Abstract
This study investigated syntrophic interactions between chlorinated benzene respiring Dehalococcoides mccartyi strain CBDB1 and fermenting partners (Desulfovibrio vulgaris, Syntrophobacter fumaroxidans, and Geobacter lovleyi) during hexachlorobenzene respiration. Dechlorination rates in syntrophic co-cultures were enhanced 2-3 fold compared to H2 fed CBDB1 pure cultures (0.23 ± 0.04 μmol Cl− day−1). Syntrophic partners were also able to supply cobalamins to CBDB1, albeit with 3–10 fold lower resultant dechlorination activity compared to cultures receiving exogenous cyanocobalamin. Strain CBDB1 pure cultures accumulated ~1 μmol of carbon monoxide per 87.5 μmol Cl− released during hexachlorobenzene respiration resulting in decreases in dechlorination activity. The syntrophic partners investigated were shown to consume carbon monoxide generated by CBDB1, thus relieving carbon monoxide autotoxicity. Accumulation of lesser chlorinated chlorobenzene congeners (1,3- and 1,4-dichlorobenzene and 1,3,5-trichlorobenzene) also inhibited dechlorination activity and their removal from the headspace through adsorption to granular activated carbon was shown to restore activity. Proteomic analysis revealed co-culturing strain CBDB1 with Geobacter lovleyi upregulated CBDB1 genes associated with reductive dehalogenases, hydrogenases, formate dehydrogenase, and ribosomal proteins. These data provide insight into CBDB1 ecology and inform strategies for application of CBDB1 in ex situ hexachlorobenzene destruction technologies.
Keywords: syntrophy, hexachlorobenzene, organohalide respiration, Dehalococcoides mccartyi strain CBDB1, carbon monoxide
Introduction
Chlorinated benzenes including hexachlorobenzene (HCB) are toxic and persistent compounds that have been studied extensively in the context of microbiological degradation (Field and Sierra-Alvarez, 2008). Whilst lesser chlorinated benzenes are susceptible to aerobic biodegradation in conventional activated sludge treatment processes (van Agteren et al., 1998) microbes can only degrade hexachlorobenzene, pentachlorobenzene, 1,2,3,4-tetrachlorobenzene, and 1,3,5-trichlorobenzene through anaerobic or reductive reactions (Adrian and Görisch, 2002). Such reductive reactions have been linked to bacterial growth (Adrian and Görisch, 2002) and therefore have potential as a biotechnology for commercial chlorinated benzene disposal. The world's largest stockpile of HCB (8,000 tons) is maintained in Sydney, Australia.
To date, only two genera, Dehalococoides and Dehalobacter are known to contain species that use HCB as an electron acceptor in a respiratory process. Dehalococcoides mccartyi strains are obligate hydrogenotrophs and are restricted to organohalides as electron acceptors. D. mccartyi strain CBDB1 grows in mineral medium with H2 as the electron donor, acetate as an organic carbon source, and chlorobenzenes as electron acceptors, in conjunction with cyanocobalamin as an essential cofactor (Adrian et al., 2000).
Syntrophic interactions between fermentative bacteria and organohalide respiring bacteria (ORB) are of interest because they underpin dechlorination reactions in organochlorine contaminated environments where H2 is generated via fermentation of organic matter. The term syntrophy has been used to describe microbial cross-feeding as a cooperation where both partners are involved in metabolic activity and cannot be replaced by adding a co-substrate or any type of nutrient (Schink, 1997). The benefits that syntrophic growth in co-culture can offer over a pure culture include the generation of a metabolic resource by one microbe serving as a resource for another and enhancing the growth/metabolism of one microbe by preventing accumulation of inhibitory compounds consumed by another (McInerney et al., 2008; Morris et al., 2013; Worm et al., 2014). Specifically, in the context of this study, the principal mode of syntrophy involves H2 produced by fermentation of reduced organic substrates serving as an electron donor for hydrogenotrophic ORB that in turn maintain low H2 partial pressures enabling fermentation to proceed (Becker et al., 2005).
Despite the importance of syntrophy in ecology and engineering settings, data on syntrophic interactions involving ORB remain limited. Breakthrough studies in this area focused on syntrophic growth of a dechlorinating organism (DCB-1), a benzoate degrader (BZ-1), and a lithotrophic methanogen (Methanospirillum strain PM-1) in dechlorination of 3-chlorobenzoate (Dolfing and Tiedje, 1986) and methanogenic incubation with Syntrophus species degrading 3-chlorobenzoate and 2-chlorophenol (Becker et al., 2005). Research more closely related to the current study focused on syntrophic growth of chlorinated ethene respiring D. mccartyi strain 195 with fermenters Desulfovibrio desulfuricans and/or Acetobacterium woodii (He et al., 2007), interspecies corrinoid transfer between Geobacter lovleyi and D. mccartyi strains BAV1 and FL2 (Yan et al., 2012, 2013) and interspecies cobamide transfer from the methanogen Methanobacterium congolense to D. mccartyi (Men et al., 2012).
Carbon monoxide (CO) was recently discovered to inhibit chlorinated ethene dechlorination of D. mccartyi strain 195 but the presence of a syntrophic partner was shown to mitigate this toxicity and enhance the growth as well as dechlorination activity (Zhuang et al., 2014; Mao et al., 2015). Additionally, organochlorine dechlorination products can inhibit dechlorination activity (Wei et al., 2016). To date, the inhibitory impacts of lesser chlorinated ethenes alone have been investigated for such inhibition (Yu and Semprini, 2004; Yu et al., 2005; Popat and Deshusses, 2011; Wang et al., 2014; Jiang et al., 2015).
In this study syntrophic interactions of HCB respiring D. mccartyi strain CBDB1 and fermentative partners during reductive dechlorination of HCB were investigated, with a view to utilizing syntrophic co-cultures or enrichment cultures in ex situ remediation technologies for HCB destruction where full-scale deployment of gaseous hydrogen is considered a safety hazard. Three syntrophic partners were examined, namely Desulfovibrio vulgaris, Geobacter lovleyi, and Syntrophobacter fumaroxidans provided with lactate, acetate and propionate respectively as carbon and energy sources. D. mccartyi cannot derive energy from these organic substrates and none of the syntrophic partners can utilize HCB creating syntrophic dependencies.
This is the first study to reveal syntrophic interactions in HCB respiration between strain CBDB1 and fermenters. CO auto-toxicity to strain CBDB1 is described along with the impact of CO consumption by syntrophic partners. Daughter product inhibition is also described along with a demonstration of the impact of removing lesser chlorinated chlorobenzenes from culture headspace. The data generated broadens our understanding of syntrophic metabolism in organohalide respiration by D. mccartyi and provides information relevant to bioreactor applications for HCB destruction.
Materials and methods
Bacterial cultures and growth conditions
D. mccartyi strain CBDB1 was grown in a basal medium with 5 mM acetate as described previously (Adrian et al., 1998). Briefly, mineral salts medium was amended with a vitamin solution, SL-9 minerals and Trace B mineral solution (Adrian et al., 1998). Final concentrations of 100 mM NaHCO3 solution was used to buffer the media to pH 6.8–7.0. Aliquots (80 mL) were dispensed into 160 mL serum flasks and flushed with N2 gas, sealed with Teflon septa and sterilized at 121°C for 20 min. After cooling Ti (III) citrate solution was amended to 1 mM and 0.2 bar of N2:CO2 (4:1, vol/vol) and 0.3 bar of H2 were added to the headspace. Hexachlorobenzene (20 mg, Sigma-Aldrich) was added as electron acceptor and H2 as electron donor with nominal concentration of 7.5 mM as described previously (Jayachandran et al., 2003).
Desulfovibro vulgaris (DSM 2119) and Syntrophobacter fumaroxidans (DSM 10017) were obtained from the German Collection of Microorganisms (DSMZ) and grown according to DSMZ recommendations. Briefly, D. vulgaris was grown in the medium described above with 10 mM lactate as carbon and energy source and 10 mM sulfate as electron acceptor. S. fumaroxidans was grown in MPOB medium with 20 mM sodium fumarate as described previously (Harmsen et al., 1998). Geobacter lovleyi was isolated by dilution to extinction from a chlorinated ethene contaminated aquifer in Sydney, Australia using acetate and Fe (III) citrate as electron donor and acceptor respectively. Purity was confirmed through microscopy and molecular fingerprinting and the species was identified by sequencing the near full length 16S rRNA gene (98% similarity to G. lovleyi strain SZ).
Co-cultures containing D. mccartyi strain CBDB1 with D. vulgaris (DSV/CBDB1), with S. fumaroxidans (SFO/CBDB1), or with G. lovleyi (GBL/CBDB1) were grown in the media described above for strain CBDB1 with the substitutions of 10 mM lactate, 30 mM propionate or 30 mM acetate, respectively as electron donor with no exogenous H2 added. Cyanocobalamin (1 μM VB12) was added as indicated. All cultures were inoculated to an total initial density of ~5 × 106 cells mL−1 and incubated in the dark at 30°C without agitation.
