expression were quantified using ImageJ, which was normalized with respective β-actin levels. Protein level remaining upon CHX treatment was plotted relative to untreated control at 0 h.
The cancer genome atlas (TCGA) analysis and statistical analysis
Global gene expression profiles by whole-transcriptome sequencing (WTS) were obtained from the TCGA data portal. Data of 50 paired HCC and non-tumorous liver samples were retrieved for subsequent analyses, as previously described [56]. Differential gene expression analysis was performed using edgeR on the raw read counts of Cripto-1. It used negative binomial models to capture variance dispersion for WTS read count data, empirical Bayes estimation for gene-specific variation, and generalized linear models applicable to general experiments. Since raw read count values of Cripto-1, AXIN2, and NANOG were skewed, expression correlation analysis was based on transformed values, calculated as log10 (count + 1) for expression levels of both genes. Promoter methylation status and expression of Cripto-1 from 373 HCC samples were retrieved from the TCGA database. The correlation was analyzed by Pearson correlation. Statistical analysis for clinicopathological correlation was performed using SPSS 19 for Windows (SPSS, Inc., Chicago, IL). Fisher’s exact test was used for categorical data. Survival analysis was performed by Kaplan–Meier method and the difference was compared by log-rank test. All experimental data were analyzed using t-test or Mann–Whitney’s U-test wherever appropriate (SigmaPlot 10.0 and SigmaStat 3.1, Jandel Scientific, San Rafael, CA; or GraphPad Prism 5.0, La Jolla, CA, USA). p < 0.05 was considered as statistically significant.
Electronic supplementary material
Supplementary figures and legends clean copy
Acknowledgements
This work is supported by General Research Fund, Research Grants Council of Hong Kong (17126216). The authors thank Dr Stephanie KY Ma for critically reviewing the manuscript. We also thank Dr Alan Kai, Dr Lo-Kong Chan, Mr Han-Wei Tan, Ms Joyce Lee and Faculty Core Facility of The University of Hong Kong for technical support. IOL Ng is Loke Yew Professor in Pathology and is supported by RGC Theme-based Research Scheme (T12-704/16-R).
Author contributions
Conception and design: RCL. Development of methodology: COL, TKL, ION. Acquisition of data: COL, RCL, KKC, TKL. Analysis and interpretation of data: RCL, COL, DWH, CMW. Writing, review, and/or revision of the manuscript: RCL, COL, T.K.Lee, ION. Supervision of the study: RCL, ION.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Regina Cheuk-Lam Lo and Carmen Oi-Ning Leung.
Edited by Y Shi
Contributor Information
Regina Cheuk-Lam Lo, Phone: +(852) 2255-2688, Email: reginalo@pathology.hku.hk.
Irene Oi-Lin Ng, Phone: +(852) 2255-2658, Email: iolng@hku.hk.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0059-x) contains supplementary material, which is available to authorized users.
References
- 1.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–61. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 2.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–9. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 3.Marquardt JU, Thorgeirsson SS. Stem cells in hepatocarcinogenesis: evidence from genomic data. Semin Liver Dis. 2010;30:26–34. doi: 10.1055/s-0030-1247130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo RC, Ng IO. Hepatic progenitor cells: their role and functional significance in the new classification of primary liver cancers. Liver Cancer. 2013;2:84–92. doi: 10.1159/000343844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee TK, Cheung VC, Ng IO. Liver tumor-initiating cells as a therapeutic target for hepatocellular carcinoma. Cancer Lett. 2013;338:101–9. doi: 10.1016/j.canlet.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Pez F, Lopez A, Kim M, Wands JR, Caron de Fromentel C, Merle P. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol. 2013;59:1107–17. doi: 10.1016/j.jhep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Wong CM, Fan ST, Ng IO. beta-Catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer. 2001;92:136–45. doi: 10.1002/1097-0142(20010701)92:1<136::AID-CNCR1301>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 8.Nejak-Bowen KN, Monga SP. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831–9. doi: 10.1158/0008-5472.CAN-07-0908. [DOI] [PubMed] [Google Scholar]
