Abstract
Campylobacter is a well-known food-borne pathogen that causes human gastroenteritis. Food products that contain Campylobacter may also be contaminated by other pathogens, however, whether this multiple contamination leads to more severe infection remains unclear. In this study, mice were gavaged with Campylobacter jejuni and other food-borne pathogenic bacteria to mimic a multiple infection. It was demonstrated that the C. jejuni load was elevated when the mice were co-infected with C. jejuni and Salmonella typhimurium, and the campylobacteriosis that followed was also enhanced, with features of decreased body weight, heavier bloody stools and more pronounced inflammatory changes to the colon. In addition, infection with C. jejuni was also promoted by co-infection with entero-invasive Escherichia coli but unaffected over time. In contrast to S. typhimurium and entero-invasive E. coli, co-infection by Listeria monocytogenes showed little effect on C. jejuni infection and even hindered its progress. In addition, the intestinal microecology was also affected by co-infection of C. jejuni with other pathogens, with an increased relative abundance of unclassified Enterobacteriaceae, decreased levels of butyric acid and changes in the abundance of several genera of gut microbe, which suggests that some food-borne pathogenic bacteria might affect the progression of C. jejuni infection in mice by influencing the composition of the gut microbiota and the resulting changes in SCFA levels. Collectively, our findings suggest that co-infection of Campylobacter with other pathogenic bacteria can impact on the progression of infection by C. jejuni in mice, which may also have implication for the etiology of Campylobacter on human health.
Keywords: Campylobacter, food-borne pathogenic bacteria, co-infection, campylobacteriosis, gut microbiota, butyric acid
Introduction
Campylobacter is a food-borne pathogen and the leading cause of human gastroenteritis around the world. The incidence of campylobacteriosis has been estimated to be 4.4 per 1000 people, with 1.3 million cases in the United States (Scallan et al., 2011), 5.8 per 1000 person–years in the Netherlands (Havelaar et al., 2012), and 1.2 per 1000 person–years in China (Huang et al., 2018). Campylobacter infection usually results in symptoms such as bloody diarrhea, abdominal pain and fever, and the course of the disease is self-limited in most cases. However, an increased risk of irritable bowel syndrome and inflammatory bowel disease was observed amongst those infected with Campylobacter (Marshall et al., 2006). Moreover, some peripheral neuropathies such as Guillain–Barré syndrome and Miller Fisher syndrome are also long-term consequences of Campylobacter infection (Poropatich et al., 2010; Sejvar et al., 2011).
Campylobacter is most frequently found in poultry, but it can also be found in other food items such as pork, beef, and raw milk. The prevalence of Campylobacter contamination in retail poultry and by-products exceeds 50% around the world, varying from 0 to 100% (Sahin et al., 2015). In many developed countries, the rate of Campylobacter contamination usually exceeds 60%, but it can be much lower in developing countries such as China and Brazil (Silva et al., 2017; Zhu et al., 2017). Notably, when food items are contaminated with Campylobacter, they may also be contaminated with other food-borne pathogens. Many reports have found food samples that harbor more than one species of pathogen. In Ireland, 10 of 25 raw chicken samples were contaminated with multiple pathogens (Gorman et al., 2002). Another study also described the presence of more than two pathogens in ready-to-eat meat products (Gibbons et al., 2006). In addition, Campylobacter strains from different sources can also be detected in the same poultry house (Hiett et al., 2002). Due to the conventional intestinal microbiota, Campylobacter cannot effectively colonize mice sufficiently to exert obvious clinical symptoms (Dorrell and Wren, 2007). Acute ileitis induced by Toxoplasma gondii can abrogate the colonization resistance of mice, after which high loads of Campylobacter can be achieved (Haag et al., 2012a). However, it remains unclear whether other co-infection of pathogenic bacteria with Campylobacter can exacerbate the campylobacteriosis. To date, few studies have focused directly on campylobacteriosis caused by Campylobacter and other pathogenic bacteria.
In this study, mice were co-infected with Campylobacter jejuni and other pathogenic bacteria [Salmonella typhimurium, entero-invasive Escherichia coli (EIEC) and Listeria monocytogenes] to determine whether this multiple infection would lead to more severe campylobacteriosis in mice. Moreover, the changes in the microbiota and the level of short-chain fatty acids (SCFAs) in feces were also monitored to determine if multiple infections also affect the intestinal microenvironment which may explain the potential mechanism by which co-infection with food-borne pathogenic bacteria influences the progression of C. jejuni infection in mice.
Materials and Methods
Bacterial Strains and Culture Conditions
Campylobacter jejuni NCTC 11168 (ATCC 700819), S. typhimurium SL1344, EIEC (ATCC 43893), and L. monocytogenes (ATCC 19114) were acquired from the Culture Collection of Food Microorganisms of Jiangnan University (Wuxi, China). Columbia blood agar base plates (Oxoid, United Kingdom) supplemented with 5% sterile sheep blood and C. jejuni selective supplement (Oxoid) were used to culture C. jejuni strains under microaerophilic conditions (5% O2, 10% CO2, 85% N2) for 48 h at 37°C. S. typhimurium, EIEC and L. monocytogenes were cultured with brain-heart infusion broth (Haibo, China) for 24 h at 37°C.
Animals and Experimental Design
Three-week-old female C57BL/6 mice were obtained from Shanghai Laboratory Animal Center (Shanghai, China) and used in all animal experiments. Six mice were housed in each cage, with a 12 h light–dark cycle in a controlled environment (temperature, 22 ± 2°C; humidity, 50 ± 5%). All experimental procedures (#JIPD2017029) were approved by the Animal Care and Use Committee at Jiangsu Institute of Parasitic Diseases. All experiments conformed to the China Ministry of Science and Technology Guide for the Care and Use of Laboratory Animals.
Mice were infected with pathogenic bacteria or parasites by gavage at a volume of 0.2 mL. C. jejuni concentration was adjusted to 2 × 109 colony-forming units (CFU)/mL in sterile phosphate-buffered saline solution (PBS), and S. typhimurium, EIEC, L. monocytogenes were used at doses of 1 × 105 CFU/mL in sterile PBS. Cysts of the T. gondii ME49 strain (from Jiangsu Institute of Parasitic Diseases Remington, Wuxi, China) were obtained from the homogenized brains of mice infected with 10 cysts for 2 months, and the mice were infected perorally with 100 T. gondii cysts. Sterile PBS was used as a naive control. The mice were divided into 11 groups; the treatment of each group is shown in Table 1. Each mouse’s body weight was recorded every 3 days, and stool samples were collected during the experimental procedure. The mice were anesthetised before sacrifice with an injection of 100 mg/kg body weight of ketamine, and plasma and colonic tissues were collected for further analysis.
Table 1.
Animal model experimental design.
| Groups | Treatment of different days | ||||
|---|---|---|---|---|---|
| Day 1 | Day 2–4 | Day 5–6 | Day 7–10 | Day 11 | |
| Naive | PBS | Normal breed | PBS | Normal breed | Sacrificed |
| Cj | PBS | C. jejuni | |||
| T.g + Cj | T. gondii | C. jejuni | |||
| S.t + Cj | S. typhimurium | C. jejuni | |||
| EIEC + Cj | EIEC | C. jejuni | |||
| L.m + Cj | L. monocytogenes | C. jejuni | |||
| T.g | T. gondii | PBS | |||
| S.t | S. typhimurium | PBS | |||
| EIEC | EIEC | PBS | |||
| L.m | L. monocytogenes | PBS | |||
Detection of C. jejuni in Feces
Stool samples were collected from all mice and transported to the laboratory on an ice pack. The feces were resuspended in sterile PBS and serially diluted. Diluted samples were spread on Columbia blood agar with C. jejuni selective supplement and incubated under microaerobic conditions at 37°C for 48 h. After incubation, C. jejuni numbers were determined by CFU.
Bloody Stool Assay
A bloody stool detection kit (Jiancheng, China) was used to assess the presence of blood in fecal samples, which acted as a clinical sign of C. jejuni-induced infection. The detection kit was used immediately after the fecal samples were collected, and the level of bloody stool was divided into four grades according to a previous study (Siegmund et al., 2001).