Application of granular activated carbon (GAC) to mitigate HCB daughter product toxicity
GAC (0.5 g, G60 powder, 100 mesh, Sigma-Aldrich) was mounted in glass Pasteur pipettes sealed at one end and placed into CBDB1 co-cultures such that the sealed end was submerged and the open end exposed to the headspace enabling exposure of GAC to the headspace but avoiding direct contact between GAC and the culture medium (Figure S3). Thus volatile organic substances (lesser chlorinated benzenes) could be adsorbed from the headspace. To quantify trichlorobenzene (TCB) and dichlorobenzene (DCB) removal from the headspace, GAC was extracted five times using 3 mL of dichloromethane (DCM). DCM was reduced to a final volume of 1 mL via evaporation prior to analysis by gas chromatography with flame ionization detection (GC-FID). Extraction recovery was determined to be 90-95% (data not shown).
Quantification of chlorinated benzenes
Chlorinated benzenes were quantified by headspace analysis gas chromatography using a Shimadzu GC-2010 Plus gas chromatograph with flame ionization detection (GC-FID) fitted with a J&W DB-5 30 m × 0.32 mm (inner diameter) × 0.25 μm column and a Shimadzu headspace auto-sampler (Koutsogiannouli et al., 2009). Culture (1 mL) was transferred into a 10 mL headspace sample vial. Prior to injection, the samples were heated to 80°C and shaken for 2 min. The injector and detector temperature were set at 250°C. The oven temperature was held at 100°C for 1 min, followed by a 25°C min−1 increase, and then held at 250°C for 0.5 min. Aqueous chlorinated benzene standards were prepared with the same gas to liquid space ratio as the cultures flasks accounting for phase partitioning according to Henry's Law. The amount of chloride released was calculated by multiplying the concentration of daughter products (mmole l−1) by the difference in Cl− number between the parent and the daughter compounds.
Carbon monoxide quantification
Carbon monoxide (CO) was quantified with a Shimadzu GC-2010 Plus GC, equipped with a pulsed discharge detector (GC-PDD) and fitted with a J&W Molecular sieve (30 m × 0.32 mm (inner diameter) × 0.25 μm column (Wurm et al., 2003). Helium was used as the carrier gas with a split ratio of 1:10. The oven was held at 30°C for 1.5 min, ramped at 20°C min−1 to 50°C, then held for 7.5 min. The samples were analyzed with the PDD at 140°C. Aliquots (100 μL) of sample headspace were manually injected with a gas tight syringe. CO standards (0.5–12 μmol) were prepared in 80 mL of CBDB1 media in 160 mL flasks.
Quantitative polymerase chain reaction (qPCR)
Liquid samples (2 ml) were harvested by centrifugation (8000 × g, 20 min at 4°C). Genomic DNA was extracted from cell pellets as described previously (Urakawa et al., 2010). qPCR was used to determine Dehalococcoides spp. and total Bacterial 16S rRNA gene copies in the cultures using Dehalococcoides specific primers Dco728F (5′-AAGGCGGTTTTCTAGGTTGTCAC- 3′) and Dco994R (5′-CTTCATGCATGTCAAAT-3′) (Smits et al., 2004), and universal bacterial primers Eub1048F (5′-GTGSTGCAYGGYTGTCGTCA-3′) and Eub1194R (5′-ACGTCRTCCMCACCTTCCTC-3′) (Horz et al., 2005). Reaction mixtures (10 μL final volume) contained 5 μL SsoFast Eva Green Supermix (BioRad), 100 nM of each forward and reverse primer, 2 μL template, 0.1 mg bovine serum albumin (Thermo Fisher Scientific) and 2.7 μL nuclease free water (Thermo Fisher Scientific). For total bacteria quantification, cycling conditions on a CF96 Real Time System (BioRad) were as follows: 3 min at 98°C, 39 cycles of 0.2 min at 95°C and 0.5 min at 62°C, followed by melt curve analysis from 60 to 99°C. For Dehalococcoides, cycling conditions were as follows: 3 min at 98°C, 44 cycles of 0.3 min at 94°C and 0.45 min at 58°C, followed by melt curve analysis from 55 to 95°C. Quantification of total Bacteria and Dehalococcoides 16S rRNA gene copies was performed by analyzing serial dilution of known quantities of plasmids containing partial Dehalococcoides spp. genes.
Microbiological cobalamin assay
To quantify extracellular cobamides in pure cultures and co-cultures, a microbiological assay using the cobamide-auxotroph Lactobacillus delbrueckii (ATCC7830) was performed as described previously (Yan et al., 2012). Cells from 1.5 mL of culture were removed by centrifugation at 14,000 xg for 1 min, and the supernatants were passed through 0.22 μm membrane filters. Cell-free supernatant (150 μl) was diluted from 2- to 20-fold with water and distributed into 96 well plates filled with 150 μl of double-strength vitamin B12 assay medium (Difco). To prepare vitamin B12-free inocula, cell pellets were washed with sterile, deionized water three times. The cells were suspended in double-strength vitamin B12 assay medium diluted with an equal amount of sterile, deionized water and then incubated at 37°C for 3 h to deplete carryover VB12 from the inoculum broth. The 96 well plate was sealed with an adhesive optical cover and incubated at 37°C in the dark. After 24 h of incubation, the optical density in each well was recorded at 630 nm using a 96 well plate reader (BioTek, Winooski, VT). A standard curve generated by adding known concentrations of vitamin B12 was included on each plate. The assay had a linear range from 5 to 50 ng/L of vitamin B12 with a detection limit of 2 ng/L. Cobamides measured using this microbiological approach were reported as vitamin B12 equivalents.
Estimation of the limits of interspecies distance for H2 transfer
The maximum interspecies distances between strain CBDB1 and syntrophic partners that would enable the observed dechlorination rates for substrates fermentation were estimated using Fick's diffusion law according to the procedure described by Mao et al. (2015):
JH2 is the flux of H2 between syntrophic cells and CBDB1, Asyn is the surface area of a syntrophic cell, DH2 is the H2 diffusion constant in water, at 35°C (6.31 × 105 cm2 s−1, Haynes, 2013). CH2−syn is the H2 concentration immediately outside syntroph cells (representing the maximum H2 concentration enabling exergonic fermentation). CH2−CBDB1 is the H2 concentration immediately outside CBDB1 cells (representing the theoretical minimum H2 concentration that strain CBDB1 can use for energy generation from HCB dechlorination). dsyn−CBDB1 is the distance between syntroph and CBDB1 cells that enables syntrophic oxidation.
Proteomics
D. mccartyi CBDB1 was grown with 5 mM acetate, 7.5 mM H2 and excess HCB as described above. Geobacter lovleyi was grown with 10 mM acetate and 20 mM Fe (III) citrate. D. mccartyi CBDB1 and G. lovleyi co-culture (CBDB1/GL) was grown in the same medium as strain CBDB1 pure culture, with 30 mM acetate and no exogenous H2. All cultures were grown in triplicate in 100 mL media. Cells were collected from pooled replicates (x3) during exponential growth when cell density reached ~108 cell/mL (~90 days of incubation). Cells were harvested using 0.2 μm filters (Millipore, Germany) as described previously (Schiffmann et al., 2016). Cells were washed from filters with 50 mM ammonium hydrogen carbonate. Three analytical replicates were analyzed for each sample.
Samples were prepared for in-solution digestion as described previously (Schiffmann et al., 2014). CBDB1 cells were lysed by three cycles of freeze (liquid nitrogen) and thaw (1 min at 40 °C). The protein lysates obtained were reduced with 50 mM dithiothreitol (Invitrogen, USA) for 1 h at 30°C and alkylated using 130 mM iodoacetamide (Sigma-Aldrich, Australia) for 1 h at 30°C. Proteolysis was performed overnight using trypsin at 37°C. The tryptic digestion was stopped with formic acid to a final concentration of 1% followed by desalting using ZipTip-μC18 tips and re-suspended in 1% formic acid, 2% acetonitrile (ACN), prior to LC–MS/MS analysis.