- 10.Yang W, Yan HX, Chen L, Liu Q, He YQ, Yu LX, et al. Wnt/beta-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res. 2008;68:4287–95. doi: 10.1158/0008-5472.CAN-07-6691. [DOI] [PubMed] [Google Scholar]
- 11.Chan DW, Chan CY, Yam JW, Ching YP, Ng IO. Prickle-1 negatively regulates Wnt/beta-catenin pathway by promoting Dishevelled ubiquitination/degradation in liver cancer. Gastroenterology. 2006;131:1218–27. doi: 10.1053/j.gastro.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Tung EK, Mak CK, Fatima S, Lo RC, Zhao H, Zhang C, et al. Clinicopathological and prognostic significance of serum and tissue Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int. 2011;31:1494–504. doi: 10.1111/j.1478-3231.2011.02597.x. [DOI] [PubMed] [Google Scholar]
- 13.Tung EK, Wong BY, Yau TO, Ng IO. Upregulation of the Wnt co-receptor LRP6 promotes hepatocarcinogenesis and enhances cell invasion. PLoS One. 2012;7:e36565. doi: 10.1371/journal.pone.0036565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung CO MW, Kai AK, Chan K, Lee TK, Ng IO, Lo RC. Sox9 confers stemness properties in hepatocellular carcinoma through Frizzled-7 mediated Wnt/beta-catenin signaling. Oncotarget. 2016;7:29371–86. doi: 10.18632/oncotarget.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bianco C, Rangel MC, Castro NP, Nagaoka T, Rollman K, Gonzales M, et al. Role of Cripto-1 in stem cell maintenance and malignant progression. Am J Pathol. 2010;177:532–40. doi: 10.2353/ajpath.2010.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minchiotti G, Parisi S, Liguori GL, D’Andrea D, Persico MG. Role of the EGF-CFC gene cripto in cell differentiation and embryo development. Gene. 2002;287:33–7. doi: 10.1016/S0378-1119(01)00892-7. [DOI] [PubMed] [Google Scholar]
- 17.Ciccodicola A, Dono R, Obici S, Simeone A, Zollo M, Persico MG. Molecular characterization of a gene of the ‘EGF family’ expressed in undifferentiated human NTERA2 teratocarcinoma cells. EMBO J. 1989;8:1987–91. doi: 10.1002/j.1460-2075.1989.tb03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin JZ, Ding J. Cripto is required for mesoderm and endoderm cell allocation during mouse gastrulation. Dev Biol. 2013;381:170–8. doi: 10.1016/j.ydbio.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, et al. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature. 1998;395:702–7. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- 20.Gong YP, Yarrow PM, Carmalt HL, Kwun SY, Kennedy CW, Lin BP, et al. Overexpression of Cripto and its prognostic significance in breast cancer: a study with long-term survival. Eur J Surg Oncol. 2007;33:438–43. doi: 10.1016/j.ejso.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Ciardiello F, Kim N, Saeki T, Dono R, Persico MG, Plowman GD, et al. Differential expression of epidermal growth factor-related proteins in human colorectal tumors. Proc Natl Acad Sci USA. 1991;88:7792–6. doi: 10.1073/pnas.88.17.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeki T, Stromberg K, Qi CF, Gullick WJ, Tahara E, Normanno N, et al. Differential immunohistochemical detection of amphiregulin and cripto in human normal colon and colorectal tumors. Cancer Res. 1992;52:3467–73. [PubMed] [Google Scholar]
- 23.Wu Z, Li G, Wu L, Weng D, Li X, Yao K. Cripto-1 overexpression is involved in the tumorigenesis of nasopharyngeal carcinoma. BMC Cancer. 2009;9:315. doi: 10.1186/1471-2407-9-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong XY, Zhang LH, Jia SQ, Shi T, Niu ZJ, Du H, et al. Positive association of upregulated Cripto-1 and downregulated E-cadherin with tumour progression and poor prognosis in gastric cancer. Histopathology. 2008;52:560–8. doi: 10.1111/j.1365-2559.2008.02971.x. [DOI] [PubMed] [Google Scholar]