Determination of Colon Histopathology
The colonic samples were fixed in 4% neutral buffered paraformaldehyde and embedded in paraffin. Sections (5 μm) were stained with haematoxylin and eosin for light microscopic examination (magnification, ×100). The degree of inflammation and damage was evaluated using a histopathological score system (Paclik et al., 2008), modified as follows:
-
simple Score 0:
Histological findings identical to those of normal mice.
-
simple Score 1:
Loss of goblet cells begins.
-
simple Score 2:
Single isolated cell infiltrates within mucosa; loss of goblet cells begins.
-
simple Score 3:
Mild scattered to diffuse cell infiltrates within mucosa and submucosa; loss of goblet cells begins.
-
simple Score 4:
Mild scattered to diffuse cell infiltrates within mucosa and submucosa; loss of goblet cells.
-
simple Score 5:
Severe inflammation; loss of goblet cells; loss of crypts.
-
simple Score 6:
Severe inflammation; loss of goblet cells; extensive ulceration.
This analysis was performed blind by a pathologist.
Cytokine Assay
Blood samples were collected and centrifuged (1200 × g, 15 min) to obtain serum. Before cytokine assays, serum samples were treated using a Milliplex MAP Kit (Merck, Germany) according to the manufacturer’s instructions. A Luminex MAGPIX system (Luminex, United States) was used to detect the levels of cytokines in the treated serum samples.
DNA Extraction, PCR, and 16S rDNA Sequencing
The fecal samples were stored at −80°C before detection, and microbial genome DNA was extracted using a FastDNA Spin Kit for Soil (MP Biomedical, United States) following the manufacturer’s instructions. The V3–V4 region of the 16S rRNA gene was amplified by PCR. The products were separated in 1.5% (w/v) agarose gel, purified with a QIAquick Gel Extraction Kit (Qiagen, Germany) and quantified with a Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies, United States). A TruSeq DNA LT Sample Preparation Kit (Illumina, United States) was used to establish libraries that were sequenced for 500 + 7 cycles on Illumina MiSeq using a MiSeq Reagent Kit. QIIME pipeline was used to analyze the sequence data of 16S rRNA as described previously (Wang et al., 2017).
Short-Chain Fatty Acid Assay
Fecal samples stored at −80°C were steeped in saturated NaCl solution for 30 min using sterile tubes and homogenized. Sulfuric acid (10%; 20 μL) was added to acidify the solution. Diethyl ether (800 μL) was added with an injection syringe to extract SCFAs. The tubes were centrifuged at 14,000 rpm for 15 min, and the supernatants collected. Anhydrous sodium sulfate was used to eliminate the remaining water, and the treated supernatants were analyzed using gas chromatography-mass spectrometry (GC-MS). The parameters for GC-MS in our experiment were established with reference to Sun et al. (2015).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5, and the data are expressed as mean ± SD. SPSS 20.0 (SPSS Inc., United States) was used for significance analysis. Comparisons between groups were made with a two-tailed Student’s t-test, and a two-sided p-value of less than 0.05 was considered to indicate statistical significance.
Results
Co-infection of S. typhimurium Elevated the C. jejuni Load in Mice
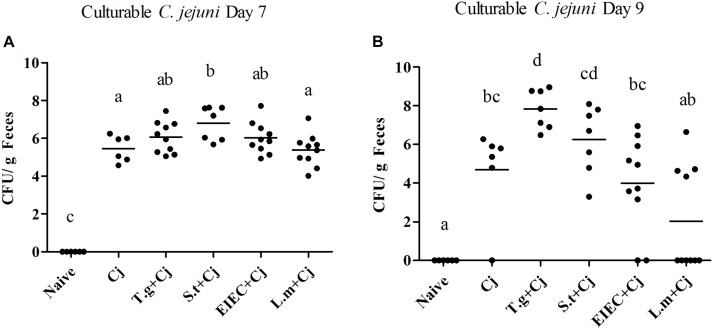
As shown in Figure 1, feces from all infected groups (C. jejuni, T. gondii+ C. jejuni, S. typhimurium + C. jejuni, EIEC + C. Jejuni, and L. monocytogenes + C. jejuni) were positive for C. jejuni, whilst C. jejuni was not detected in the mice from the control group (naive, T. gondii, S. typhimurium, EIEC, and L. monocytogenes). T. gondii infection led to significantly higher C. jejuni loads on day 9 than C. jejuni infection alone (P < 0.05). In addition, in the T. gondii coinfection group, more culturable C. jejuni was detected at day 9 than at day 7, and the number of viable bacteria reached 109 CFU per gram of feces. S. typhimurium infection also aggravated C. jejuni colonization on day 7 in comparison with C. jejuni alone (P < 0.05), which was more effective than T. gondii, with the number of C. jejuni reaching 108 CFU per gram of feces. On day 9, although C. jejuni loads in a few mice co-infected with S. typhimurium decreased, the loads remained mostly stable in other mice, where C. jejuni loads could reach 108 CFU per gram of feces. In contrast, co-infection with EIEC or L. monocytogenes did not significantly enhance the C. jejuni loads on days 7 or 9 (P > 0.05). Compared with the overall increase of C. jejuni loads by T. gondii, the effects on C. jejuni colonization by three food-borne pathogens were distinct and pathogen specific. More individuals co-infected with S. typhimurium maintained the high C. jejuni loads. A few individuals co-infected with EIEC still had a higher C. jejuni load. However, the C. jejuni in most individuals co-infected with L. monocytogenes cleared, indicating the potential negative impact of L. monocytogenes on C. jejuni colonization.
FIGURE 1.
Culturable C. jejuni in feces when mice were infected by C. jejuni alone or together with other pathogen. (A) Culturable C. jejuni on day 7. (B) Culturable C. jejuni on day 9. Means with no common letters differ significantly (P < 0.05).
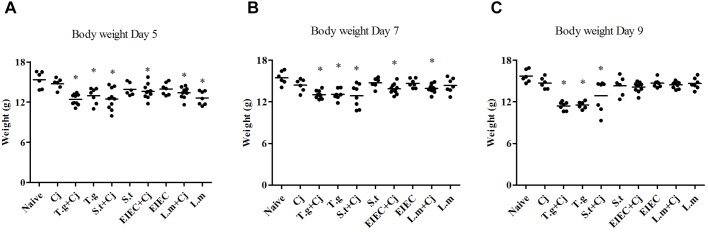
S. typhimurium Decreased the Body Weight of Mice When Co-infected With C. jejuni
Severe pathogenic infection can cause intestinal inflammation in mice, followed by a decrease in body weight. Figure 2 shows that no significant decrease occurred in the body weights of the mice in the C. jejuni infection group. Consistent with previous reports (Grainger et al., 2013), the mice in the T. gondii only group and those in the T. gondii+ C. jejuni group showed significant decreases in body weight compared with the naive control group (P < 0.05), indicating that T. gondii infection seriously damaged their health status. Infection with three food-borne pathogens alone caused little change in the mice’s body weights. However, after co-infection with C. jejuni, the mice with S. typhimurium showed significant decreases in body weight compared with healthy mice; mice in the EIEC+ C. jejuni group only showed significant weight loss (P < 0.05) on days 5 and 7; mice in the L. monocytogenes and L. monocytogenes + C. jejuni groups only showed slight weight loss on day 5, but this effect completely reversed by day 9. It is noteworthy that, unlike the overall decrease in body weight in the T. gondii+ C. jejuni group, the decrease in mice body weight in the S. typhimurium + C. jejuni group showed obvious individual differences. Similar differences can also be seen in body weight gain (Supplementary Figure S1 and Supplementary Table S1). On day 5, except for the mice in the C. jejuni group, all of the mice in the pathogen infection groups showed significantly reduced weight gain. The rate of weight gain recovered in all of the groups infected or co-infected with EIEC or L. monocytogenes. Although there were no significant differences between the weight gain of the S. typhimurium infected (and co-infected) groups and that of the naive group on day 9, obvious individual differences in the weight gain of the mice within this group were observed. The weight gain of each mouse corresponded to the C. jejuni load. In addition, several mice died in the S. typhimurium + C. jejuni, and in the T. gondii+ C. jejuni groups.