Digested peptides were separated by nano-LC using an Ultimate nano RSLC UPLC and auto-sampler system (Dionex, Amsterdam, Netherlands) as described previously (Jugder et al., 2016). Samples (2.5 mL) were concentrated and desalted onto a micro C18 pre-column (300 mm × 5 mm, Dionex) with 2% ACN in water with 0.1% triflouroacetic acid (TFA) at 15 ml/min. After a 4 min wash the pre-column was switched (Valco 10 port UPLC valve, Valco, Houston, Tx) into line with a fritless nano column (75 m × ~15 cm) containing C18AQ media (1.9 μ, 120 Å Dr Maisch, Ammerbuch-Entringen Germany). Peptides were eluted using a linear gradient of 2%−36% ACN in water containing 0.1% (v/v) formic acid at 200 nl/min over 30 min. High voltage (2000 V) was applied to low volume Titanium union (Valco) with the column oven heated to 45°C (Sonation, Biberach, Germany) and the tip positioned ~0.5 cm from the heated capillary (T = 300°C) of a QExactive Plus (Thermo Electron, Bremen, Germany) mass spectrometer. Positive ions were generated by electrospray and the QExactive operated in data dependent acquisition mode (Yang et al., 2010). A survey scan m/z 350-1750 was acquired (resolution = 70,000 at m/z 200, with an accumulation target value of 1,000,000 ions) and lockmass enabled (m/z 445.12003). Up to the 10 most abundant ions (> 80,000 counts, underfill ratio 10%) with charge states > +2 and < +7 were sequentially isolated (width m/z 2.5) and fragmented by HCD (NCE = 30) with an AGC target of 100,000 ions (resolution = 17,500 at m/z 200). M/z ratios selected for MS/MS were dynamically excluded for 30 s. Peak lists generated were submitted to the database search program MASCOT (version 2.5.1, Matrix Science). All MS/MS spectra were searched against a custom database consisting of all proteins in D. mccartyi strain CBDB1 and G. lovleyi genomes from Uniprot. Search parameters were: Precursor tolerance 4 ppm and product ion tolerances ±0.05 Da; Met(O) carboxyamidomethyl-Cys specified as variable modification, enzyme specificity was trypsin, 1 missed cleavage was possible and the non-redundant protein database from Uniprot (Jan 2015) searched. The results were exported to both XML and ROV file format prior to loading into Scaffold for further analysis.
Label free quantification was carried out in Scaffold Q1 software (version Scaffold 4.7.2, Proteome Software) according to the precursor intensity-based method, where differences in protein abundance (defined here as proteins differentially expressed relative to total cell protein) between pure culture and co-culture samples were found based on the average precursor intensity acquired. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
Statistical tests
The data was assumed to be normally distributed. One-way analysis of variance (ANOVA) tests and T-test were calculated to determine the significance of results using an alpha level of 0.05. Multiple comparisons were calculated with Dunnet's test, where applicable. All statistical tests were performed using GraphPad Prism 6 software.
Results
Syntrophic growth of D. mccartyi strain CBDB1 and syntrophic partners
Three bacterial strains were selected as hydrogen producing syntrophic partners for D. mccartyi strain CBDB1 based on the diversity of substrates used. The partner organisms included Deltaproteobacteria Desulfovibrio vulgaris (DSV) and Geobacter lovleyi (GBL) and Firmicute Syntrophomonas fumaroxidans (SFO). These organisms were supplied with lactate, acetate and propionate respectively as electron donors. Syntrophic growth of D. mccartyi strain CBDB1 and H2 producing partners was established enabling reductive dechlorination of hexachlorobenzene (HCB) to 1,3,5-trichlorobenzene (1,3,5-TCB), 1,3-dichlorobenzene (1,3-DCB), and 1,4-dichlorobenzene (1,4-DCB) without exogenous H2 provision.
Dechlorination rates were comparable with the different partners with the highest observed in the D. vulgaris and CBDB1 (DSV/CBDB1) co-culture (0.59 ± 0.05 μmol Cl− day−1) with the rates in the other co-cultures at 0.39 ± 0.15 and 0.45 ± 0.09 μmol Cl− day−1 in the S. fumaroxidans and CBDB1 (SFO/CBDB1) and G. lovleyi and CBDB1 (GBL/CBDB1) co-cultures, respectively (Table 1). The dechlorination rates in all co-cultures were significantly higher (2-3 fold, t-test, P < 0.05) compared to pure H2/acetate fed CBDB1 culture (0.23 ± 0.04 μmol Cl− day−1).
Table 1.
Maximum dechlorination rate and cell yield of D. mccartyi in syntrophic growth with syntrophic partners.
| Co-culture |
Dechlorination rate (μmol Cl−day−1) |
CBDB1 cell yield (cells μmol−1 Cl−day−1) |
P-value (T-test) |
|---|---|---|---|
| DSV/CBDB1 | 0.59 ± 0.05*** | 3.7 (±0.3) × 108 | 0.0005 |
| SFO/CBDB1 | 0.39 ± 0.12* | 2.7 (±0.1) × 108 | 0.048 |
| GBL/CBDB1 | 0.45 ± 0.09** | 3.8 (±0.8) × 108 | 0.003 |
| CBDB1 | 0.23 ± 0.04 | 2.3 (±0.3) × 108 |
P-values for rates. Significantly higher than CBDB1 culture alone at
P < 0.05,
P < 0.01,
P < 0.001. Values are given as means ± the standard deviation from triplicate cultures.
Acetate and H2 accumulated to a maximum of 1.8 (±0.06) mM and 2.0 (±0.09) mM respectively in the DSV/CBDB1 co-culture, while in the SFO/CBDB1 co-culture accumulation was slightly lower with 1.1 (±0.02) mM of acetate and 1.2 (±0.03) mM of H2 observed (Figure 1). Thereafter, acetate and H2 concentrations decreased, presumably as a function of consumption by CBDB1. In the acetate fed GBL/CBDB1 co-culture, acetate consumption corresponded with H2 concentration peaking at 0.5 (±0.01) mM followed by consumption and stabilization of H2 concentration and ongoing acetate consumption (Figure 1). After two months incubation, 5.1 (±0.41), 9.4 (±0.98), and 13.7 (±0.94) mM lactate, propionate and acetate had been consumed respectively.
Figure 1.
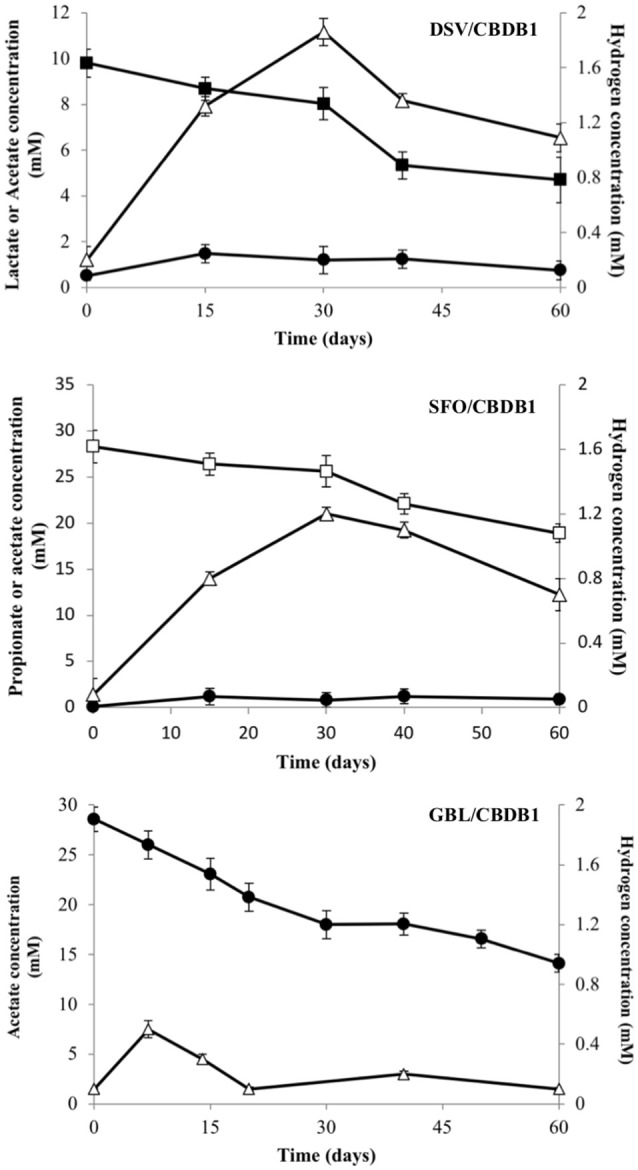
Electron donor production and consumption in HCB reducing syntrophic co- cultmes DSV/CBDB1, SFO/CBDB1 and GBL/CBDB1. H2 (triangles), Acetate (circles), Lactate (closed squares), Propionate (open squares). Error bars represents standard deviation of the mean (n = 3).
Growth of D. mccartyi strain CBDB1 in co-culture was measured by quantitation of 16S rRNA genes using universal bacterial and Dehalococcoides specific primers. Cell numbers increased over 90 days. Initial 16S rRNA gene copy concentrations were 2.0 (±0.15) × 106 and 1.2 (±0.4) × 106 copies/mL for total bacteria and Dehalococcoides respectively. Deltaproteobacterial co-cultures DSV/CBDB1 and GBL/CBDB1 had the largest increase in both total bacterial gene copies and Dehalococcoides gene copies (110–125 fold) up to 2.2 (±1.2) × 108 and 1.5 (±1.8) × 108 copies /mL, respectively. The SFO/CBDB1 co-culture showed the lowest Dehalococcoides cell growth up to 8.4 (±0.3) × 107 copies/mL (70-fold increase) congruent with dechlorination activity data.