- 25.Xu CH, Sheng ZH, Hu HD, Hao KK, Wang QB, Yu LK. Elevated expression of Cripto-1 correlates with poor prognosis in non-small cell lung cancer. Tumour Biol. 2014;35:8673–8. doi: 10.1007/s13277-014-2039-1. [DOI] [PubMed] [Google Scholar]
- 26.Chen F, Hou SK, Fan HJ, Liu YF. MiR-15a-16 represses Cripto and inhibits NSCLC cell progression. Mol Cell Biochem. 2014;391:11–9. doi: 10.1007/s11010-014-1981-y. [DOI] [PubMed] [Google Scholar]
- 27.Wang JH, Wei W, Xu J, Guo ZX, Xiao CZ, Zhang YF, et al. Elevated expression of Cripto-1 correlates with poor prognosis in hepatocellular carcinoma. Oncotarget. 2015;6:35116–28. doi: 10.18632/oncotarget.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo J, Lau EY, Ching RH, Cheng BY, Ma MK, Ng IO, et al. Nuclear factor kappa B-mediated CD47 up-regulation promotes sorafenib resistance and its blockade synergizes the effect of sorafenib in hepatocellular carcinoma in mice. Hepatology. 2015;62:534–45. doi: 10.1002/hep.27859. [DOI] [PubMed] [Google Scholar]
- 29.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–5. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 30.Merle P, de la Monte S, Kim M, Herrmann M, Tanaka S, Von Dem Bussche A, et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–22. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Merle P, Kim M, Herrmann M, Gupte A, Lefrancois L, Califano S, et al. Oncogenic role of the frizzled-7/beta-catenin pathway in hepatocellular carcinoma. J Hepatol. 2005;43:854–62. doi: 10.1016/j.jhep.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 32.Nambotin SB, Lefrancois L, Sainsily X, Berthillon P, Kim M, Wands JR, et al. Pharmacological inhibition of Frizzled-7 displays anti-tumor properties in hepatocellular carcinoma. J Hepatol. 2011;54:288–99. doi: 10.1016/j.jhep.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 33.Fiorenzano A, Pascale E, D’Aniello C, Acampora D, Bassalert C, Russo F, et al. Cripto is essential to capture mouse epiblast stem cell and human embryonic stem cell pluripotency. Nat Commun. 2016;7:12589. doi: 10.1038/ncomms12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaoka T, Karasawa H, Turbyville T, Rangel MC, Castro NP, Gonzales M, et al. Cripto-1 enhances the canonical Wnt/beta-catenin signaling pathway by binding to LRP5 and LRP6 co-receptors. Cell Signal. 2013;25:178–89. doi: 10.1016/j.cellsig.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Qin Z, Yang K, Liu R, Xu Y. Cripto-1 promotes epithelial-mesenchymal transition in prostate cancer via Wnt/beta-catenin signaling. Oncol Rep. 2017;37:1521–8. doi: 10.3892/or.2017.5378. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe K, Meyer MJ, Strizzi L, Lee JM, Gonzales M, Bianco C, et al. Cripto-1 is a cell surface marker for a tumorigenic, undifferentiated subpopulation in human embryonal carcinoma cells. Stem Cells. 2010;28:1303–14. doi: 10.1002/stem.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francescangeli F, Contavalli P, De Angelis ML, Baiocchi M, Gambara G, Pagliuca A, et al. Dynamic regulation of the cancer stem cell compartment by Cripto-1 in colorectal cancer. Cell Death Differ. 2015;22:1700–13. doi: 10.1038/cdd.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q, Cui X, Yu X, Bian BS, Qian F, Hu XG, et al. Cripto-1 acts as a functional marker of cancer stem-like cells and predicts prognosis of the patients in esophageal squamous cell carcinoma. Mol Cancer. 2017;16:81. doi: 10.1186/s12943-017-0650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamada S, Watanabe K, Hirota M, Bianco C, Strizzi L, Mancino M, et al. beta-Catenin/TCF/LEF regulate expression of the short form human Cripto-1. Biochem Biophys Res Commun. 2007;355:240–4. doi: 10.1016/j.bbrc.2007.01.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou YF, Xie CW, Yang SX, Xiong JP. AMPK activators suppress breast cancer cell growth by inhibiting DVL3-facilitated Wnt/beta-catenin signaling pathway activity. Mol Med Rep. 2017;15:899–907. doi: 10.3892/mmr.2016.6094. [DOI] [PubMed] [Google Scholar]
