Abstract
Campylobacter spp. is one of the main cause of bacterial gastroenteritis in the world. The increase of antibiotic resistance in this species is a threat to public health. A Campylobacter spp. surveillance study was performed in Italy in the 2013–2016 period by the Enter-Net Italia network. The most prevalent Campylobacter species identified causing gastroenteritis was Campylobacter jejuni (73.4%) and 45% of all the annual cases of campylobacteriosis were reported in the summer period. High rates of ciprofloxacin and tetracycline resistance in Campylobacter spp. have been observed. An increasing percentage of Campylobacter coli strains simultaneously resistant to ciprofloxacin, tetracycline and erythromycin has been found. Molecular mechanisms of resistance have been investigated and the role of efflux pumps evaluated. Antibiotic resistance in Campylobacter spp. is an increasing serious threat that requires coordinated action to minimize the emergence and spread of antimicrobial resistant strains from animals to humans throughout the food chain.
Keywords: Campylobacter jejuni, Campylobacter coli, ciprofloxacin, tetracycline, erythromycin, resistance
Introduction
Campylobacter spp. is considered one of the most common cause of bacterial gastroenteritis in the world, and Campylobacter jejuni and Campylobacter coli are the most common bacterial species isolated from human stool samples (Havelaar et al., 2009; Haagsma et al., 2013; Gibbons et al., 2014; European Food Safety Authority and European Centre for Disease Prevention and Control, 2017). In 2016, 246,307 laboratory-confirmed cases of campylobacteriosis were reported in EU with a notification rate of 66.3 per 100,000 population, 6.1% higher than in 2015 (European Centre for Disease Prevention and Control, European Food Safety Authority, and European Medicines Agency, 2017). Campylobacter spp. infections are self-limiting diseases, and treatment with antimicrobials is not recommended. However, antibiotic therapy is indicated for patients with persistent fever, bloody or persistent diarrhea. HIV-positive or immunocompromised individuals normally receive antibiotic treatment if infected by Campylobacter spp. (Cha et al., 2016; European Food Safety Authority and European Centre for Disease Prevention and Control, 2017).
Poultry is described as the main reservoir and common source of transmission of campylobacteriosis to humans (Kaakoush et al., 2015). Other risk factors include the consumption of meat, unpasteurized milk and water, contact with animals and international travels, but very rarely human-to-human transmission is reported (Iovine, 2013).
Campylobacter spp. exhibits intrinsic resistance to novobiocin, bacitracin, vancomycin, and polymyxins, presumably due to the absence of appropriate targets and/or low affinity binding sites for these drugs (Iovine, 2013). Azithromycin or erythromycin are the first line of choice for antibiotic treatment of campylobacteriosis. Ciprofloxacin and tetracycline are alternatives but not used for treatment of children. In the last decade, ciprofloxacin and tetracycline resistance rapidly increased in Campylobacter spp. in humans in Europe, while erythromycin resistance levels remained relatively low (European Food Safety Authority and European Centre for Disease Prevention and Control, 2017). The major mechanism of tetracycline resistance in Campylobacter spp. is the binding and protection of ribosomal A site by the protein TetO. Ciprofloxacin resistance in C. jejuni and C. coli is mainly due to point mutations in the quinolone resistance-determining region (QRDR) of the GyrA protein (Iovine, 2013). Erythromycin resistance in Campylobacter spp. has been associated with mutations in the 23S rRNA and in the large loop of the L4 and L22 50S ribosomal proteins (Hao et al., 2013). Multidrug efflux can also contribute to reduce the intracellular concentration of several antibiotics, including tetracycline, ciprofloxacin, and erythromycin (Iovine, 2013).
In Italy, enteric bacterial pathogen surveillance is carried out by a voluntary, passive, laboratory-based, surveillance system named Enter-Net Italia (www.iss.it/site/rmi/enternet) that collects Escherichia coli, Listeria, Salmonella, Campylobacter, Shigella, and Yersinia records and strains. It is constituted by a network of 27 regional reference laboratories and clinical centers of the national health system. Twenty centers are from Northern (covering 23.8 million people), 4 from Central (8.3 millions) and 3 from Southern (11.5 millions) Italy and they are coordinated by the Istituto Superiore di Sanità (ISS). Enter-Net Italia contributes data to the Foodborne and Waterborne (FWD) European surveillance system coordinated by the European Centre for Disease Control (ECDC).
Results of the Campylobacter spp. Italian surveillance activities in the period 2013–2016 are herein presented together with results of antimicrobial resistance testing and the investigation of the resistance mechanisms.
Materials and methods
Data source
The Enter-Net Italia database was analyzed for age, sex, date of disease onset, and travels for both pediatric and adult patients with a positive Campylobacter spp. isolation occurred from January 2013 to December 2016. Patient identity was anonymised. No informed consent was requested because this retrospective study was only focused on the bacteria and did not have impact on the patients.
Bacterial strains and culture conditions and antimicrobial susceptibility testing
Along the 4 years of surveillance, the National reference center at ISS received 647 Campylobacter spp. strains, all accompanied by epidemiological records submitted by strain providers in the Enter-Net Italia database. Strains were grown on Columbia agar plates supplemented with 5% defibrinated horse blood (Oxoid, Hampshire, UK) and Vitox (Oxoid Limited, Hampshire, UK) at 42°C under microaerobic conditions (10% CO2, 5% O2, and 85% N2) for 48 h. When not reported in the record, Campylobacter species was determined at ISS by multiplex PCRs for C. jejuni and C. coli and by simplex PCR for the other species, as previously described (Linton et al., 1996; Denis et al., 1999).