FIGURE 2.
Body weight of mice over the course of infection. (A) Day 5. (B) Day 7. (C) Day 9. Asterisk (∗) indicates means that differ significantly from the naive group (P < 0.05).
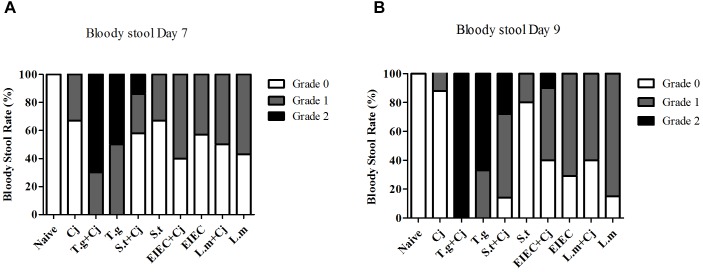
Co-infection With S. typhimurium and C. jejuni Promoted Bloody Stools in Mice
Mice infected with C. jejuni had mild blood-positive feces by day 7, and the symptoms were partly relieved by day 9 (Figure 3). In T. gondii+ C. jejuni and T. gondii only groups, up to 50% of infant mice presented severe blood-positive (Grade 2) feces samples by days 7 and 9. In general, bloody stools were significantly more frequent in all co-infected groups than in groups infected with a single pathogen (either C. jejuni or another pathogenic bacteria), except that the bloody stools in the L. monocytogenes + C. jejuni group were less frequent than those in the L. monocytogenes group by day 9. Severe blood-positive (Grade 2) fecal samples were also observed in the S. typhimurium co-infection group (days 7 and 9) and in the EIEC coinfection group (day 9). It is noteworthy that although the frequency of bloody stools in the S. typhimurium only group was similar to or even lower than that in the EIEC and L. monocytogenes groups, the bloody stools in the S. typhimurium + C. jejuni group were more serious than those in all of the other co-infected groups, whilst the frequency of bloody stool caused by S. typhimurium infection was reduced to a relatively low level between 7 and 9 day.
FIGURE 3.
Occurrence of blood in fecal samples of mice over the course of infection. (A) Bloody stool rate on day 7. (B) Bloody stool rate on day 9.
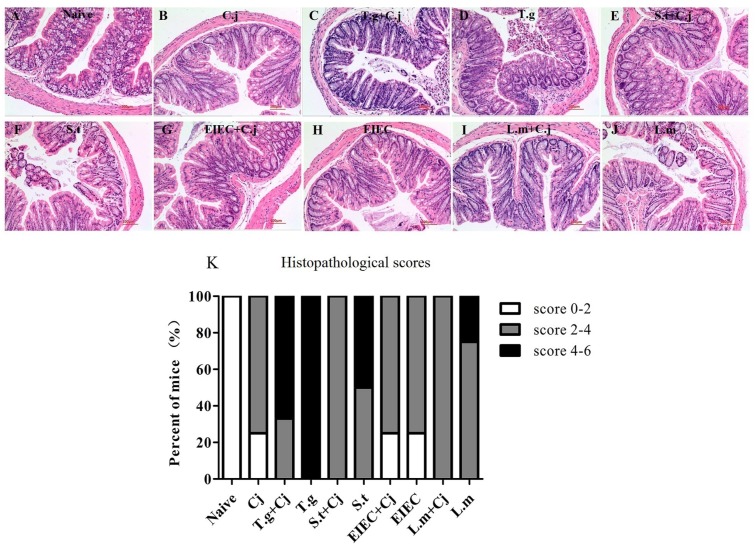
C. jejuni Relieved the Damage to Colonic Tissue Caused by Co-infected Food-Borne Pathogenic Bacteria
After the mice were killed (day 11), colon samples were collected from each group for histological analysis. As shown in Figure 4A, the mice from the naive group displayed normal villi, crypts, and muscular layer with a large number of goblet cells. Mice infected with C. jejuni alone had fewer goblet cells, whilst the structure of villi, crypts, and muscular layer was normal (Figure 4B). Infection with T. gondii alone resulted in severe colonic pathological changes, including a loss of goblet cells, damage to the crypt and villi architecture and inflammatory cell infiltration (Figure 4D), indicating that T. gondii infection induced serious inflammation and tissue injury. However, the histopathological scores in Figure 4K indicate that co-infected C. jejuni partly relieved the damage to the colonic tissue. S. typhimurium infection alone caused the highest histopathological scores in the groups of food-borne pathogenic bacteria infection alone, whilst EIEC caused the lowest scores, similar to that of C. jejuni (Figures 4F,H,J). It is noteworthy that, like the T. gondii+ C. jejuni group (Figure 4C), co-infected C. jejuni partly relieved the damage to the colonic tissue caused by S. typhimurium and L. monocytogenes, whilst it had no effects on the damage caused by EIEC (Figures 4E,G,I).
FIGURE 4.
Histopathological changes in haematoxylin and eosin–stained colonic paraffin sections. (A) Naive group. (B) C. jejuni group. (C) T. gondii + C. jejuni group. (D) T. gondii group. (E) S. typhimurium + C. jejuni group. (F) S. typhimurium group. (G) EIEC + C. jejuni group. (H) EIEC group. (I) L. monocytogenes + C. jejuni group. (J) L. monocytogenes group. (K) Histopathological scores of colonic paraffin sections.
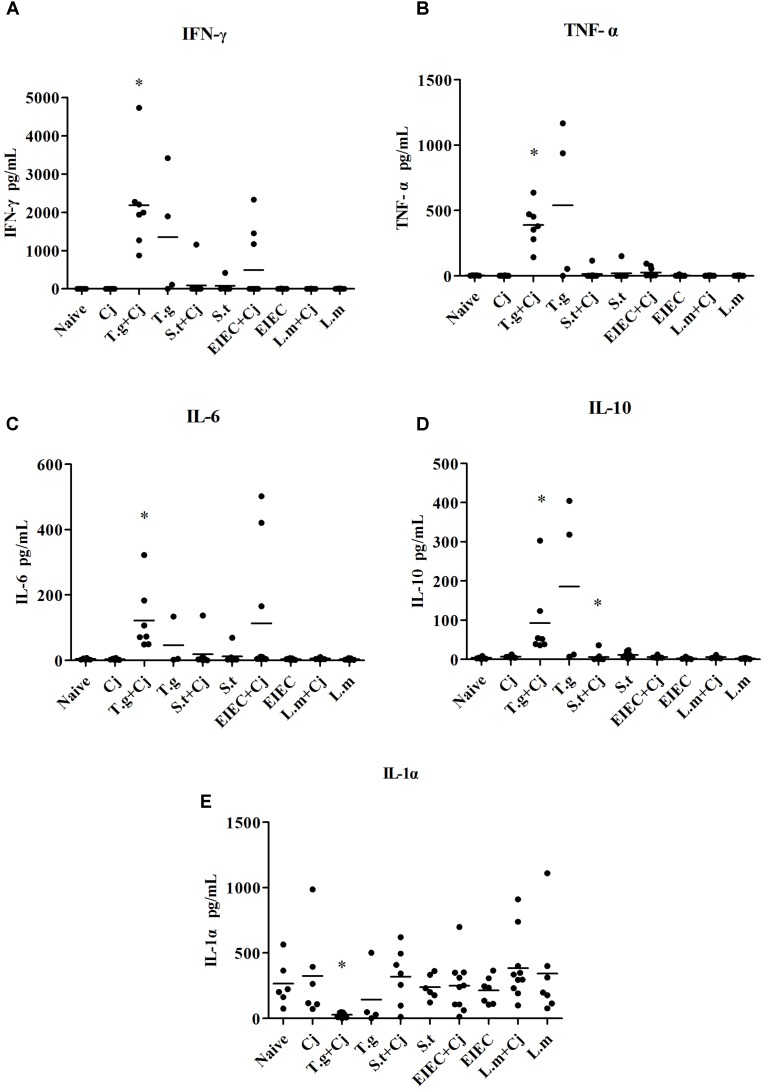
Effects of Co-infected Food-Borne Pathogenic Bacteria on Inflammation
The concentration of inflammatory factors in blood serum was also investigated. As shown in Figure 5, infection with C. jejuni did not induce any of the inflammatory cytokines. However, the concentrations of interferon (IFN) γ, tumour necrosis factor (TNF) α, interleukin (IL) 6, and IL-10 were significantly elevated due to infection with T. gondii alone, whilst the levels of IL-1α were down-regulated compared with other groups. In the T. gondii+ C. jejuni group, the concentrations of IFN-γ and IL-6 were higher than those in the T. gondii only group, whilst the concentrations of TNF-α, IL-10, and IL-1α were lower than those in the T. gondii group. As in the T. gondii+ C. jejuni group, the levels of IFN-γ, TNF-α, and IL-6 were also increased in some mice co-infected with C. jejuni in the EIEC + C. jejuni group, although no changes in any of the mice in the EIEC group. Moreover, IFN-γ, TNF-α, IL-10, and IL-6 in some mice in the S. typhimurium and S. typhimurium + C. jejuni groups also showed slight increases, although these were not significant. No significant variation could be detected in other cytokines, such as IL-1β, IL-2, IL-4, IL-12, and IL-17 (data not shown).