Cell to cell distance and thermodynamic considerations in syntrophic co-cultures
The maximum interspecies distance for molecular hydrogen transfer between cells of strain CBDB1 and syntrophic partners can be calculated using Fick's diffusion law as previously described for other D. mccartyi strains (Mao et al., 2015). This distance was calculated for the syntrophic co-cultures of strain CBDB1 and partners with lactate, propionate and acetate as substrates. In the randomly dispersed cells of DSV/CBDB1, GBL/CBDB1 and SFO/CBDB1 co-cultures, the average distance enabling interspecies H2 transfer was calculated to be 178, 10 and 19 μm, respectively (Table 2). The values for cell-cell distances in DSV/CBDB1, GBL/CBDB1 and SFO/CBDB1 co-cultures calculated using 16S qPCR data for bacteria as proxy were 16.8, 19.5, and 24.4 μm, respectively. The measured value for the DSV/CBDB1 co-culture was well below the theoretical maximum whilst the GBL/CBDB1 and SFO/CBDB1 co-cultures had measured values marginally above the theoretical maximum. This potentially explains the superior activity of the lactate fed DSV/CBDB1 co-culture across all quantified metrics (dechlorination activity, growth and acetate and H2 production).
Table 2.
Estimation of the cell-cell distance in syntrophic co-culture at day 60 of incubation.
| D. vulgaris with strain CBDB1 on lactate | G. lovleyi with strain CBDB1 on acetate | S. fumaroxidans with strain CBDB1 on propionate | |
|---|---|---|---|
| Total number of cells/ml (a) | 2.11 × 108 | 1.34 × 108 | 6.89 × 107 |
| Mean cell-cell distance (μm) (b) | 16.8 | 19.5 | 24.4 |
| Maximum interspecies distance for H2 transfer (μm) (c) | 178 | 10.1 | 18.6 |
(a) Total cell number for strain CBDB1 and syntrophic partners in co-cultures using 16S copies as proxy; (b) Calculated using 16S copies as a proxy for cell number; (c) Calculated using Fick's diffusion law for H2 transfer from syntrophic partners to strain CBDB1. Detailed calculations for these values can be found in the Supplementary Materials Table S1.
Effect of syntrophic partners on CO concentrations in CBDB1 co-cultures
Carbon monoxide (CO) has previously been shown to be inhibitory to D. mccartyi strain 195 (Zhuang et al., 2014). Consequently, CO toxicity, production and consumption was examined in strain CBDB1. Figure 2 shows that CO concentrations above 1 μmol flask−1 reduce the dechlorination activity of strain CBDB1 and 6 μmol flask−1 is completely inhibitory. The half maximal inhibitory concentration (IC50) was 2.24 ± 0.09 μmol flask−1. Figure 2 also shows that after 60 days incubation, CO accumulation in strain CBDB1 pure culture reached the threshold whereby CO toxicity manifests (0.9 ± 0.1 μmol flask−1). After 60 days, 17% of the initial HCB amendment (70 μmol) was degraded to release 11.2 ± 0.2 μmol Cl−. Hence, complete dechlorination of 70 μmol HCB could accumulate ~5.6 μmol CO in pure culture. In syntrophic co-culture, regardless of the fermentative partner, the CO concentration remained below those shown to confer toxicity at 0.4 μmol flask−1 in all co-cultures after 60 days (Figure 2).
Figure 2.
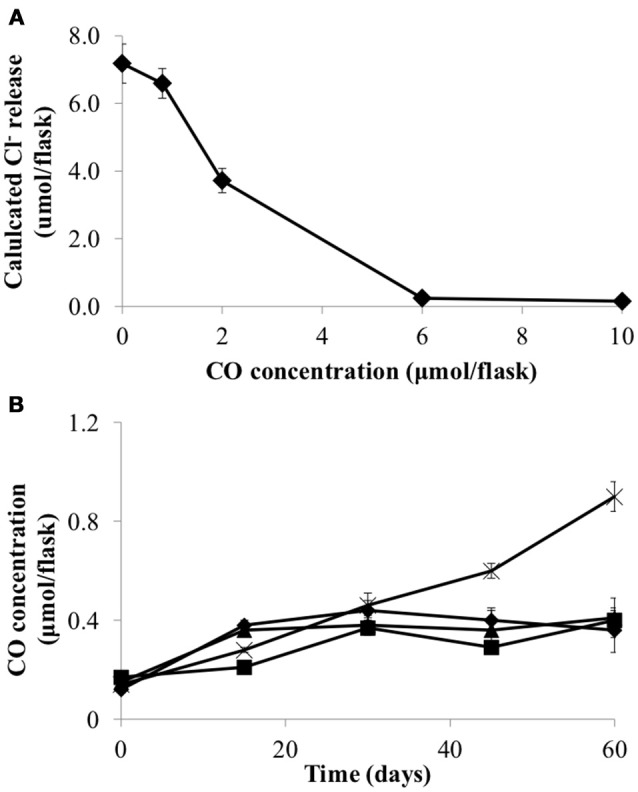
(A) CO inhibits HCB dechlorination activity of D. mccartyi strain CBDB1 shown as Cl- ion release at the end of incubation (60 days) and (B) CO accumulation during HCB dechlorination by strain CBDB1 in pure culture (crosses) and in co-cultures with D. vulgaris (DSV/CBDB1, diamonds), S. fumaroxidans (SFO/CBDB1, squares) and G. lovleyi (GBL/CBDB1, triangles). After 60 days strain CBDB1 produced enough CO to confer autotoxicity. In co-cultures, CO concentration remained below that showing toxicity. Data points are averages of hiplicate cultures. Error bars represent the standard deviation (n = 3).
Growth of CBDB1 with syntrophic partners excluding exogenous cobalamin supply
To examine whether the syntrophic partners could supply cobalamin to CBDB1, microcosms were established without exogenous cyanocobalamin. Data was collected after four transfers (10% v/v inoculation) of the parent culture representing a dilution of cobalamin from 0.5 μg/L to 0.05 ng/L. The total concentration of HCB dechlorination products were on average 28.3 ± 1.8 μM in all three co-cultures, compared to 72.4 ± 3.5 μM in pure CBDB1 culture (supplied with cyanocobalamin) at day 60 (Figure 3). Dechlorination activity in co-cultures was ten-fold less than in co-cultures amended with cyanocobalamin and three-fold less than a pure CBDB1 culture. Dechlorination was not observed in a control where CBDB1 pure cultures were not supplied exogenous cobalamin (data not shown).
Figure 3.
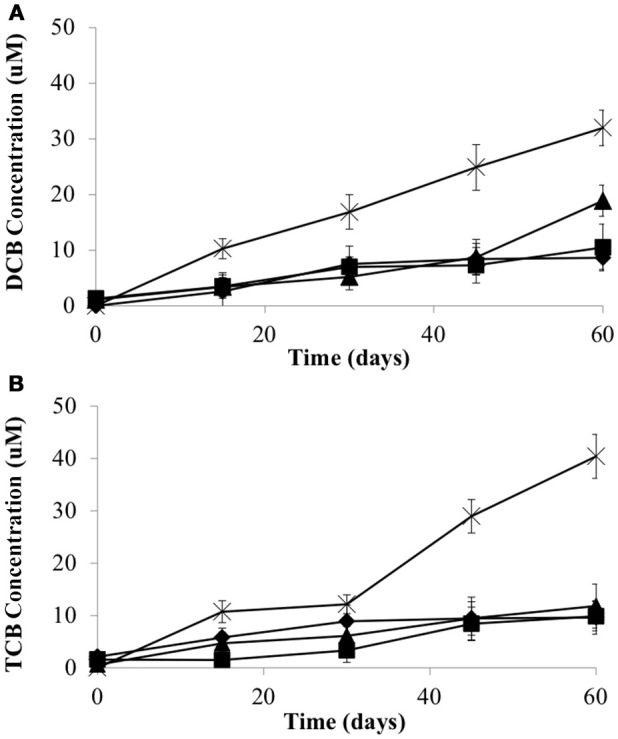
Production of (A) 1,3-Dichlorobenzene and 1,4-Dichlorobenzene (sum total) and (B) 1,3,5-Trichlorobenzene from HCB by Dehalococcoides mccartyi strain CBDB1 in pure culture with exogenous provision of acetate, H2 and Vitamin B12 (crosses), and in co-culture with D. vulgaris (diamonds), S. fumaroxidans (squares) or G. lovleyi (triangles) without exogenous provision of acetate, H2 and Vitamin B12. Data points are averages of triplicate cultures. Error bars represent standard deviation.