Antimicrobial susceptibility was determined by the disc diffusion method on Mueller–Hinton agar with 5% defibrinated horse blood (Oxoid Limited, Hampshire, UK) and 20 mg/L β-NAD (MH-F) (Oxoid Limited, Hampshire, UK) and microaerobic environment, as recommended by European Committee on Antimicrobial Susceptibility Testing (EUCAST). The following antimicrobials (Becton, Dickinson and Company, NJ, USA) were tested: ciprofloxacin (CIP) (5 μg), tetracycline (TET) (30 μg), erythromycin (ERY) (15 μg) and gentamicin (GEN) (10 μg). C. jejuni strain ATCC33560 was used as quality control. Breakpoints were determined following the EUCAST v8.1 guidelines (http://www.eucast.org/clinical_breakpoints/) for CIP, TET and ERY, and the CLSI 2018 guidelines for GEN (CLSI, 2018).
PAβN efflux pump testing
For 12 selected isolates, disc diffusion antimicrobial susceptibility tests were performed as described above adding in parallel two plates, containing 20 or 40 mg/L efflux pump inhibitor (EPI) PAβN (Sigma–Aldrich, Saint Louis, USA), respectively (Payot et al., 2004). C. jejuni ATCC33560 fully susceptible strain was used as control.
Antibiotic resistance mechanism investigation
A loop of bacteria growth on the plate was resuspended in 1 ml of autoclaved water and chromosomal DNA was extracted and purified using the Chelex 100 molecular Biology grade resin (Bio-Rad Laboratories, Milano, Italy), according to manufacturer's instructions. C. jejuni and C. coli gyrA, rplD (coding for the 50S L4 protein), rplV (coding for the 50S L22 protein) genes, and the cmeR-cmeA intergenic region (CmeR-BOX) were amplified by PCR using previously described primer pairs (Zirnstein et al., 1999, 2000; Cagliero et al., 2006; Pérez-Boto et al., 2010; Hao et al., 2013). The amplicons were purified using the Wizard PCR Preps DNA purification system (Promega, Madison, WI, USA) and fully sequenced by fluorescent dye-labeled deoxyribonucletide method with an ABI 3730 automatic DNA sequencer (Perkin-Elmer, Foster City, CA). Multiple sequence alignments of nucleotide and/or amino acid sequences were performed by DNAMAN v.5.2.10 software. The sequences were also analyzed using BLASTN and BLASTP software at the NCBI GenBank sequence database (https://www.ncbi.nlm.nih.gov) and compared in silico to those of C. jejuni NCTC11168 strain (GenBank accession n. AL111168), considered as the wild type reference sequences.
The V region of 23S rRNA gene (rrnB operon) was amplified by PCR, the amplicons, purified using the ISOLATE II PCR and Gel Kit (Bioline, London, UK) were digested with BsaI (5U) and BceAI (1U) (New England Biolabs, Beverly, UK) restriction enzymes, respectively, as previously described (Vacher et al., 2003). The fragments were separated by electrophoresis on a 2% agarose gel, with ethidium bromide, and visualized under UV light. The results were interpreted as follows: Digestion by BsaI of the 316-bp amplicon containing the A2075G mutation led to two subproducts of 201 and 115 bp in contrast to the uncut wild type and A2074C amplicons. The BceAI digestion, produced three fragments of 41, 24, and 251 bp on the wild type and on the A2075G amplified sequences (Vacher et al., 2003).
All ERY and TET resistant strains were screened by PCR for the presence of ermB and tet(O) genes, respectively (Pratt and Korolik, 2005; Zhou et al., 2016). The cmeR–cmeA intergenic region and the conserved CmeR-Box (5′-TGTAATAAAT [or A] ATTACA-3′) were analyzed in 72 strains with the aim to identify mutations potentially associated with ERY resistance, as previously described (Cagliero et al., 2007; Pérez-Boto et al., 2010; Grinnage-Pulley and Zhang, 2015; Zhang et al., 2017)
Statistical analysis
Statistical analysis was performed using GraphPad Software (GraphPad Software,La Jolla, USA). Fisher's exact test was used to compare differences in the ratios of infection between patient gender and in the ratios of resistance between the two main Campylobacter species. The overall rates of resistance of the isolates and their 95% confidence intervals (95% CIs) were calculated. P < 0.05 were considered as significantly different.
Results
Surveillance results
From January 2013 to December 2016, 4672 records of Campylobacter spp. isolation were included in the Enter-Net Italia database, representing 21% of all data collected in this period of surveillance. In most of these records, Campylobacter spp. was isolated from feces (n. 4626, confirmed campylobacteriosis cases) but some notifications reported different sources: 26 strains were from blood, one from urine and 19 from unidentified source. C. jejuni was the most frequent species (73.4%), followed by C. coli (8%), C. upsaliensis (0.4%), C. lari (0.1%), C. fetus (0.09%), and C. concisus (0.04%). In 835/4672 (17.9%) isolates the species was not determined and strains were not sent to ISS for identification.