FIGURE 5.
Cytokine production in infected mice. (A) IFN-γ. (B) TNF-α. (C) IL-6. (D) IL-10. (E) IL-1α. Asterisk (∗) indicates means that differ significantly from the naive group (P < 0.05).
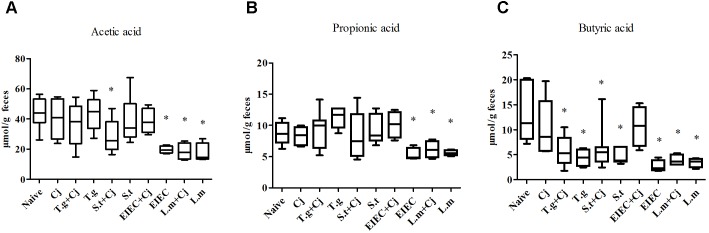
Co-infection of Food-Borne Pathogenic Bacteria Alter the Composition of SCFAs in Feces
The contents of acetic acid, propionic acid and butyric acid in feces were analyzed by GC-MS to evaluate the metabolism of the intestinal microbiota. Figure 6 shows that C. jejuni infection alone did not cause significant changes in the composition of SCFAs in mouse feces compared to that in the naive group. In contrast, infection by EIEC or L. monocytogenes alone resulted in distinct decreases in the levels of acetic acid, propionic acid and butyric acid in mouse feces (P < 0.05). Interestingly, co-infection with C. jejuni led to recovery of the SCFA level in the EIEC group but showed no effects on the SCFA level in the L. monocytogenes group, indicating the specific alteration of microbial metabolism. Infection with T. gondii or S. typhimurium alone only decreased the level of butyric acid (P < 0.05). Co-infection with C. jejuni showed no significant effects on the SCFAs level influenced by T. gondii or S. typhimurium except for a further decrease in the acetic acid level in the S. typhimurium + C. jejuni group (P < 0.05).
FIGURE 6.
Level of SCFAs in feces of infected mice. (A) Acetic acid level. (B) Propionic acid level. (C) Butyric acid level. Asterisk (∗) indicates means that differ significantly from the naive group (P < 0.05).
Co-infection of Food-Borne Pathogenic Bacteria Alter the Diversity and Relative Abundance of the Gut Microbiota
The estimated richness (Chao-1) and Shannon index were used to evaluate the community diversity of each sample. Supplementary Figure S2 shows that infection by C. jejuni alone seemed to cause no obvious differences in the diversity of gut microbiota. Infection by T. gondii, S. typhimurium, EIEC, or L. monocytogenes alone caused significant decreases in gut microbiota diversity. In addition, there was a tendency toward further reduction in the diversity of the gut microbiota in mice co-infected with C. jejuni, although individual differences existed between different mice. At phylum level (Figure 7), Firmicutes, Bacteroidetes, and Proteobacteria were dominant amongst the experimental groups. Tenericutes, Actinobacteria, TM7, Verrucomicrobia, and Cyanobacteria were also found in the feces of mice with a relative abundance of less than 1%. Mice co-infected with T. gondii and C. jejuni and those infected with EIEC alone showed a decrease in the relative abundance of Firmicutes compared with the naive group (P < 0.05). Each experimental group had between 20 and 40% Bacteroidetes in feces; no statistical differences were seen (P > 0.05). Notably, infection by different pathogens induced evident changes in Proteobacteria. The relative abundance of Proteobacteria in the naive control group was less than 1% but increased significantly in the T. gondii + C. jejuni, the S. typhimurium + C. jejuni, the S. typhimurium, EIEC, and the L. monocytogenes groups. The abundances of Proteobacteria in the T. gondii + C. jejuni, the S. typhimurium + C. jejuni, the S. typhimurium and the EIEC groups were positively correlated with the corresponding pathogen loads in the intestine. Although L. monocytogenes is not a member of the Proteobacteria, it significantly increased the abundance of Proteobacteria in the gut. Interestingly, although EIEC and L. monocytogenes increased the abundance of Proteobacteria, the presence of C. jejuni significantly decreased the abundance of this phylum, indicating an acceleration in the elimination of pathogenic bacteria from the host in the late period of co-infection.
FIGURE 7.
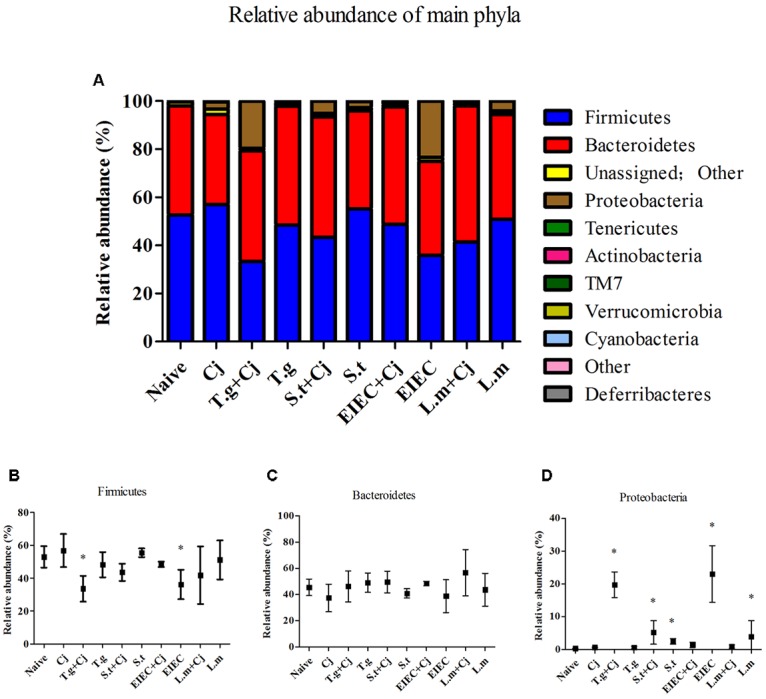
Microbial composition at phylum level in different groups. (A) Relative abundance of main phyla in different groups. (B) Firmicutes. (C) Bacteroidetes. (D) Proteobacteria. Asterisk (∗) indicates means that differ significantly from the naive group (P < 0.05).