Measurement of cobamides in co-culture supernatants was achieved using a biological assay involving the growth of the cobamide auxotroph Lactobacillus delbrueckii. Supernatants were analyzed at the end of the experiment (i.e., day 60). In GBL/CBDB1 cultures 6.0 ± 1.0 ng/L extracellular cobamide was detected. In the DSV/CBDB1 and SFO/CBDB1 co-cultures extracellular cobalamins were below the limits of detection. This suggests that none of the partners could produce sufficient cobalamin to support maximum HCB dechlorination activity by D. mccartyi strain CBDB1 (ie., activity observed with cyanocobalamin amendment).
HCB respiration products inhibit D. mccartyi strain CBDB1 growth and activity
To determine if 1,3- and 1,4-dichlorobenzene (DCB) and 1,3,5-trichlorobenzene (TCB) had an inhibitory impact on HCB respiration by D. mccartyi strain CBDB1, pure cultures were established with increasing concentrations of DCB or TCB. Figure 4 shows that DCB and TCB have an inhibitory effect on HCB dechlorination activity and cell growth of CBDB1, with lower IC50 and LD50 values for TCB compared to DCB (45 μM and 70 μM, respectively).
Figure 4.
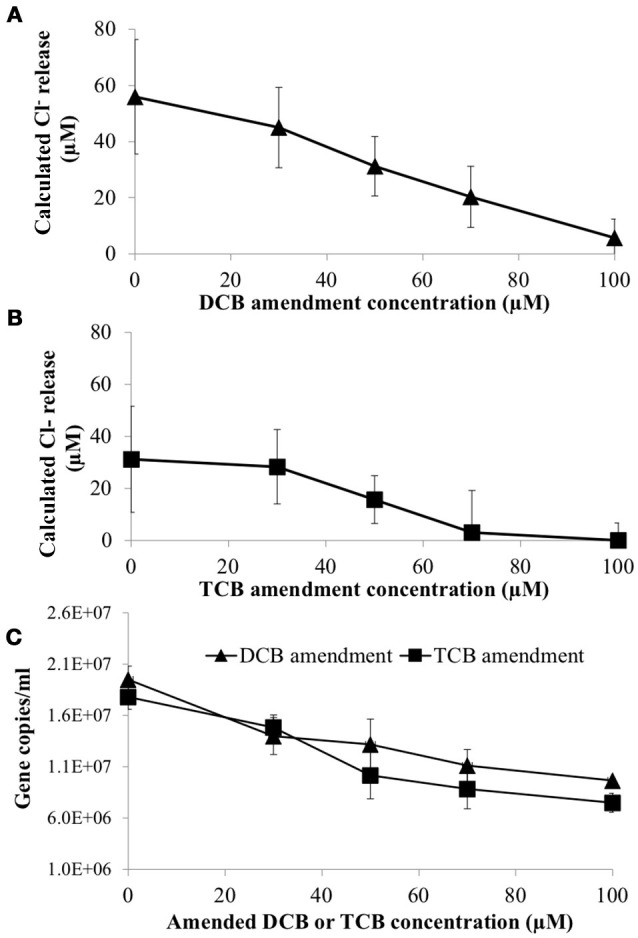
Impact of daughter products from reductive dechlorination of HCB (Dichlorobenzene or Trichlorobenzene) on HCB reduction activity of Dehalococcoides mccartyi CBDB1 after 30 days incubation. (A) Chloride ion release calculated from concentrations of DCB and TCB produced from HCB in the presence of increasing amended DCB concentrations. (B) Chloride ion release calculated from concentrations of DCB and TCB produced from HCB in the presence of increasing TCB concentrations. (C) Impact of DCB and TCB on CBDB1 gene copy concentration after 30 days incubation. Error bars represent SD (n = 3).
DCB and TCB are volatile compounds so Granular Activated Carbon (GAC) was mounted in culture flask headspace (Figure S3) in an attempt to mitigate the inhibitory impacts of daughter product formation from HCB reduction. First order rate constants for DCB and TCB adsorption on GAC were −0.14 (±95% CI) day−1 and −0.16 (±90% CI) day−1, respectively (Figure S1). In comparison, first order rate constants of DCB and TCB production in strain CBDB1 culture were 0.1 (±99% CI) day−1 and 0.07 (±97% CI) day−1, respectively. Hence, trapping DCB and TCB on GAC could mitigate the inhibitory effect of DCB and TCB accumulation CBDB1 in co-cultures.
To test the mitigation strategy, DSV/CBDB1, SFO/CBDB1 and GBL/CBDB1 co-cultures were grown with and without 0.5 g GAC mounted in the culture headspace. At day 70, HCB daughter products were analyzed both in the culture medium and adsorbed to GAC. The latter was analyzed by dichloromethane (DCM) extractions of the GAC. DCB and TCB production were significantly higher (2-fold, t-test, P <0 .05) in co-cultures amended with GAC for all three syntrophic partners (Figure 5). For example, treatments with GAC in DSV/CBDB1 co-cultures had a dechlorination rate of 1.1 ± 0.2 μmol Cl− day−1, compared to a rate of 0.6 ± 0.2 μmol Cl− day−1 in the GAC-free treatments. Total bacterial growth in treatments with and without GAC was determined via 16S rRNA gene quantitative PCR using universal bacterial primers at day 0, 30 and 70 of incubation. After 70 days the gene copies in the presence of GAC were 2.1-2.7 fold higher (t-test, P < 0.05) than in the absence of GAC (Figure 5).
Figure 5.
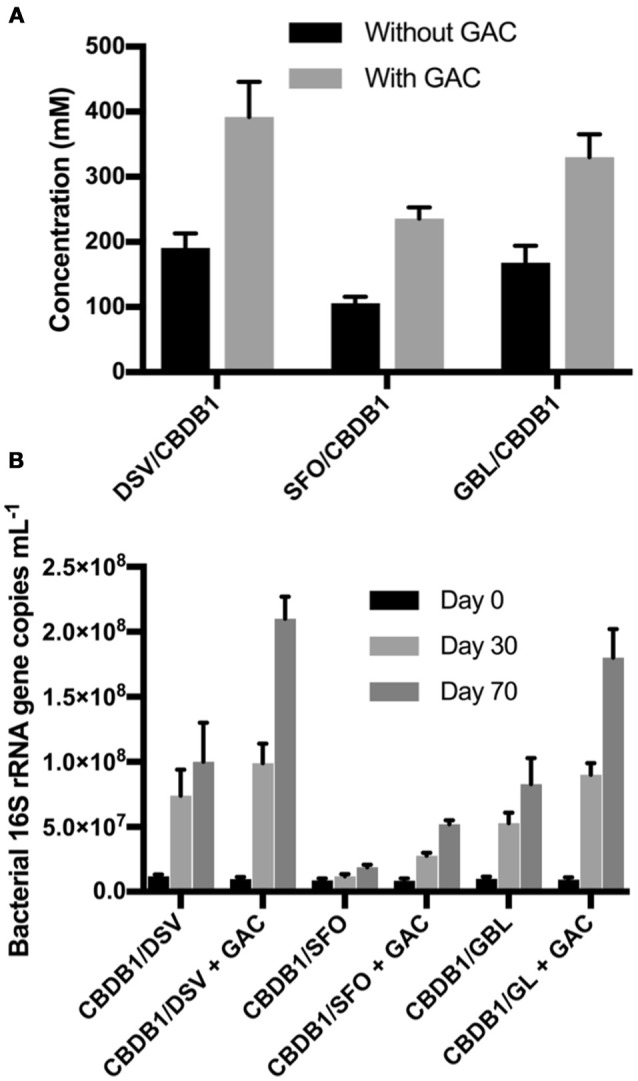
(A) Total concentrations of HCB dechlorination products (1,3- and 1,4-DCB and 1,3,5-TCB) in different co-cultures of D. mccartyi strain CBDB1 with and without GAC after 70 days of incubation. Data points are averages of triplicate cultures. Error bars represent the standard deviation (n = 3). (B) Quantification of 16S rRNA gene copies in syntrophic co-cultures of D. mccartyi strain CBDB1 and partners with and without GAC after 70 days of incubation. All cultures were provided with cyanocobalamin, Ti (III) citrate, and without exogenous H2. Data points are averages of triplicate cultures. Error bars represent the standard deviation.
Proteomic analysis in GL/CBDB1 co-culture
Whole cell proteomic analyses of CBDB1 grown alone and in co-culture with syntrophic partner G. lovleyi was undertaken to identify changes in gene expression in response to co-culture. In pure culture G. lovleyi was grown with Fe (III) citrate as electron acceptor. Equal amounts of total protein (~30 μg) from late exponential phase samples (~3 months incubation) were digested. Each sample was prepared via the filter aided sample preparation (FASP) method and subjected to LFQ by nano LC-MS/MS. A total of 363 D. mccartyi proteins and 404 G. lovleyi proteins were identified in pure and co-cultures (Figure S2). Of the D. mccartyi proteins, 119 were identified in co-culture with 62 of these exclusively observed in co-culture. Of the G. lovleyi proteins, 102 were observed in co-culture with 15 of these exclusive to the co-culture.