Among 647 (647/4672, 14%) Campylobacter spp. strains received at ISS, 583 (90.1%) were classified as C. jejuni, 62 (9.6%) C. coli, 1 (0.15%) C. fetus, and 1 (0.15%) C. lari.
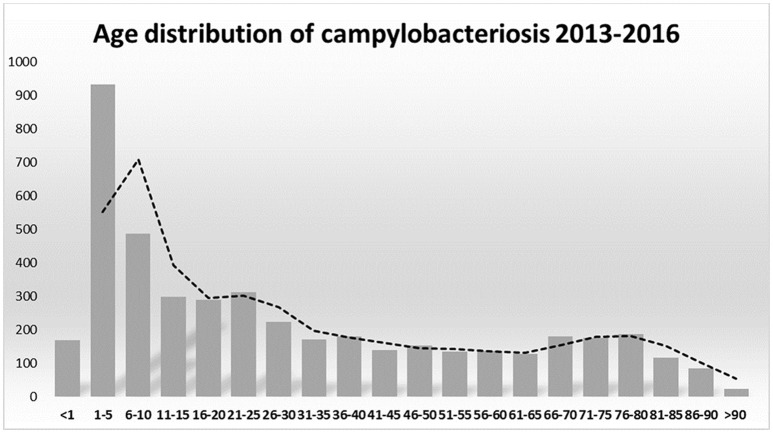
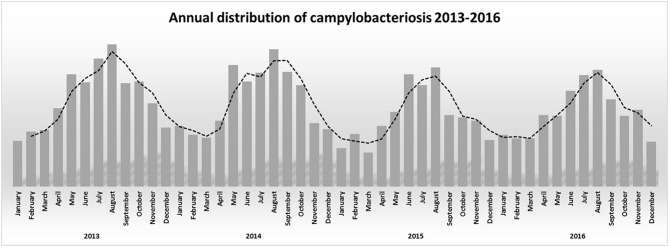
The association between campylobacteriosis and gender was statistically significant (P < 0.0001), with 2,294 male (49%), 1746 female (37%), and 632 unknown patients reported as positive for Campylobacter spp. isolation, respectively. The highest number of Campylobacter spp. isolates was obtained from 1 to 5 years old infants with a total of 932 cases (20%) (Figure 1). Campylobacter spp. isolation showed a summer peak, with an average of 45% of all the annual cases reported in the June-August trimester (Figure 2). Travels were not significantly associated with a positive Campylobacter spp. isolation (217/4672, 5%).
Figure 1.
Campylobacter spp. notifications by ages 2013–2016.
Figure 2.
Annual distribution of campylobacteriosis 2013–2016.
Campylobacter antimicrobial resistance
Antimicrobial susceptibility was determined in 218 of the 647 (34%) Campylobacter spp. strains, whose 176 (176/583, 30%) were C. jejuni, 41 (41/62, 66%) C. coli and 1 C. fetus. Strains were selected to represent almost one third of the C. jejuni and more than half of the C. coli received. C. coli was more extensively studied because multi-drug resistance (MDR, defined as resistance to at least three antimicrobial classes) has been described in Europe in this species (European Food Safety Authority and European Centre for DiseasePrevention and Control, 2018). Results showed that 46 (21%) strains (38 C. jejuni, 7 C. coli, 1 C. fetus) were susceptible to all antibiotics tested, and all strains but two C. jejuni (0,91%) were susceptible to gentamicin. Differently, 76% of the C. jejuni and 70% of the C. coli strains were CIP-resistant (CIP-R). Many strains were TET-resistant (TET-R, 64%) and less ERY-resistant (ERY-R, 12%). TET-R and ERY-R strains were more frequently C. coli (76% TET-R, 32% ERY-R) than C. jejuni (61% TET-R, 7% ERY-R), with P < 0.0869 for TET-R and P < 0.00001 for ERY-R significant values, respectively.
Among 176 C. jejuni isolates, 49.4% were resistant to two, 5.7% to three and 0.6% to four antimicrobial agents, respectively. Of the 41 C. coli isolates, 15 (36.6%) strains were resistant to two antimicrobial agents and 15 (36.6%) to three drugs, respectively. The most common resistance patterns were CIP-TET for C. jejuni and CIP-TET-ERY for C. coli.
Antimicrobial resistance mechanisms
Seventy-five strains were screened for antimicrobial resistance mechanisms that conferred resistance to CIP, TET and ERY, respectively. Fifty-two C. jejuni (5 susceptible, 10 resistant to CIP, 32 to CIP-TET, 3 to CIP-TET-ERY, 1 to TET and 1 to CIP-TET-GEN) and 23 C. coli (2 resistant to CIP, 12 to CIP-TET, 1 to CIP-ERY and 8 to CIP-TET-ERY) were tested. All ciprofloxacin-susceptible (CIP-S) strains had wild type GyrA protein sequence, as expected. Among CIP-R strains, the single point mutation Thr86Ile was detected in all but 8 strains. Among them, one showed the Thr86Ala mutation and 3 showed two mutations, Thr86Ile+Asp90Asn, while 4 CIP-R resistant strains did not show amino acid changes in the GyrA deduced protein sequence.
All TET-R strains showed the acquisition of the tet(O) gene.