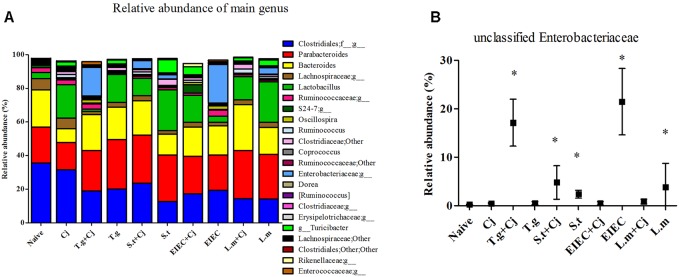
The microbial composition was further analyzed at the genus level, to further explore the differences observed in the Proteobacteria phylum. Figure 8A shows all genera with a relative abundance of more than 1%. Unclassified genera within Enterobacteriaceae is the only genus in the genera listed in Figure 8A that belongs to Proteobacteria phylum. The relative abundance of unclassified Enterobacteriaceae in the experimental groups was then checked, and significant increases in unclassified Enterobacteriaceae were found in the T. gondii + C. jejuni, S. typhimurium + C. jejuni, the S. typhimurium, the EIEC, and the L. monocytogenes groups (P < 0.05), which exhibited the same tendency as Proteobacteria phylum (Figure 8B). This indicates that all four food-borne pathogenic bacteria can increase the abundance of unclassified Enterobacteriaceae. In addition to changes in the levels of unclassified Enterobacteriaceae, decreases in the abundance of unclassified Clostridiales and Lachnospiraceae corresponded to exposure of pathogenic bacteria, except no significant reduction in abundance was observed in the group infected with C. jejuni alone (Supplementary Figure S3). The abundance of these two genera also showed positive correlations with the diversity of the gut microbiota and the SCFA level. In addition, although low C. jejuni loads in the C. jejuni group caused no significant changes in the different indexes, the abundances of Bacteroides and Lactobacillus were significantly changed by infection with C. jejuni alone. The abundance of Turicibacter was also relatively high in the two co-infection groups with high C. jejuni loads. Moreover, although large individual differences were observed, the abundances of Dorea and unclassified S24-7 showed significant increases only in some of the mice in the EIEC + C. jejuni group, and were highly correlated with the recovery of SCFA level in this group.
FIGURE 8.
Microbial composition at genus level in different groups. (A) Relative abundance of main genus in different groups. (B) Unclassified Enterobacteriaceae. Asterisk (∗) indicates means that differ significantly from the naive group (P < 0.05).
Discussion
Although many efforts have been made to prevent C. jejuni infections, it remains the most common food-borne pathogen from a global perspective (Haagsma et al., 2013). Humans can become infected with C. jejuni via contaminated water and food and by direct contact (Domingues et al., 2012). However, these pathways are compatible for other food-borne pathogenic bacteria as well, and the shared pathways lead to cross contamination. Contamination by multiple pathogenic bacteria, including different strains of Campylobacter from different sources (Konkel et al., 2007; Atterby et al., 2018), may cause symptoms that are more complex and serious than those caused by infection by only one pathogen. This study was performed to investigate whether co-infection with other common pathogenic bacteria affects the symptoms caused by C. jejuni. The results of our study show that co-infection with S. typhimurium significantly increased the C. jejuni load and resulted in more severe symptoms of C. jejuni infection. EIEC promoted infection by C. jejuni to some extent, but the promotion effects subsided with time. Co-infection with L. monocytogenes had no effects on C. jejuni load and even showed some reduction. Moreover, during co-infection with pathogenic bacteria, variations in the abundance of gut microbes such as unclassified Enterobacteriaceae, Clostridiales, and Lachnospiraceae, and corresponding changes in the level of SCFAs in the gut were observed (Supplementary Tables S2–S6).
Campylobacter jejuni has the disadvantage of sporadic colonization and barely triggers disease-defining clinical manifestations in mice due to their well established robust intestinal microbiota. In this study, 3-week-old female C57BL/6 mice with reported susceptibility to C. jejuni were used (Field et al., 1981). However, C. jejuni colonization levels remained low and even decreased beyond the infection day, which is consistent with the colonization resistance reported previously (Heimesaat et al., 2013). In addition, no significant pathological symptoms were found with C. jejuni alone, which may be related to the low colonization by C. jejuni. We therefore used T. gondii infection model (Liesenfeld, 2002; Haag et al., 2012b), that make the host more susceptible to C. jejuni infection as a positive control to assess the impact of food-borne pathogens on C. jejuni infections.
Because infection with high doses of S. typhimurium, EIEC, and L. monocytogenes can result in loss of weight, enteritis, and death in a mouse model (Miller and Burns, 1970; Medici et al., 2005; Martins et al., 2013; Franca et al., 2015), these food-borne pathogens are expected to promote the infection of C. jejuni. However, our study showed that only co-infection with S. typhimurium can promote C. jejuni colonization, and induce weight loss and bloody stools, even with a low infectious dose (3 × 104 CFU/mouse) of S. typhimurium. Bloody stools and weight loss can be caused by C. jejuni alone to some extent (Haag et al., 2012a; Heimesaat et al., 2014). In the presence of S. typhimurium, C. jejuni was found to promote weight loss, bloody stools and even death of the mice. These results suggest that the presence of S. typhimurium can enhance C. jejuni numbers and exacerbate the corresponding symptoms. Although no significant difference in weight loss was found between the EIEC + C. jejuni group and the EIEC group, the bloody stools in the EIEC + C. jejuni group suggest that the presence of EIEC may promote C. jejuni infection to a limited extent. L. monocytogenes is a poor colonizer of the mouse intestine due to lack of recognition by mouse intestinal cell receptors (Lecuit et al., 1999) and germ free mice (Manohar et al., 2001) and mice pre-treated with streptomycin can increase the pathogen load (Takeuchi et al., 2006). However, even low-dose L. monocytogenes gavage still caused damage to the gut health of the mice. Unexpectedly, it seems that the symptoms in the L. monocytogenes + C. jejuni group were milder than those in the L. monocytogenes group. Taking the C. jejuni loads into consideration, it seems that the co-existence of these two pathogenic bacteria impedes each other’s infection process. Corresponding with the above results, histological analysis show that the mice infected with C. jejuni alone displayed mild pathological changes in the colon, likely due to sporadic colonization of C. jejuni. However, different to the results of weight loss and bloody stool, except for the EIEC group, it seems that the co-infected C. jejuni instead reduces the degree of intestinal tissue lesions. This finding may be related to the colonization area of the pathogens, and further investigation is needed.
Although the major site of T. gondii replication is different from that of the other four food-borne pathogenic bacteria (Dubey et al., 1997), T. gondii promoted infection with C. jejuni in mice. This indicates that the synergistic effect may not be confined to the site of infection but rather a systemic effect. T. gondii infection leads to Th1-type immunopathology in mice, which causes an elevated IFN-γ concentration (Heimesaat et al., 2007; Zhou and Wang, 2017). The effects of one pathogen on the host’s immune system may be the cause of the host’s susceptibility to or tolerance of other pathogens. IL-10-/- mice that show heavier pathogen loads and more severe clinical signs are usually used to establish models of C. jejuni infection (Bereswill et al., 2011; Sun et al., 2012). This indicates that changes in the host’s immune status may have an important impact in C. jejuni infection. It has been reported that TNF-α, IL-1, IL-4, and IL-10 are all elevated in mice after C. jejuni infection (Al-Banna et al., 2012). In this study, co-infection of T. gondii and C. jejuni further increased the levels of IFN-γ and IL-6 caused by T. gondii, much in the same way as the changes in cytokines induced by C. jejuni in gnotobiotic mice (Bereswill et al., 2011), which suggests that T. gondii infection promotes C. jejuni infection symptoms. In addition, from the level of cytokines, it seemed that EIEC does promote C. jejuni infection to a certain extent, whilst L. monocytogenes infection without any promotion on C. jejuni infection, which is also confirmed by the disease indicators described above. However, unexpectedly, although S. typhimurium can promote C. jejuni infection and its related symptoms in mice from pathogen loads and disease index, and S. typhimurium may activate local inflammatory processes in the colon with elevated levels of IFN-γ, TNF-α, and IL-6 (Castillo et al., 2011), S. typhimurium infection, whether alone or together with C. jejuni, did not result in statistical changes in cytokine levels in this study, despite slight increases in IFN-γ and IL-6 in a few mice. This indicates that infection promotion on C. jejuni by S. typhimurium differs somewhat from that by T. gondii.