Table 3 reports D. mccartyi and G. lovleyi proteins for which expression was upregulated greater than five-fold (see Table S2 for lesser fold upregulation and Table S3 for the whole data set). Three reductive dehalogenases were identified from D. mccartyi including cbdbA84 (accession number CAI82345), cbdbA80 (accession number CAI82340) and cbdbA1453 (accession number CAI83480), which encode 1,2,3- and 1,2,4-TCB reductive dehalogenases. Among them, cbdbA84 and cbdbA80 were more abundant in the co-culture than in the pure CBDB1 culture suggestive of dependence of regulation on the presence of another organism (in this case a syntrophic partner).
Table 3.
Identified proteins with greater than five-fold increase in abundance in co-culture.
| NCBI locus | Predicted functions | D. mccartyi | G. lovleyi |
|---|---|---|---|
| cbdbA84 | Putative reductive dehalogenase | 33 | |
| cbdbA80 | Putative reductive dehalogenase | 14 | |
| cbdbA195 | Formate dehydrogenase, major subunit | 8.2 | |
| cbdbA960 | Translation elongation factor Tu | 5.6 | |
| Glov_0477 | Conserved hypothetical protein | – | 8.6 |
| Glov_1625 | Malate dehydrogenase | – | 6.6 |
| Glov_0475 | Alkyl hydroperoxide reductase | – | 5.5 |
Translation elongation factor Tu (locus cbdbA960), was among the high coverage proteins that were more abundant in the co-culture than in CBDB1 culture alone. Formate dehydrogenase (locus cbdbA195) was also shown to be more abundant during syntrophic growth than in the CBDB1 isolate. Several other conserved protein complexes involved in respiratory electron transfer were found in the CBDB1 isolate, including Rdh complexes, four hydrogenase complexes (Hup, Ech, Hyc, and Hym), NADH dehydrogenase complexes and two oxidoreductase complexes. Three hydrogenases, VhuA, HymB, and HymC, encoded by cbdbA597, cbdbA684, and cbdbA685, respectively, were found to be marginally more abundant in syntrophic growth than in the isolate (Table S2). The co-culture also possessed 16 ribosomal proteins that were detected with high expression compared to the CBDB1 isolate, congruent with the more robust growth of CBDB1 in co-culture. None of the cobalamin biosynthesis proteins were detected in either the co-culture or the CBDB1 isolate. Conversely, two types of cobalamin transporter, ABC-type cobalamin/Fe3+-siderophores transport system ATPase component (locus cbdbA633) and periplasmic component (locus cbdbA636) were detected, with no difference in expression between the co-culture and CBDB1 isolate, both of which were amended with VB12.
Among the 87 G. lovleyi proteins detected in both G. lovleyi culture alone and in co-culture (Figure S2), thiamine pyrophosphate protein (locus Glov_1628), succinate dehydrogenase or fumarate reductase (locus Glov_2213), as well as elongation factor and ribosomal proteins were among the proteins with the highest coverage. Proteins related to acetate oxidation, including malate dehydrogenase (locus Glov_1625), isocitrate dehydrogenase (locus Glov_1624) and citrate synthase (locus Glov_1379), were expressed more abundantly during syntrophic growth than in isolate cultures (Table S2). Two molybdopterinoxidoreductase proteins (loci Glov_3132 and Glov_0661) were detected less abundantly in the co-culture than in the G. lovleyi isolate, with fold change ratios of 0.7 and 0.1 respectively.
There were two proteins detected exclusively in the co-culture: peptidylprolylisomerase (locus Glov_2546) and histidinol dehydrogenase (encoded Glov_0822). Interestingly, there were two types of flagellin detected only during G. lovleyi growth with Fe (III) citrate, which were the flagellin domain protein (locus Glov_3371) and a flagellin basal associated-protein FilL (locus Glov_3294). In addition, three chemotaxis sensory transducers (encoded Glov_ 2776, Glov_ 1733 and Glov_1239) were found with less abundance during syntrophic growth than during pure culture growth on iron. Cobalamin-dependent proteins were identified in both G. lovleyi isolate and co-culture including ribonucleotide reductase, methionine synthase and methylmalonyl-CoA mutase. Among these proteins, only methylmalonyl-CoA mutase (encoded Glov_3260) was more abundant in co-culture than in the isolate, with a fold change of 2.0 (Table S2). Nicotinate-nucleotide/dimethylbenzimidazolephosphoribosyltransferase (CobT-Glov_3082) was the only cobalamin biosynthesis protein detected, which was from G. lovleyi in isolation.
Discussion
In this present study, lactate, propionate and acetate were supplied as organic carbon and energy sources for D. mccartyi strain CBDB1 in co-culture with Desulfovibrio, Syntrophobacter or Geobacter. Oxidation of these substrates by the syntrophic partners of CBDB1 was expected to provide H2 for the reductive dechlorination of HCB. Table 4 provides the ΔG0' for the H2 producing and H2 consuming reactions revealing that H2 production in the absence of the H2 consuming reductive dechlorination reaction is marginally favorable or unfavorable thus representing the energetic dependency of the syntrophic interactions reported in this study. The enhanced dechlorination observed in this study between D. mccartyi strain CBDB1 and partners supports previous observations on the impact of syntrophic partners in organochlorine respiring cultures (He et al., 2007; Men et al., 2012; Yan et al., 2012). The fact that strain CBDB1 shows higher activity in syntrophic partnership with Desulfovibrio, Syntrophobacter, and Geobacter lineages in the presence of exogenous VB12 represents a proof of concept for a bioreactor in which HCB could be reduced to less persistent oxidisable compounds (DCB, TCB) using lactate, propionate or acetate as energy sources which are cost effective and safe compared to H2.
Table 4.
Hydrogen-releasing and hydrogen-consuming reactions occurring in co-cultures.
| HYDROGENOGENIC REACTIONS | |
| C3H6+ 2 H2O → CH3COO −++ H+ + 2H2 | ΔG0′ = − 8.4 kJ/reaction |
| CH3CH2COO−+ 3 H2O → CH3COO−++ 3 H2 | ΔG0′ = + 76.1 kJ/reaction |
| CH3COO−+ 2 H2O → 2 + 4 H2 + H+ | ΔG0′ = + 104.6 kJ/reaction |
| HYDROGEN-CONSUMING REACTIONS | |
| C6Cl6 + H2 → C6HCl5 + H+ + Cl− | ΔG0′ = − 171.4 kJ/reaction |
| C6Cl6 + 4 H2 → C6H4Cl2 + 4 H+ + 4 Cl− | ΔG0′ = − 447 kJ/reaction |
From previous work, in addition to supplying electron donor (H2), the robust growth of D. mccartyi strain 195 in syntrophic co-culture appears to stem from acetate supply by D. vulgaris, along with potential benefit from proton translocation, cobalamin-salvaging and amino acid biosynthesis as evidenced by gene expression analysis (Men et al., 2012). The impact of an incomplete Wood-Ljungdahl pathway i.e. the absence of CO dehydrogenase in the D. mccartyi genome, may also explain the more robust growth in co-culture and therefore further underpins the syntrophic interaction (Zhuang et al., 2014; Mao et al., 2015). D. mccartyi strain 195 was inhibited in pure culture by CO accumulating from acetyl-CoA cleavage due to the lack of CO dehydrogenase in D. mccartyi strain 195 (Zhuang et al., 2014).
In this study the inhibitory effect of CO on both dechlorination activity and cell growth of D. mccartyi strain CBDB1 was described. The adverse effect of CO toxicity on strain CBDB1 was mitigated by the presence of any of the tested syntrophic partners, (i.e. Desulfovibro vulgaris, Syntrophobacter fumaroxidans, or Geobacter lovleyi), which are capable of consuming CO (Davidova et al., 1994; Ragsdale, 2004; Diender et al., 2015). Other work using 13C-labeling and bioinformatic analysis confirmed that Bacteria and Archaea exist in consortia as CO-oxidizing organisms capable of gaining additional energy via coexistence with D. mccartyi while simultaneously enhancing growth of D. mccartyi (Zhuang et al., 2014). The results from the present study support and broaden previous observations of metabolic exchange between D. mccartyi and syntrophic partners in dechlorinating microbial environments (Zhuang et al., 2014; Mao et al., 2015). In a pure culture bioreactor based on CBDB1 activity an alternative approach to removal of CO would need to be devised.