All ERY-R strains were negative for the presence of the ermB gene (Table 1). The conserved CmeR-Box (5′-TGTAATAAAT [or A] ATTACA-3′) analysis showed that 20 out of 72 (28%) strains, presented one or more substitutions, insertions and/or deletions respect to the reference sequence of the wild type C. jejuni NCTC11168 strain (Table 2). However, most of these mutations were not associated to ERY resistance, being also identified in several ERY-S strains (Table 2). In one ERY-R strain the deletion of the 8th to 11th nt of the 16 bp CmeR-BOX was observed, but its implication in ERY resistance has not yet been proven. More informative results about potential ERY resistance mechanisms were obtained by BsaI and BceAI restriction of the 316-bp PCR amplicon of the 23S rDNA. This analysis demonstrated the presence of the A2075G mutation in 9 ERY-R strains (Vacher et al., 2003).
Table 1.
Characteristics of antimicrobial resistance mechanisms in 12 the erythromycin-resistant and 3 erythromycin-susceptible Campylobacter spp. strains isolated between 2013 and 2016.
| Strain | Year | Species | Resistance | E-EPI | GyrA | Tet(O) | 23S rRNA | L4 | L22 |
|---|---|---|---|---|---|---|---|---|---|
| 91/1/13 | 2013 | jejuni | Suscept | nd | wt | nd | wt | wt | wt |
| 19/4/14 | 2014 | coli | CIP TET | nd | T86I | POS | wt | V196A | I65V,A74G,S109T,E111A,T114A, A126V, A135V,V137A,K138E* |
| 49/1/14 | 2014 | coli | CIP TET | nd | T86I | POS | wt | V196A | I65V,A74G,S109T,E111A,T114A, A126V, A135V,V137A,K138E* |
| 57/1/13 | 2013 | coli | CIP TET ERY | neg | T86I | POS | wt | V196A | I65V,A74G,S109T,E111A,T114A, A126V, A135V,V137A,K138E* |
| 74/1/15 | 2015 | coli | CIP TET ERY | neg | T86I | POS | wt | V196A | I65V,A74G,A103V,S109A,A135V,V137A,K138E |
| 14/20/15 | 2014 | coli | CIP TET ERY | neg | T86I+D90N | POS | wt | V196A | I65V,A74G,A103V,S109A,A135V,V137A,K138E |
| 32/2/15 | 2015 | coli | CIP TET ERY | POS | T86I+D90N | POS | A2075G | V196A | I65V,A74G,S109T,E111A,T114A, A126V, A135V,V137A,K138E* |
| 7/7/16 | 2015 | coli | CIP TET ERY | neg | T86I | POS | A2075G | V196A | I65V,A74G,S109T,E111A,T114A, A126V, A135V,V137A,K138E* |
| 42/2/15 | 2015 | coli | CIP TET ERY | neg | T86I | POS | A2075G | V196A | I65V,A74G,S109T,E111A,T114A, A126V, A135V,V137A,K138E* |
| 55/3/15 | 2015 | coli | CIP TET ERY | neg | T86I | POS | A2075G | V196A | I65V,A74G,S109T,E111A,T114A, A126V, A135V,V137A,K138E* |
| 3/14/14 | 2013 | coli | CIP TET ERY | neg | T86I | POS | A2075G | V196A | I65V,A74G,S109T,E111A,T114A, A126V, A135V,V137A,K138E* |
| 57/5/14 | 2014 | coli | CIP TET ERY | neg | T86I | POS | A2075G | V121A,V176I,T177S,V184I,M192I,V196A | I65V,A74G,A103V,S109T,E111A, T114A,A126V, A135V,V137A,K138E* |
| 6/17/16 | 2015 | jejuni | CIP TET ERY | POS | T86I | POS | A2075G | V121A,M192I,V196A | wt |
| 67/1/16 | 2016 | jejuni | CIP TET ERY | POS | T86I | POS | A2075G | V121A,M192I,V196A | wt |
| 89/6/13 | 2013 | jejuni | CIP TET ERY | neg | T86I | POS | A2075G | V196A | wt |
CIP, ciprofloxacin; TET, tetracycline; ERY, erythromycin; Suscept, Susceptible strain; E-EPI, Erythromycin-Efflux Pump Inhibition test; nd, not determined; POS, positive; neg, negative; wt, wild type (GyrA like ATCC 33560 and 23S rRNA/L4/L22 like NCTC 11168).
Deletion from 120 to 124 nt (TTTKA) and 130 nt (T).
Table 2.
Partial multiple alignment of the 16 nucleotide intergenic region between cmeR and cmeA identified in erythromycin-susceptible (ERY-S) and resistant (ERY-R) C. jejuni and C. coli strains.
| cmeR-BOX | ERY-R | ERY-S | Referencesa |
|---|---|---|---|
| TGTAATAAATATTACAb | 8 | 19 | Pérez-Boto et al., 2010; Zhou et al., 2016; Zhang et al., 2017 |
| TGTAATAAAaATTACAb | 1 | 24 | Pérez-Boto et al., 2010; Grinnage-Pulley and Zhang, 2015; Zhou et al., 2016; Zhang et al., 2017 |
| cGTAATAAATATTACA | 1 | 1 | |
| TGTAATAAAa-TTACA | 0 | 5 | Zhou et al., 2016 |
| TGTAATAAAaATTAtA | 0 | 1 | Cagliero et al., 2007; Pérez-Boto et al., 2010; Zhou et al., 2016 |
| TGTAATAAATATcACA | 0 | 2 | |
| TGTAATAAATATTgCA | 1 | 3 | Grinnage-Pulley and Zhang, 2015; Zhou et al., 2016; Zhang et al., 2017 |
| TGTAATA----TTACA | 1 | 0 | |
| TGTAATAAATATT--- | 0 | 1 | Grinnage-Pulley and Zhang, 2015 |
| TGTAATAAATATTACtattacA | 0 | 3 | Pérez-Boto et al., 2010 |
| TGTAATAtgtgtaataAATATTACA | 0 | 1 |
Polymorphisms are shown as lower case. Nucleotide deletions are shown as dash. The IR regions are underlined.