Studies have shown that in addition to the immune system, the colonization of C. jejuni in the intestine is closely related to the gut microbiota (Bereswill et al., 2011). Even in the C57/6J mice that were proven to be less susceptible to C. jejuni colonization, a small number of individuals with high C. jejuni loads also showed significant differences in the composition of their caecal microbiota. C. jejuni colonization did not incite visible pathologic changes, but was associated with increased abundances of Coriobacteriaceae, Lachnospiraceae, and Ruminococcaceae (Lone et al., 2013). This suggests that the gut microbiota might play an important role in determining the extent of which C. jejuni can colonize the mice gut. An investigation in humans also indicated that low diversity of gut microbiota may result in C. jejuni infection (Kampmann et al., 2016). In addition, infection with S. typhimurium, EIEC and other pathogens can lead to a decline in the diversity of the gut microbiota and a decrease in the abundance of some intestinal microbes (Chassaing et al., 2014; Mon et al., 2015; Borton et al., 2017; Zhang et al., 2017; Du et al., 2018). In this study, because of the low load of C. jejuni, no significant change in the diversity of gut microbiota was found in the group infected with only C. jejuni. However, all other groups treated with pathogens showed significant decreases in the diversity of the gut microbiota. Co-infection with C. jejuni generally resulted in a tendency toward further reduction of the diversity of the gut microbiota, suggesting that infection with some food-borne pathogens may increase the colonization rate of C. jejuni by decreasing the diversity of gut microbiota.
Firmicutes and Bacteroidetes are the two dominant phyla in the murine intestinal microbiota, and the abundance of these phyla is related to host health (Hansen et al., 2010). In this study, the two groups whose Firmicutes/Bacteroidetes ratio decreased also showed an increase in the relative abundance of Proteobacteria, a phylum including a wide variety of pathogens, such as Escherichia, Salmonella, Vibrio, Helicobacter, Yersinia, Legionella, and many other notable genera (Madigan et al., 2018). In addition, although no significant decrease was seen in the level of Firmicutes, the S. typhimurium, S. typhimurium+ C. Jejuni, and L. monocytogenes groups also showed significant increases in Proteobacteria. This might reflect the effective colonization of this three pathogen in the mice. Unexpectedly, the increase in Proteobacteria abundance caused by infection with EIEC or L. monocytogenes disappeared due to the presence of C. jejuni. This was further confirmed by the variation in abundance of unclassified Enterobacteriaceae at the genus level. Infection with some pathogens has been reported to increase the level of Enterobacteriaceae in the gut (Corridoni et al., 2012; David et al., 2014). The lack of change in unclassified Enterobacteriaceae abundance in the T. gondii group but significant increase in the T. gondii+ C. jejuni group in this study suggests that the increase in unclassified Enterobacteriaceae is a manifestation of C. jejuni infection (Sakaridis et al., 2018). The increased abundance of unclassified Enterobacteriaceae may also predicts the possibility of an elevated rate of Salmonella colonization in association with C. jejuni infection. However, for EIEC and L. monocytogenes, there appears to be an antagonistic relationship that makes C. jejuni and these two pathogens unable to co-exist at high levels in the host’s gut. This may be due to the different effects of these bacteria on the immune system, or differential effects on the composition of the gut microbiota.
Changes in the abundances of unclassified Clostridiales and Lachnospiraceae also showed similar patterns with respect to gut microbiota diversity. As the Clostridiales includes a portion of the intestinal bacteria producing butyric acid (Liu et al., 2010; Myszka et al., 2012; Cai et al., 2013), these decreases can also explain the general decline of intestinal butyric acid levels in infected mice. It has been reported that butyric acid has potent anti-inflammatory effects and can efficiently maintain the integrity of the intestinal mucosa (Mishiro et al., 2013). Therefore, infection with these pathogenic bacteria may cause intestinal damage by affecting the abundance of SCFA-producing bacteria in the intestinal tract. The decrease in unclassified Lachnospiraceae observed in this study was also consistent with previous studies (Kampmann et al., 2016), but no correlations of Coprococcus and Dorea abundances were found in our study. Furthermore, similar to Dorea, unclassified S24-7 also showed a significant increase in only some mice in the EIEC + C. jejuni group, consistent with the recovery of SCFAs in this group. Both of these genera have been reported to produce acetic, propionic and butyric acids (Taras et al., 2002; Ormerod et al., 2016; Meisel et al., 2017; Bishehsari et al., 2018). Therefore, the specific regulation of gut microbiota by EIEC and C. jejuni, resulting in the restoration of SCFA levels may also be a cause of infection remission. In addition, similar to the findings of a previous study (Borewicz et al., 2015), the levels of Turicibacter in this study also increased due to S. typhimurium or T. gondii infection, which correlates with the promotion of C. jejuni colonization by these two pathogens. It has been reported there was a significantly lower proportion of Turicibacter in Tnf-/- compared to WT mice both prior to and after colitis induction (Jones-Hall et al., 2015). According to the levels of TNF-α in this study, the changes in cytokine and Turicibacter abundance showed a certain degree of coincidence. Whether the increase in Turicibacter abundance and TNF-α levels contribute to colonization by C. jejuni still needs further study. Besides, the changes in the abundances of Parabacteroides and Lactobacillus showed similar tendency in this study. Taking the effects on the host by these genera into account (Sanchez et al., 2010; Clarke et al., 2011), these changes appear to be a self-protection by the host to alleviate further infections and injuries. However, this protection seems to be limited because the abundances of these genera decrease in seriously co-infected mice. Therefore, changes in the gut microbiota caused by pathogens would further affect aspects of the gut environment such as metabolites, nutrients, and immune factors, influencing the infection progression of subsequent pathogens.
Conclusion
This study demonstrates that different food-borne pathogenic bacteria co-infected with C. jejuni exert different effects on the progress of C. jejuni infection in mice. Co-infection with S. typhimurium can significantly increase the C. jejuni burden in mice and lead to more severe campylobacteriosis. Moreover, co-infection with EIEC promotes infection by C. jejuni to some extent, but this promotion disappears over time. In contrast, co-infection with L. monocytogenes has little effect on C. jejuni infection and even hinders its progress. In addition, an increase in the relative abundance of Enterobacteriaceae and a decreased level of butyric acid were also observed during co-infection of C. jejuni with other pathogenic bacteria. Changes in the abundance of some intestinal microbes may be directly related to the progression of C. jejuni infection and they might thus be used as indicators of C. jejuni infection. Given the possibility of co-infection, it is clear from this study that some food-borne pathogenic bacteria might play an important role in the progression of C. jejuni infection.