- 41.Sun C, Sun L, Jiang K, Gao DM, Kang XN, Wang C, et al. NANOG promotes liver cancer cell invasion by inducing epithelial-mesenchymal transition through NODAL/SMAD3 signaling pathway. Int J Biochem Cell Biol. 2013;45:1099–108. doi: 10.1016/j.biocel.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 42.Yin D, Tian L, Ye Y, Li K, Wang J, Cheng P, et al. Nanog and beta-catenin: a new convergence point in EpSC proliferation and differentiation. Int J Mol Med. 2012;29:587–92. doi: 10.3892/ijmm.2011.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Lou Y, Wang H, Zheng X, Dong Q, Sun J, et al. Wnt signaling regulates the stemness of lung cancer stem cells and its inhibitors exert anticancer effect on lung cancer SPC-A1 cells. Med Oncol. 2015;32:95. doi: 10.1007/s12032-014-0462-1. [DOI] [PubMed] [Google Scholar]
- 44.Yong X, Tang B, Xiao YF, Xie R, Qin Y, Luo G, et al. Helicobacter pylori upregulates Nanog and Oct4 via Wnt/beta-catenin signaling pathway to promote cancer stem cell-like properties in human gastric cancer. Cancer Lett. 2016;374:292–303. doi: 10.1016/j.canlet.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 45.Munoz P, Iliou MS, Esteller M. Epigenetic alterations involved in cancer stem cell reprogramming. Mol Oncol. 2012;6:620–36. doi: 10.1016/j.molonc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–27. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–59. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao C, Sun W, Tan M, Glazer CA, Bhan S, Zhong X, et al. Integrated, genome-wide screening for hypomethylated oncogenes in salivary gland adenoid cystic carcinoma. Clin Cancer Res. 2011;17:4320–30. doi: 10.1158/1078-0432.CCR-10-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon MJ, Shin YK. Epigenetic regulation of cancer-associated genes in ovarian cancer. Int J Mol Sci. 2011;12:983–1008. doi: 10.3390/ijms12020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun G, Yan SS, Shi L, Wan ZQ, Jiang N, Fu LS, et al. MicroRNA-15b suppresses the growth and invasion of glioma cells through targeted inhibition of cripto-1 expression. Mol Med Rep. 2016;13:4897–903. doi: 10.3892/mmr.2016.5126. [DOI] [PubMed] [Google Scholar]
- 51.Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, et al. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012;40:4615–25. doi: 10.1093/nar/gkr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng C, Li J, Wang Q, Liu W, Zhou J, Liu R, et al. MicroRNA-195 functions as a tumor suppressor by inhibiting CBX4 in hepatocellular carcinoma. Oncol Rep. 2015;33:1115–22. doi: 10.3892/or.2015.3734. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Yang Y, Yang T, Yuan S, Wang R, Pan Z, et al. Double-negative feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1 regulates hepatocellular carcinoma tumor growth and metastasis. Hepatology. 2015;61:561–73. doi: 10.1002/hep.27491. [DOI] [PubMed] [Google Scholar]
- 54.Yang H, Zheng W, Shuai X, Chang RM, Yu L, Fang F, et al. MicroRNA-424 inhibits Akt3/E2F3 axis and tumor growth in hepatocellular carcinoma. Oncotarget. 2015;6:27736–50. doi: 10.18632/oncotarget.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu XM, Wu LJ, Xu J, Yang R, Wu FS. Let-7c microRNA expression and clinical significance in hepatocellular carcinoma. J Int Med Res. 2011;39:2323–9. doi: 10.1177/147323001103900631. [DOI] [PubMed] [Google Scholar]
- 56.Ho DW, Kai AK, Ng IO. TCGA whole-transcriptome sequencing data reveals significantly dysregulated genes and signaling pathways in hepatocellular carcinoma. Front Med. 2015;9:322–30. doi: 10.1007/s11684-015-0408-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and legends clean copy