References of previously reported mutations.
Sequences previously described as wild type or “conserved sequences.”
In 12 ERY-R (9 C. coli and 3 C. jejuni) and 3 ERY-S strains (2 C. coli and 1 C. jejuni), the rplD and rplV genes coding for L4 and L22 50S ribosomal proteins, respectively, were studied. One C. jejuni, susceptible to all antimicrobials tested, presented wild type L4 and L22 proteins. The L4 protein showed the V196A mutation in 2 ERY-S and 9 ERY-R strains, respectively. Three ERY-R strains presented additional mutations in the L4 protein, possibly associated with the resistant phenotype (Table 1). Three ERY-R strains exhibited a wild type L22 protein. The other 11 strains showed different mutations in L22, any of them certainly associated with the ERY-R phenotype (Table 1).
PAβN efflux pump study
The EPI PAβN used at 40 mg/L concentration reduced resistance levels to ERY and TET in some strains, despite the drug did not restore full susceptibility in any of them. The inhibitory effect was dose dependent, being 20 mg/L PAβN less efficacious than 40 mg/L. EPI effect increased the diameter of the inhibition zone for ERY of 9/10 mm in 3 strains (6/17/16, 32/2/15, 67/1/16) and 4 mm in 2 strains (74/1/15, 7/7/16). For TET, an increment of 4/5 mm of the diameter of the inhibition zone was observed in 2 strains (57/1/13, 14/20/15). EPI did not affect ciprofloxacin resistance and bacterial growth at the two concentration tested.
Discussion
Despite 18% of the records notified in the Enter-Net Italia database did not contain information on species, and strains were not sent to ISS for identification, the study showed a clear prevalence of C. jejuni as the main species responsible for campylobacteriosis, followed by C. coli. Nevertheless other minor species were reported in the database, but their prevalence can be underestimated because of the lack of identification of numerous strains.
In developing countries, Campylobacter spp. infections are more frequent in children under 2 years of age, sometimes resulting in fatal outcomes of complicated infections. In industrialized countries there is a bimodal age distribution for campylobacteriosis, the largest peak at 5 years of age and the second peak at 20–29 years of age (Evans and Brachman, 1998; Butzler, 2004). In Italy Campylobacter spp. infection occurred in all age groups, but 1–5 years old children were the most susceptible. A weak increase of campylobacteriosis was also present in young adults and in over 65 years old people (Figure 1). Our epidemiological records reported higher morbidity in males than in females. Other studies also showed a similar gender difference (Butzler, 2004; Schielke et al., 2014; Walter et al., 2018). Seasonality was also observed for campylobacteriosis in Italy, the incidence strongly increased in the summer period, being an independent variable by the year of notification and age groups (Figure 2). Other zoonotic enteric diseases of bacterial origin, e.g., salmonellosis, are also characterized by seasonality, reflecting differences in the risk for exposure to infectious agents during the summer period (Lal et al., 2012; European Food Safety Authority and European Centre for Disease Prevention and Control, 2017).
Currently, macrolides are the drugs of choice for severe campylobacteriosis treatment. Fluoroquinolones are also recommended as the first-line therapy for empirical treatment of undiagnosed diarrheal cases, whereas tetracyclines are considered a second-line treatment. Very high to extremely high resistance levels to ciprofloxacin were reported in human campylobacter isolates in Europe in 2016 (European Food Safety Authority and European Centre for Disease Prevention and Control, 2017). Quinolone resistance in C. jejuni from humans in Europe was significantly associated to resistance in C. jejuni from food-producing animals, and this resistance was significantly associated to the consumption of quinolones in such animals (European Centre for Disease Prevention and Control, European Food Safety Authority, and European Medicines Agency, 2017). For this reason, fluoroquinolones are currently considered as an inappropriate empirical treatment of human Campylobacter spp. infections (Collignon et al., 2016; European Food Safety Authority and European Centre for Disease Prevention and Control, 2017). Resistance to ERY in C. jejuni in Europe is generally low (1.5%), while it is higher in C. coli (14.4%) that can rise to 24.2–54.5% in several countries. Studies performed in Europe showed that the consumption of macrolides in food-producing animals and resistance in C. coli from both food-producing animals and humans was statistically-significant. Equally, resistance in C. jejuni from humans was strongly associated with the consumption of macrolides in food-producing animals (European Centre for Disease Prevention and Control, European Food Safety Authority, and European Medicines Agency, 2017; European Food Safety Authority and European Centre for Disease Prevention and Control, 2017). In Italy and in other EU countries, gentamicin resistance was extremely low with 0.9% resistant strains (European Food Safety Authority and European Centre for Disease Prevention and Control, 2017). However, our study showed that in Italy most of the strains (>60%) are CIP-R or TET-R, but also a significant percentage is resistant to macrolides (13%). Furthermore, multidrug resistant Campylobacter spp. strains are emerging. It is of public health concern that a high percentage of Campylobacter spp. was CIP-TET-R (48% of C. jejuni and 41% of C. coli isolates) and 29% of the Italian human C. coli isolates were CIP-TET-ERY-R.