Author Contributions
GW, YZ, and WC conceived and designed the experiments. GW, YH, XJ, and XC performed the experiments. JZ and HZ analyzed the data. YZ, JZ, HZ, and WC contributed reagents, materials, and the analysis tools. GW and YH wrote the paper. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (No. 31671839), the National Natural Science Foundation of China Key Program (No. 31530056), the National Natural Science Foundation of China (Nos. 31601444 and 31301407), the Fundamental Research Funds for the Central Universities (JUSRP51501), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the national first-class discipline program of Food Science and Technology (JUFSTR20180102), the Program of Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province, and the project/research supported by Scientific Research Fund of Hunan Provincial Education Department (15B034).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01977/full#supplementary-material
References
- Al-Banna N. A., Raghupathy R., Albert M. J. (2012). Induction of cytokines in different organs after intranasal inoculation of Campylobacter jejuni in mice. Gut Pathog. 4:23. 10.1186/1757-4749-4-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atterby C., Mourkas E., Meric G., Pascoe B., Wang H., Waldenstrom J., et al. (2018). The potential of isolation source to predict colonization in avian hosts: a case study in Campylobacter jejuni strains from three bird species. Front. Microbiol. 9:591. 10.3389/fmicb.2018.00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereswill S., Fischer A., Plickert R., Haag L. M., Otto B., Kuhl A. A., et al. (2011). Novel murine infection models provide deep insights into the “menage a trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6:e20953. 10.1371/journal.pone.0020953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishehsari F., Engen P. A., Preite N. Z., Tuncil Y. E., Naqib A., Shaikh M., et al. (2018). Dietary fiber treatment corrects the composition of gut microbiota, promotes SCFA production, and suppresses colon carcinogenesis. Genes 9:E102. 10.3390/genes9020102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borewicz K. A., Kim H. B., Singer R. S., Gebhart C. J., Sreevatsan S., Johnson T., et al. (2015). Changes in the porcine intestinal microbiome in response to infection with Salmonella enterica and Lawsonia intracellularis. PLoS One 10:e0139106. 10.1371/journal.pone.0139106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borton M. A., Sabag-Daigle A., Wu J., Solden L. M., O’Banion B. S., Daly R. A., et al. (2017). Chemical and pathogen-induced inflammation disrupt the murine intestinal microbiome. Microbiome 5:47. 10.1186/s40168-017-0264-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Jin B., Monis P., Saint C. (2013). A genetic and metabolic approach to redirection of biochemical pathways of Clostridium butyricum for enhancing hydrogen production. Biotechnol. Bioeng. 110 338–342. 10.1002/bit.24596 [DOI] [PubMed] [Google Scholar]
- Castillo N. A., Perdigon G., de Moreno de Leblanc A. (2011). Oral administration of a probiotic Lactobacillus modulates cytokine production and TLR expression improving the immune response against Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol. 11:177. 10.1186/1471-2180-11-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B., Koren O., Carvalho F. A., Ley R. E., Gewirtz A. T. (2014). AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut 63 1069–1080. 10.1136/gutjnl-2013-304909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. M., Topping D. L., Christophersen C. T., Bird A. R., Lange K., Saunders I., et al. (2011). Butyrate esterified to starch is released in the human gastrointestinal tract. Am. J. Clin. Nutr. 94 1276–1283. 10.3945/ajcn.111.017228 [DOI] [PubMed] [Google Scholar]
- Corridoni D., Pastorelli L., Mattioli B., Locovei S., Ishikawa D., Arseneau K. O., et al. (2012). Probiotic bacteria regulate intestinal epithelial permeability in experimental ileitis by a TNF-dependent mechanism. PLoS One 7:e42067. 10.1371/journal.pone.0042067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A., Materna A. C., Friedman J., Campos-Baptista M. I., Blackburn M. C., Perrotta A., et al. (2014). Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15:R89. 10.1186/gb-2014-15-7-r89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues A. R., Pires S. M., Halasa T., Hald T. (2012). Source attribution of human salmonellosis using a meta-analysis of case-control studies of sporadic infections. Epidemiol. Infect. 140 959–969. 10.1017/S0950268811002172 [DOI] [PubMed] [Google Scholar]
- Dorrell N., Wren B. W. (2007). The second century of Campylobacter research: recent advances, new opportunities and old problems. Curr. Opin. Infect. Dis. 20 514–518. 10.1097/QCO.0b013e3282a56b15 [DOI] [PubMed] [Google Scholar]
- Du C. T., Gao W., Ma K., Yu S. X., Li N., Yan S. Q., et al. (2018). MicroRNA-146a deficiency protects against Listeria monocytogenes infection by modulating the gut microbiota. Int. J. Mol. Sci. 19:E993. 10.3390/ijms19040993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey J. P., Speer C. A., Shen S. K., Kwok O. C., Blixt J. A. (1997). Oocyst-induced murine toxoplasmosis: life cycle, pathogenicity, and stage conversion in mice fed Toxoplasma gondii oocysts. J. Parasitol. 83 870–882. 10.2307/3284282 [DOI] [PubMed] [Google Scholar]
- Field L. H., Underwood J. L., Pope L. M., Berry L. J. (1981). Intestinal colonization of neonatal animals by Campylobacter fetus subsp. jejuni. Infect. Immun. 33 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca R. C., Conceicao F. R., Mendonca M., Haubert L., Sabadin G., de Oliveira P. D., et al. (2015). Pichia pastoris X-33 has probiotic properties with remarkable antibacterial activity against Salmonella Typhimurium. Appl. Microbiol. Biotechnol. 99 7953–7961. 10.1007/s00253-015-6696-9 [DOI] [PubMed] [Google Scholar]
- Gibbons I. S., Adesiyun A., Seepersadsingh N., Rahaman S. (2006). Investigation for possible source(s) of contamination of ready-to-eat meat products with Listeria spp. and other pathogens in a meat processing plant in Trinidad. Food Microbiol. 23 359–366. 10.1016/j.fm.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Gorman R., Bloomfield S., Adley C. C. (2002). A study of cross-contamination of food-borne pathogens in the domestic kitchen in the Republic of Ireland. Int. J. Food Microbiol. 76 143–150. 10.1016/s0168-1605(02)00028-4 [DOI] [PubMed] [Google Scholar]
- Grainger J. R., Wohlfert E. A., Fuss I. J., Bouladoux N., Askenase M. H., Legrand F., et al. (2013). Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nat. Med. 19 713–721. 10.1038/nm.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag L. M., Fischer A., Otto B., Grundmann U., Kuhl A. A., Gobel U. B., et al. (2012a). Campylobacter jejuni infection of infant mice: acute enterocolitis is followed by asymptomatic intestinal and extra-intestinal immune responses. Eur. J. Microbiol. Immunol. 2 2–11. 10.1556/EuJMI.2.2012.1.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag L. M., Fischer A., Otto B., Plickert R., Kuhl A. A., Gobel U. B., et al. (2012b). Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PLoS One 7:e35988. 10.1371/journal.pone.0035988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagsma J. A., Geenen P. L., Ethelberg S., Fetsch A., Hansdotter F., Jansen A., et al. (2013). Community incidence of pathogen-specific gastroenteritis: reconstructing the surveillance pyramid for seven pathogens in seven European Union member states. Epidemiol. Infect. 141 1625–1639. 10.1017/S0950268812002166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Gulati A., Sartor R. B. (2010). The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr. Opin. Gastroenterol. 26 564–571. 10.1097/MOG.0b013e32833f1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelaar A. H., Haagsma J. A., Mangen M. J., Kemmeren J. M., Verhoef L. P., Vijgen S. M., et al. (2012). Disease burden of foodborne pathogens in the Netherlands, 2009. Int. J. Food Microbiol. 156 231–238. 10.1016/j.ijfoodmicro.2012.03.029 [DOI] [PubMed] [Google Scholar]
- Heimesaat M. M., Fischer A., Alutis M., Grundmann U., Boehm M., Tegtmeyer N., et al. (2014). The impact of serine protease HtrA in apoptosis, intestinal immune responses and extra-intestinal histopathology during Campylobacter jejuni infection of infant mice. Gut Pathog. 6:16. 10.1186/1757-4749-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimesaat M. M., Fischer A., Jahn H. K., Niebergall J., Freudenberg M., Blaut M., et al. (2007). Exacerbation of murine ileitis by Toll-like receptor 4 mediated sensing of lipopolysaccharide from commensal Escherichia coli. Gut 56 941–948. 10.1136/gut.2006.104497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimesaat M. M., Plickert R., Fischer A., Gobel U. B., Bereswill S. (2013). Can microbiota transplantation abrogate murine colonization resistance against Campylobacter jejuni? Eur. J. Microbiol. Immunol. 3 36–43. 10.1556/EuJMI.3.2013.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiett K. L., Stern N. J., Fedorka-Cray P., Cox N. A., Musgrove M. T., Ladely S. (2002). Molecular subtype analyses of Campylobacter spp. from Arkansas and California poultry operations. Appl. Environ. Microbiol. 68 6220–6236. 10.1128/AEM.68.12.6220-6236.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zang X., Zhai W., Guan C., Lei T., Jiao X. (2018). Campylobacter spp. in chicken-slaughtering operations: a risk assessment of human campylobacteriosis in East China. Food Control 86 249–256. 10.1016/j.foodcont.2017.11.026 [DOI] [Google Scholar]