The theoretical maximum interspecies distances for strain CBDB1 and partners were calculated based on the observed substrate oxidation rate. These estimates showed that proximity is more essential for syntrophic acetate and propionate oxidation compared to lactate oxidation. In the present study, cell aggregation was not observed in co-culture. When strain CBDB1 grew with D. vulgaris in co-culture, the maximum interspecies distance (178 μm) allowed H2 transfer between these bacteria without multi-lineage aggregate (flocculate) formation, whilst the other two co-cultures were close to the theoretical limit for H2 transfer over distance suggesting a reduction in distance between cells would lead to higher dichlorination activity. This result is consistent with observations by Mao et al. (2015) that aggregation was not established in co-cultures of strain 195 and Desulfovibrio vulgaris while it did occur in Syntrophomonas wolfei and strain 195 co-culture. Previous studies reported that syntrophic co-cultures form cell aggregates during the growth for optimal H2 transfer (Schink and Thauer, 1988; Stams et al., 2012; Felchner-Zwirello et al., 2013; Mao et al., 2015) or created biofilms in membrane bioreactors (Chung and Rittmann, 2008; Ziv-El et al., 2012). The cell aggregates reduce interspecies H2 transfer distance. In a bioreactor context, these data suggest approaches to stimulate flocculation or suspended biofilm formation would be beneficial.
In co-culture with CBDB1, G. lovleyi was hypothesized to provide cobalamin to support dechlorination of HCB as in previous studies on co-cultures of G. lovleyi and D. mccartyi strain 195 (Yan et al., 2012). However, there was only 0.006 ± 0.01 μg/L of extracellular cobalamin detected in CBDB1/GBL co-culture, while in the other co-cultures cobalamins were under the limit detection of the assay used. The amount of cobalamin detected was much lower than the requirement of cobalamin (50 μg/L) for maximal reductive dechlorination rates in pure CBDB1 culture (Adrian et al., 2000). Hence, syntrophic growth of D. mccartyi strain CBDB1 still requires exogenous cyanocobalamin supply for maximum HCB respiration rates. This suggests cyanocobalamin would be required as an amendment for a bioreactor application with significant implications with respect to cost.
Proteomics plays a key role in exploring adaptive responses of microbes to environments or interactions among microorganisms (Wang et al., 2016). Previous proteomic studies have evaluated the effects of long-term syntrophic growth in oxidation of butyrate (Schmidt et al., 2013; Sieber et al., 2015) or methanogenic consortia in degradation of terephthalate (Wu et al., 2013). Additionally, Men et al. (2012) explored protein expression during syntrophic interactions between D. mccartyi strain 195 and Desulfovibrio vulgaris strain Hildenborough dechlorinating trichloroethene.
In the present study, almost all ribosomal proteins in strain CBDB1 were found to be more abundant in the co-culture than in the isolate. Ribosomal proteins are related to cell growth indicating a higher rate of protein synthesis, resulting in a faster growth rate in the co-culture compared with the pure culture. This was congruent with our growth data. Similar to the observations of Men et al. (2012), the higher abundance of three CBDB1 hydrogenase proteins (VhuA, HymB, and HymC) and lower abundance of proteins related to electron transport such as NADH, oxidoreductase complexes as well as HymS in co-culture compared to in isolation suggested that syntrophic growth involves different H2 transfer systems than those used when H2 is supplied exogenously.
Another CBDB1 protein found with high abundance in co-culture was formate dehydrogenase, encoded by the cbdbA195 gene. This protein is expected to have another function other than formate utilization found in both D. mccartyi strains CBDB1 and 195 (Adrian et al., 2007; Morris et al., 2007). Recently, Kublik et al. (2016) revealed that this formate dehydrogenase-like enzyme highly expressed in D. mccartyi is a putative molybdopterin containing oxidoreductase that co-localizes with the active subunit of the reductive dehalogenase (RdhA). This may explain the high abundance of these two proteins detected in co-cultures where favorable growth conditions lead to more respiration. These results support the only previous proteomic analysis of ORB syntrophic growth with D. mccartyi strain 195 and D. vulgaris with TCE dechlorination (Men et al., 2012).
When comparing proteins detected in G. lovleyi in isolation and GBL/CBDB1 co-culture, there were two proteins detected exclusively in the latter: peptidylprolylisomerase and histidinol dehydrogenase. Histidinol dehydrogenase is responsible for catalyzing the reaction: L-histidinol + NAD+ = L-histidine + NADH + H+, while peptidylprolylisomerase plays the role of PceT, identified to be a trigger factor-like protein that functions as a dedicated chaperone for PceA (Maillard et al., 2011). The reason why these two proteins were only detected in co-culture and not expressed in G. lovleyi pure culture is unclear.
In the present study, no cobalamin biosynthesis proteins from G. lovleyi were detected and no differences in expression of cobalamin transport proteins were detected when comparing the co-culture with G. lovleyi in pure culture. These results might explain why cobalamin production by G. lovleyi was unable to support the GBL/CBDB1 co-culture. The observation that proteins related to acetate oxidation through the citric acid cycle (malate dehydrogenase, isocitratedehyrogenase and citrate synthase) were more abundant in co-culture than in the isolate is consistent with the stimulation of acetate oxidation by syntrophic growth. The lack of flagellin and lower expression of chemotaxis sensory transducer in co-culture supports the idea that syntrophic growth of CBDB1 and G. lovleyi occurs via interspecies H2 transfer.
Cross-inhibition of co-contaminants and self-inhibition by biodegradation products in the environment has been widely reported (Chan et al., 2011; Schiffmacher et al., 2016). DCB and TCB amended in pure CBDB1 cultures were found to be inhibitory. The presence of GAC in CBDB1 co-culture headspace reduced the inhibitory effect of HCB daughter products on cell growth and dechlorination activity. Application of GAC to remove chlorinated compounds has been studied widely from water to soil in contaminated environments (Pavoni et al., 2006; Choi et al., 2009; Kjellerup et al., 2014; Dang et al., 2016). The data presented here suggests this approach could serve as an effective strategy in bioreactor applications with GAC mounted in a reactor headspace serving to continuously remove DCB and TCB to prevent daughter product inhibition.
Conclusions
Despite the ubiquity of syntrophic processes in anoxic environments, little is known about the mechanisms by which syntrophic consortia regulate their metabolism, such as the cross-supply of cobalamins or proteomic insights into syntrophic growth during organohalide respiration. Here we provide the first description of syntrophic growth of D. mccartyi strain CBDB1 and partners (Desulfovibrio vulgaris, Syntrophobacter fumaroxidans and Geobacter lovleyi) for exogenous provision of H2, acetate and cobalamin for the reductive dechlorination of HCB. HCB dechlorination rates were enhanced two- to three-fold compared to pure cultures of D. mccartyi strain CBDB1. In co-cultures, acetate and H2 were supplied from substrate oxidation by syntrophic partners, whilst amendment of exogenous cobalamin was necessary in all cultures to achieve maximal rates. Superior growth and dechlorination by strain CBDB1 in co-culture may be explained by the consumption of CO, preventing the adverse effect of CO toxicity by the presence of any of its syntrophic partners. Additionally, proteomic analysis in co-culture revealed an abundance of ribosomal proteins, reductive dehalogenase, hydrogenase proteins, and formate dehydrogenase, and down-regulation of proteins related to electron transport. The GAC application and results obtained from this study were the first investigation on GAC application in attempt to mitigate the inhibitory effect of HCB daughter products and thus enhance HCB dechlorination activity. This approach is relevant to bioreactor applications for HCB remediation with a potential low cost and easy application.
Author contributions
AC conceived, designed, analysed and executed the experimental work as well as drafted the manuscript. ML, LA, and MM conceived, designed, performed the data analysis, and the interpretation. ML, LA, and MM drafted the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AC is thankful to the Australian Awards Scholarships (Department of Foreign Affairs and Trade) for provision of PhD scholarship. The authors would like to acknowledge the valuable assistance from the Ramaciotti Centre for Genomics and the Bio-analytical Mass Spectrometry Facility at UNSW. We would like to thank Tammy Sihui Tang for provision of the Geobacter lovleyi culture.