Several studies performed in Campylobacter spp. isolates from broilers and turkeys reared at the slaughters in Northern Italy, showed high resistance levels to quinolones, tetracycline and macrolides. The occurrence of resistant campylobacters in food-producing animals has been associated with the use of antibiotics in animal farming and it is currently considered an important threat to human health because of the high risk of transmission through the food chain of these resistant bugs to humans (Smith and Fratamico, 2010; Giacomelli et al., 2014; Manfreda et al., 2016).
Resistance mechanisms in Italian Campylobacter spp. isolates were not univocally identified for all the phenotypes observed. TET resistance was clearly associated to the presence of the tet(O) gene, as previously described in most of the Campylobacter spp. isolated around the world (Iovine, 2013). Differences of TET resistance between C. jejuni and C. coli were not statistically significant. Fluoroquinolone resistance in our strains was mainly due to the presence of the Thr86Ile GyrA mutation. This mutation is the most prevalent in clinical and also in veterinary isolates (Iovine, 2013; Wieczorek and Osek, 2013). Previously described mutations in the GyrA protein were also found in two strains of our collection (Wang et al., 1993). Interestingly, four CIP-R strains showed a wild type GyrA sequence. This genotype has been also previously reported in other CIP-R strains (Giacomelli et al., 2014; Tang et al., 2017). It has been hypothesized that in these strains, CIP resistance could be mediated by a chromosomally encoded multidrug efflux pump, which reduces the intracellular concentration of fluoroquinolones (Iovine, 2013; Wieczorek and Osek, 2013). However, we did not measure any effect of the efflux pump inhibitor on wild-type GyrA CIP-R strains, leaving undetermined the mechanism of resistance.
The previously described A2075G mutation in the 23S rRNA gene, was found in nine out of 12 ERY-R strains tested (3 C. jejuni and 6 C. coli). Other studies have also indicated that this mutation is usually responsible for high-level ERY resistance (Mamelli et al., 2003; Vacher et al., 2003; Payot et al., 2004; Gibreel et al., 2005; Corcoran et al., 2006). However, ERY resistance mechanism remained undefined in most of our isolates. It has been proposed that modifications in the large loop of the L4 protein (amino acids 55 to 77) and in the highly conserved large loop of the L22 protein (aa 78 to 98) can be associated with macrolide resistance in various bacteria (Cagliero et al., 2006; Hao et al., 2013). In the present study, no variations were found in loop regions of the L4 or L22 proteins. However, the V196A modification, which is located in another region than the large loop of L4, was detected in both ERY-S and ERY-R Campylobacter spp. strains, confirming previous reports that did not recognize in this mutation any association with ERY resistance, such as other L4 mutations located outside the large loop (Cagliero et al., 2006; Corcoran et al., 2006). The L22 protein showed several amino acid substitutions and a high variable C-terminal region (amino acids 109 to 138), but these modifications were found in both ERY-R and ERY-S strains and were not associated to macrolide resistance (Cagliero et al., 2006; Pérez-Boto et al., 2010; Zhou et al., 2016). As previously demonstrated, the efflux pump inhibitor PAβN at 20 mg/L had no effect on ERY resistance (Mamelli et al., 2003; Gibreel et al., 2005; Corcoran et al., 2006), whereas a higher concentration of the inhibitor (40 mg/L) led to two- to four-fold decrease in ERY resistance level (Payot et al., 2004). In two ERY-R strains of our collection the pump efflux inhibitor PAβN at 40 mg/L did not restore ERY susceptibility but induced a significant decrement of ERY inhibition halos of the antibiograms, leading to an intermediate inhibition zone. These strains also presented the 23S rDNA mutation A2075G, suggesting that this mutation may contribute to ERY resistance in those strains, together with activation of the efflux pumps. High variability in the CmeR-Box region was found, identifying 11 different sequence variants in our strains. Among them, some were previously described as significantly associated with macrolides resistance (Cagliero et al., 2007; Pérez-Boto et al., 2010; Grinnage-Pulley and Zhang, 2015; Zhou et al., 2016; Zhang et al., 2017). In our strains these modifications were not related to ERY resistance, being also identified in ERY-S strains, and some ERY-R strains showed no mutations in this locus. To our knowledge four of the CmeR-Box variants identified in our study were not previously described. One of them consisted of a deletion of four nucleotides, from nt8 to nt11, into the CmeR binding region, and was detected in one ERY-R strain (89/6/13). This deletion could be associated with deregulation of the cmeABC operon expression causing ERY resistance (Table 2). However, this and the other ERY-R strains were negative when they were tested for PAβN inhibition, therefore efflux did not seem relevant for conferring resistance in these strains.