- Jones-Hall Y. L., Kozik A., Nakatsu C. (2015). Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PLoS One 10:e0119441. 10.1371/journal.pone.0119441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann C., Dicksved J., Engstrand L., Rautelin H. (2016). Composition of human faecal microbiota in resistance to Campylobacter infection. Clin. Microbiol. Infect. 22 61.e1–61.e68. 10.1016/j.cmi.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Christensen J. E., Dhillon A. S., Lane A. B., Hare-Sanford R., Schaberg D. M., et al. (2007). Campylobacter jejuni strains compete for colonization in broiler chicks. Appl. Environ. Microbiol. 73 2297–2305. 10.1128/AEM.02193-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M., Dramsi S., Gottardi C., Fedor-Chaiken M., Gumbiner B., Cossart P. (1999). A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18 3956–3963. 10.1093/emboj/18.14.3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesenfeld O. (2002). Oral infection of C57BL/6 mice with Toxoplasma gondii: a new model of inflammatory bowel disease? J. Infect. Dis. 185 S96–S101. 10.1086/338006 [DOI] [PubMed] [Google Scholar]
- Liu Q., Liu H., Deng Y., Hu G., Zhang H. (2010). Identification and metabolism characterization of a Clostridium lituseburense strain isolated from high-altitude soil. Wei Sheng Wu Xue Bao 50 1135–1140. [PubMed] [Google Scholar]
- Lone A. G., Selinger L. B., Uwiera R. R., Xu Y., Inglis G. D. (2013). Campylobacter jejuni colonization is associated with a dysbiosis in the cecal microbiota of mice in the absence of prominent inflammation. PLoS One 8:e75325. 10.1371/journal.pone.0075325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan M. T., Bender K. S., Buckley D. H., Sattley W. M., Stahl D. A. (2018). Brock Biology of Microorganisms. New York, NY: Pearson. [Google Scholar]
- Manohar M., Baumann D. O., Bos N. A., Cebra J. J. (2001). Gut colonization of mice with actA-negative mutant of Listeria monocytogenes can stimulate a humoral mucosal immune response. Infect. Immun. 69 3542–3549. 10.1128/IAI.69.6.3542-3549.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. K., Thabane M., Garg A. X., Clark W. F., Salvadori M., Collins S. M., et al. (2006). Incidence and epidemiology of irritable bowel syndrome after a large waterborne outbreak of bacterial dysentery. Gastroenterology 131 445–450. 10.1053/j.gastro.2006.05.053 [DOI] [PubMed] [Google Scholar]
- Martins F. S., Vieira A. T., Elian S. D., Arantes R. M., Tiago F. C., Sousa L. P., et al. (2013). Inhibition of tissue inflammation and bacterial translocation as one of the protective mechanisms of Saccharomyces boulardii against Salmonella infection in mice. Microbes Infect. 15 270–279. 10.1016/j.micinf.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Medici M., Vinderola C. G., Weill R., Perdigon G. (2005). Effect of fermented milk containing probiotic bacteria in the prevention of an enteroinvasive Escherichia coli infection in mice. J. Dairy Res. 72 243–249. 10.1017/S0022029905000750 [DOI] [PubMed] [Google Scholar]
- Meisel M., Mayassi T., Fehlner-Peach H., Koval J. C., O’Brien S. L., Hinterleitner R., et al. (2017). Interleukin-15 promotes intestinal dysbiosis with butyrate deficiency associated with increased susceptibility to colitis. ISME J. 11 15–30. 10.1038/ismej.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. K., Burns J. (1970). Histopathology of Listeria monocytogenes after oral feeding to mice. Appl. Microbiol. 19 772–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiro T., Kusunoki R., Otani A., Ansary M. M., Tongu M., Harashima N., et al. (2013). Butyric acid attenuates intestinal inflammation in murine DSS-induced colitis model via milk fat globule-EGF factor 8. Lab. Invest. 93 834–843. 10.1038/labinvest.2013.70 [DOI] [PubMed] [Google Scholar]
- Mon K. K., Saelao P., Halstead M. M., Chanthavixay G., Chang H. C., Garas L., et al. (2015). Salmonella enterica serovars enteritidis infection alters the indigenous microbiota diversity in young layer chicks. Front. Vet. Sci. 2:61. 10.3389/fvets.2015.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka K., Leja K., Olejnik-Schmidt A. K., Czaczyk K. (2012). Isolation process of industrially useful Clostridium bifermentans from natural samples. J. Biosci. Bioeng. 113 631–633. 10.1016/j.jbiosc.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Ormerod K. L., Wood D. L., Lachner N., Gellatly S. L., Daly J. N., Parsons J. D., et al. (2016). Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 4:36. 10.1186/s40168-016-0181-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paclik D., Berndt U., Guzy C., Dankof A., Danese S., Holzloehner P., et al. (2008). Galectin-2 induces apoptosis of lamina propria T lymphocytes and ameliorates acute and chronic experimental colitis in mice. J. Mol. Med. 86 1395–1406. 10.1007/s00109-007-0290-2 [DOI] [PubMed] [Google Scholar]
- Poropatich K. O., Walker C. L. F., Black R. E. (2010). Quantifying the association between Campylobacter infection and Guillain-Barre syndrome: a systematic review. J. Health Popul. Nutr. 28 545–552. 10.3329/jhpn.v28i6.6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin O., Kassem I. I., Shen Z., Lin J., Rajashekara G., Zhang Q. (2015). Campylobacter in poultry: ecology and potential interventions. Avian. Dis. 59 185–200. 10.1637/11072-032315-Review [DOI] [PubMed] [Google Scholar]
- Sakaridis I., Ellis R. J., Cawthraw S. A., van Vliet A. H. M., Stekel D. J., Penell J., et al. (2018). Investigating the association between the caecal microbiomes of broilers and Campylobacter burden. Front. Microbiol. 9:927. 10.3389/fmicb.2018.00927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez E., Donat E., Ribes-Koninckx C., Calabuig M., Sanz Y. (2010). Intestinal Bacteroides species associated with coeliac disease. J. Clin. Pathol. 63 1105–1111. 10.1136/jcp.2010.076950 [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.P1110110.3201/eid1701.091101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejvar J. J., Baughman A. L., Wise M., Morgan O. W. (2011). Population incidence of Guillain-Barre syndrome: a systematic review and meta-analysis. Neuroepidemiology 36 123–133. 10.1159/000324710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund B., Rieder F., Albrich S., Wolf K., Bidlingmaier C., Firestein G. S., et al. (2001). Adenosine kinase inhibitor GP515 improves experimental colitis in mice. J. Pharmacol. Exp. Ther. 296 99–105. [PubMed] [Google Scholar]
- Silva W. C., Targino B. N., Mendonça R. S., Sant’Ana A. S., Hungaro H. M. (2017). Campylobacter: an overview of cases, occurrence in food, contamination sources, and antimicrobial resistance in Brazil. Food Rev. Int. 34 364–389. 10.1080/87559129.2017.1298125 [DOI] [Google Scholar]
- Sun J., Furio L., Mecheri R., van der Does A. M., Lundeberg E., Saveanu L., et al. (2015). Pancreatic beta-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity 43 304–317. 10.1016/j.immuni.2015.07.013 [DOI] [PubMed] [Google Scholar]
- Sun X., Threadgill D., Jobin C. (2012). Campylobacter jejuni induces colitis through activation of mammalian target of rapamycin signaling. Gastroenterology 142 86.e5–95.e5. 10.1053/j.gastro.2011.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Mytle N., Lambert S., Coleman M., Doyle M. P., Smith M. A. (2006). Comparison of Listeria monocytogenes virulence in a mouse model. J. Food Prot. 69 842–846. 10.4315/0362-028X-69.4.842 [DOI] [PubMed] [Google Scholar]
- Taras D., Simmering R., Collins M. D., Lawson P. A., Blaut M. (2002). Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 52(Pt 2), 423–428. 10.1099/00207713-52-2-423 [DOI] [PubMed] [Google Scholar]
- Wang L., Pan M., Li D., Yin Y., Jiang T., Fang S., et al. (2017). Metagenomic insights into the effects of oligosaccharides on the microbial composition of cecal contents in constipated mice. J. Funct. Foods 38 486–496. 10.1016/j.jff.2017.09.045 [DOI] [Google Scholar]
- Zhang W., Zhu Y. H., Yang G. Y., Liu X., Xia B., Hu X., et al. (2017). Lactobacillus rhamnosus GG affects microbiota and suppresses autophagy in the intestines of pigs challenged with Salmonella infantis. Front. Microbiol. 8:2705. 10.3389/fmicb.2017.02705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang L. (2017). SAG4 DNA and peptide vaccination provides partial protection against T. gondii infection in BALB/c mice. Front. Microbiol. 8:1733. 10.3389/fmicb.2017.01733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yao B., Song X., Wang Y., Cui S., Xu H., et al. (2017). Prevalence and quantification of Campylobacter contamination on raw chicken carcasses for retail sale in China. Food Control 75 196–202. 10.1016/j.foodcont.2016.12.007 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.