In conclusion, the Enter-Net surveillance improves knowledge and provides novel information on campylobacteriosis in Italy. The present study showed that antibiotic resistance in Campylobacter spp. in humans is a worrisome and an underreported problem. The high rates of CIP resistance in Campylobacter spp. and the increasing levels of ERY resistance in C. coli have converted Campylobacter spp. in one of the species included in the priority list of antibiotic-resistant bacteria, created by WHO, to research and develop new and effective drugs (Tacconelli et al., 2018). Prudent use of antimicrobials in food-producing animals, particularly reducing those critically important for human treatment, is important for food safety and public health. Further surveillance and monitoring studies to control the prevalence and antimicrobial resistance in Campylobacter spp. must be performed. The application of innovative approaches, such as genomics and proteomics, will offer new insights into the molecular mechanisms responsible of antimicrobial resistance in Campylobacter spp.
Author contributions
Design of the work was developed and overseen by AG-F. Experimental assays were performed by AG-F, AD, SA, and YI-T. AC and IL were responsible for re-drafting the work and revising it critically for important intellectual content. All authors approved the final submitted draft and had opportunity for editing the document.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by the Italian Ministry of Health (Project: Surveillance of Diseases Transmitted by Food and Water [Enter-Net]: adaptation of the Italian system to the European normative framework and management and progressive transfer of surveillance).
References
- Butzler J. P. (2004). Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10, 868–876. 10.1111/j.1469-0691.2004.00983.x [DOI] [PubMed] [Google Scholar]
- Cagliero C., Maurel M. C., Cloeckaert A., Payot S. (2007). Regulation of the expression of the CmeABC efflux pump in Campylobacter jejuni: Identification of a point mutation abolishing the binding of the CmeR repressor in an in vitro-selected multidrug-resistant mutant. FEMS Microbiol. Lett. 267, 89–94. 10.1111/j.1574-6968.2006.00558.x [DOI] [PubMed] [Google Scholar]
- Cagliero C., Mouline C., Cloeckaert A., Payot S. (2006). Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 50, 3893–3896. 10.1128/AAC.00616-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha W., Mosci R., Wengert S. L., Singh P., Newton D. W., Salimnia H., et al. (2016). Antimicrobial susceptibility profiles of human Campylobacter jejuni isolates and association with phylogenetic lineages. Front. Microbiol. 7:589. 10.3389/fmicb.2016.00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing. 28th Edn. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Collignon P. C., Conly J. M., Andremont A., McEwen S. A., Aidara-Kane A., Griffin P. M., et al. (2016). World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin. Infect. Dis. 63, 1087–1093. 10.1093/cid/ciw475 [DOI] [PubMed] [Google Scholar]
- Corcoran D., Quinn T., Cotter L., Fanning S. (2006). An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int. J. Antimicrob. Agents 27, 40–45. 10.1016/j.ijantimicag.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Denis M., Soumet C., Rivoal K., Ermel G., Blivet D., Salvat G., et al. (1999). Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 29, 406–410. 10.1046/j.1472-765X.1999.00658.x [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control European Food Safety Authority, and European Medicines Agency. (2017). ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence ofantimicrobial resistance in bacteria from humans and food-producing animals. Joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA) Report. EFSA J. 15:4872 10.2903/j.efsa.2017.4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority and European Centre for Disease Prevention and Control (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 15:5077 10.2903/j.efsa.2017.5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority and European Centre for DiseasePrevention and Control (2018). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 16:5182 10.2903/j.efsa.2018.5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A. S., Brachman P. S. (eds.). (1998). Bacterial Infections of Humans: Epidemiology and Control. 2nd Edn New York, NY: Plenum Publishing, Springer US. [Google Scholar]
- Giacomelli M., Salata C., Martini M., Montesissa C., Piccirillo A. (2014). Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli from poultry in Italy. Microb. Drug Resist. 20, 181–188. 10.1089/mdr.2013.0110 [DOI] [PubMed] [Google Scholar]
- Gibbons C. L., Mangen M. J. J., Plass D., Havelaar A. H., Brooke R. J., Kramarz P., et al. (2014). Measuring underreporting and under-ascertainment in infectious disease datasets: a comparison of methods. BMC Public Health 14:147. 10.1186/1471-2458-14-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibreel A., Kos V. N., Keelan M., Trieber C. A., Levesque S., Michaud S., et al. (2005). Molecular mechanism and stability of the resistance phenotype. Society 49, 2753–2759. 10.1128/AAC.49.7.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnage-Pulley T., Zhang Q. (2015). Genetic basis and functional consequences of differential expression of the CmeABC efflux pump in Campylobacter jejuni isolates. PLoS ONE 10:131534. 10.1371/journal.pone.0131534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagsma J. A., Geenen P. L., Ethelberg S., Fetsch A., Hansdotter F., Jansen A., et al. (2013). Community incidence of pathogen-specific gastroenteritis: reconstructing the surveillance pyramid for seven pathogens in seven European Union member states. Epidemiol. Infect. 141, 1625–1639. 10.1017/S0950268812002166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Yuan Z., Shen Z., Han J., Sahin O., Liu P., et al. (2013). Mutational and transcriptomic changes involved in the development of macrolide resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 57, 1369–1378. 10.1128/AAC.01927-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelaar A. H., Van Pelt W., Ang C. W., Wagenaar J. A., Van Putten J. P. M., Gross U., et al. (2009). Immunity to campylobacter: its role in risk assessment and epidemiology. Crit. Rev. Microbiol. 35, 1–22. 10.1080/10408410802636017 [DOI] [PubMed] [Google Scholar]
- Iovine N. M. (2013). Resistance mechanisms in Campylobacter jejuni. Virulence 4, 230–240. 10.4161/viru.23753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush N. O., Castaño-Rodríguez N., Mitchell H. M., Man S. M. (2015). Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 28, 687–720. 10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Hales S., French N., Baker M. G. (2012). Seasonality in human zoonotic enteric diseases: a systematic review. PLoS ONE 7:e31883. 10.1371/journal.pone.0031883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D., Owen R. J., Stanley J. (1996). Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147, 707–718. 10.1016/S0923-2508(97)85118-2 [DOI] [PubMed] [Google Scholar]
- Mamelli L., Amoros J. P., Pagès J. M., Bolla J. M. (2003). A phenylalanine-arginine β-naphthylamide sensitive multidrug efflux pump involved in intrinsic and acquired resistance of Campylobacter to macrolides. Int. J. Antimicrob. Agents 22, 237–241. 10.1016/S0924-8579(03)00199-7 [DOI] [PubMed] [Google Scholar]
- Manfreda G., Parisi A., De Cesare A., Mion D., Piva S., Zanoni R. G. (2016). Typing of Campylobacter jejuni isolated from turkey by genotypic methods, antimicrobial susceptibility, and virulence gene patterns: a retrospective study. Foodborne Pathog. Dis. 13, 93–100. 10.1089/fpd.2015.2048 [DOI] [PubMed] [Google Scholar]
- Payot S., Avrain L., Magras C., Praud K., Cloeckaert A., Chaslus-Dancla E. (2004). Relative contribution of target gene mutation and efflux to fluoroquinolone and erythromycin resistance, in French poultry and pig isolates of Campylobacter coli. Int. J. Antimicrob. Agents 23, 468–472. 10.1016/j.ijantimicag.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Pérez-Boto D., López-Portolés J. A., Simón C., Valdezate S., Echeita M. A. (2010). Study of the molecular mechanisms involved in high-level macrolide resistance of Spanish Campylobacter jejuni and Campylobacter coli strains. J. Antimicrob. Chemother. 65, 2083–2088. 10.1093/jac/dkq268 [DOI] [PubMed] [Google Scholar]
- Pratt A., Korolik V. (2005). Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J. Antimicrob. Chemother. 55, 452–460. 10.1093/jac/dki040 [DOI] [PubMed] [Google Scholar]
- Schielke A., Rosner B. M., Stark K. (2014). Epidemiology of campylobacteriosis in Germany - insights from 10 years of surveillance. BMC Infect. Dis. 14:30. 10.1186/1471-2334-14-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L., Fratamico P. M. (2010). Fluoroquinolone Resistance in Campylobacter. Available online at: http://jfoodprotection.org/doi/pdf/10.4315/0362-028X-73.6.1141?code=fopr-site (Accessed May 3, 2018). [DOI] [PubMed]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Tang Y., Sahin O., Pavlovic N., LeJeune J., Carlson J., Wu Z., et al. (2017). Rising fluoroquinolone resistance in Campylobacter isolated from feedlot cattle in the United States. Sci. Rep. 7:494. 10.1038/s41598-017-00584-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher S., Ménard A., Bernard E., Me A., Me F. (2003). PCR-restriction fragment length polymorphism analysis for detection of point mutations associated with macrolide resistance in Campylobacter spp. Antimicrob. Agents Chemother. 47, 1125–1128. 10.1128/AAC.47.3.1125-1128.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter F., Ott J. J., Claus H., Krause G. (2018). Sex- and age patterns in incidence of infectious diseases in Germany: analyses of surveillance records over a 13-year period (2001–2013). Epidemiol. Infect. 146, 372–378. 10.1017/S0950268817002771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Huang W. M., Taylor D. E. (1993). Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37, 457–463. 10.1128/AAC.37.3.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek K., Osek J. (2013). Antimicrobial resistance mechanisms among Campylobacter. Biomed. Res. Int. 2013:340605. 10.1155/2013/340605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Cheng Y., Luo Q., Lu Q., Dong J., Zhang R., et al. (2017). Correlation between gyrA and CmeR box polymorphism and fluoroquinolone resistance in Campylobacter jejuni isolates in China. Antimicrob. Agents Chemother. 61:e00422-17. 10.1128/AAC.00422-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhang M., Yang W., Fang Y., Wang G., Hou F. (2016). A seventeen-year observation of the antimicrobial susceptibility of clinical Campylobacter jejuni and the molecular mechanisms of erythromycin-resistant isolates in Beijing, China. Int. J. Infect. Dis. 42, 28–33. 10.1016/j.ijid.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Zirnstein G., Helsel L., Li Y., Swaminathan B., Besser J. (2000). Characterization of gyrA mutations associated with fluoroquinolone resistance in Campylobacter coli by DNA sequence analysis and MAMA PCR. FEMS Microbiol. Lett. 190, 1–7. 10.1111/j.1574-6968.2000.tb09253.x [DOI] [PubMed] [Google Scholar]
- Zirnstein G., Li Y., Swaminathan B., Angulo F. (1999). Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J. Clin. Microbiol. 37, 3276–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]