Footnotes
Funding. The authors acknowledge the Australian Research Council and Orica for funding (LP130100454) and the Australian Awards Scholarships (Department of Foreign Affairs and Trade) for supporting AC.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01927/full#supplementary-material
References
- Adrian L., Görisch H. (2002). Microbial transformation of chlorinated benzenes under anaerobic conditions. Res. Microbiol. 153, 131–137. 10.1016/S0923-2508(02)01298-6 [DOI] [PubMed] [Google Scholar]
- Adrian L., Rahnenführer J., Gobom J., Hölscher T. (2007). Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. Strain CBDB1. Appl. Environ. Microbiol. 73, 7717–7724. 10.1128/AEM.01649-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian L., Szewzyk U., Gorisch H. (1998). Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3- and 1,2,4-trichlorobenzene. Appl. Environ. Microbiol. 64, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian L., Szewzyk U., Wecke J., Gorisch H. (2000). Bacterial dehalorespiration with chlorinated benzenes. Nature 408, 580–583. 10.1038/35046063 [DOI] [PubMed] [Google Scholar]
- Becker J. G., Berardesco G., Rittmann B. E., Stahl D. A. (2005). The role of syntrophic associations in sustaining anaerobic mineralization of chlorinated organic compounds. Environ. Health Perspect. 113, 310–316. 10.1289/ehp.6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. W. M., Grostern A., Löffler F. E., Edwards E. A. (2011). Quantifying the effects of 1,1,1-trichloroethane and 1,1-dichloroethane on chlorinated ethene reductive dehalogenases. Environ. Sci. Technol. 45, 9693–9702. 10.1021/es201260n [DOI] [PubMed] [Google Scholar]
- Choi H., Al-Abed S. R., Agarwal S. (2009). Catalytic role of palladium and relative reactivity of substituted chlorines during adsorption and treatment of PCBs on reactive activated carbon. Environ. Sci. Technol. 43, 7510–7515. 10.1021/es901298b [DOI] [PubMed] [Google Scholar]
- Chung J., Rittmann B. E. (2008). Simultaneous bio-reduction of trichloroethene, trichloroethane, and chloroform using a hydrogen-based membrane biofilm reactor. Water Sci. Technol. 58, 495–536. 10.2166/wst.2008.432 [DOI] [PubMed] [Google Scholar]
- Dang Y., Holmes D. E., Zhao Z., Woodard T. L., Zhang Y., Sun D., et al. (2016). Enhancing anaerobic digestion of complex organic waste with carbon-based conductive materials. Bioresour. Technol. 220, 516–522, 10.1016/j.biortech.2016.08.114 [DOI] [PubMed] [Google Scholar]
- Davidova M. N., Tarasova N. B., Mukhitova F. K., Karpilova I. U. (1994). Carbon monoxide in metabolism of anaerobic bacteria. Can. J. Microbiol. 40, 417–425, 10.1139/m94-069 [DOI] [PubMed] [Google Scholar]
- Diender M., Stams A. J. M., Sousa D. Z. (2015). Pathways and bioenergetics of anaerobic carbon monoxide fermentation. Front. Microbiol. 6:1275. 10.3389/fmicb.2015.01275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfing J., Tiedje J. M. (1986). Hydrogen cycling in a three-tiered food web growing on the methanogenic conversion of 3-chlorobenzoate. FEMS Microbiol. Lett. 38, 293–298. 10.1111/j.1574-6968.1986.tb01740.x [DOI] [Google Scholar]
- Felchner-Zwirello M., Winter J., Gallert C. (2013). Interspecies distances between propionic acid degraders and methanogens in syntrophic consortia for optimal hydrogen transfer. Appl. Microbiol. Biotechnol. 97, 9193–9205. 10.1007/s00253-012-4616-9 [DOI] [PubMed] [Google Scholar]
- Field J. A, Sierra-Alvarez R. (2008). Microbial degradation of chlorinated benzenes. Biodegradation 19, 463–480. 10.1007/s10532-007-9155-1 [DOI] [PubMed] [Google Scholar]
- Harmsen H., Van Kuijk B., Plugge C., Akkermans A., De Vos W., Stams A. (1998). Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate-degrading sulfate-reducing bacterium. Int. J. Syst. Bacteriol. 48, 1383–1138. 10.1099/00207713-48-4-1383 [DOI] [PubMed] [Google Scholar]
- Haynes W. M. (2013). CRC Handbook of Physics and Chemistry, 95th Edn. Boca Raton, FL: CRC Press, Inc. [Google Scholar]
- He J., Holmes V. F., Lee P. K. H., Alvarez-Cohen L. (2007). Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73:2847. 10.1128/AEM.02574-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horz H. P., Vianna M. E., Gomes B. P. F. A., Conrads G. (2005). Evaluation of universal probes and primer sets for assessing total bacterial load in clinical samples: general implications and practical use in endodontic antimicrobial therapy. J. Clin. Microbiol. 43, 5332–5337. 10.1128/JCM.43.10.5332-5337.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran G., Gorisch H., Adrian L. (2003). Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch Microbiol. 180, 411–416. 10.1007/s00203-003-0607-7 [DOI] [PubMed] [Google Scholar]
- Jiang L., Wang Q., Liu H., Yao J. (2015). Influence of degradation behavior of coexisting chlorobenzene congeners pentachlorobenzene, 1,2,4,5-tetrachlorobenzene, and 1,2,4-trichlorobenzene on the anaerobic reductive dechlorination of hexachlorobenzene in dye plant contaminated soil. Water. Air. Soil Pollut. 226, 1–9. 10.1007/s11270-015-2559-3 [DOI] [Google Scholar]
- Jugder B. E., Ertan H., Wong Y. K., Braidy N., Manefield M., Marquis C. P., et al. (2016). Genomic, transcriptomic and proteomic analyses of Dehalobacter UNSWDHB in response to chloroform. Environ. Microbiol. Rep. 8, 814–824. 10.1111/1758-2229.12444 [DOI] [PubMed] [Google Scholar]
- Kjellerup B. V., Naff C., Edwards S. J., Ghosh U., Baker J. E., Sowers K. R. (2014). Effects of activated carbon on reductive dechlorination of PCBs by ORB indigenous to sediments. Water. Res. 52, 1–10, 10.1016/j.watres.2013.12.030 [DOI] [PubMed] [Google Scholar]
- Koutsogiannouli E. A., Moutou K. A., Sarafidou T., Stamatis C., Spyrou V., Mamuris Z. (2009). Major histocompatibility complex variation at class II DQA locus in the brown hare (Lepus europaeus). Mol. Ecol. 18, 4631–4649. 10.1111/j.1365-294X.2009.04394.x [DOI] [PubMed] [Google Scholar]
- Kublik A., Deobald D., Hartwig S., Schiffmann C. L., Andrades A., von Bergen M., et al. (2016). Identification of a multi-protein reductive dehalogenase complex in Dehalococcoides mccartyi strain CBDB1 suggests a protein-dependent respiratory electron transport chain obviating quinone involvement. Environ. Microbiol. 18, 3044–3056. 10.1111/1462-2920.13200 [DOI] [PubMed] [Google Scholar]
- Maillard J., Genevaux P., Holliger C. (2011). Redundancy and specificity of multiple trigger factor chaperones in Desulfitobacteria. Microbiology 157, 2410–2421. 10.1099/mic.0.050880-0 [DOI] [PubMed] [Google Scholar]
- Mao X., Stenuit B., Polasko A., Alvarez-Cohen L. (2015). Efficient metabolic exchange and electron transfer within a syntrophic trichloroethene-degrading coculture of Dehalococcoides mccartyi 195 and Syntrophomonas wolfei. Appl. Environ. Microbiol. 81, 2015–2024. 10.1128/AEM.03464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Struchtemeyer C. G., Sieber J., Mouttaki H., Stams A. J., Schink B., et al. (2008). Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125, 58–72. 10.1196/annals.1419.005 [DOI] [PubMed] [Google Scholar]
- Men Y., Feil H., Verberkmoes N. C., Shah M. B., Johnson D. R., Lee P. K., et al. (2012). Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J. 6, 410–421. 10.1038/ismej.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris B. E. L., Henneberger R., Huber H., Moissl-Eichinger C. (2013). Microbial syntrophy: interaction for the common good. FEMS Microbiol. Rev. 37, 384–406. 10.1111/1574-6976.12019 [DOI] [PubMed] [Google Scholar]
- Morris R. M., Fung J. M., Rahm B. G., Zhang S., Freedman D. L., Zinder S. H., et al. (2007). Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73, 320–326. 10.1128/AEM.02129-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Pavoni B., Drusian D., Giacometti A., Zanette M. (2006). Assessment of organic chlorinated compound removal from aqueous matrices by adsorption on activated carbon. Water Res. 40, 3571–3579. 10.1016/j.watres.2006.05.027 [DOI] [PubMed] [Google Scholar]
- Popat S. C., Deshusses M. A. (2011). Kinetics and inhibition of reductive dechlorination of trichloroethene, cis-1,2-dichloroethene and vinyl chloride in a continuously fed anaerobic biofilm reactor. Environ. Sci. Technol. 45, 1569–1578. 10.1021/es102858t [DOI] [PubMed] [Google Scholar]
- Ragsdale S. W. (2004). Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 39, 165–195. 10.1080/10409230490496577 [DOI] [PubMed] [Google Scholar]
- Schiffmacher E. N., Becker J. G., Lorah M. M., Voytek M. A. (2016). The effects of co-contaminants and native wetland sediments on the activity and dominant transformation mechanisms of a 1,1,2,2-tetrachloroethane (TeCA)-degrading enrichment culture. Chemosphere 147, 239–247. 10.1016/j.chemosphere.2015.12.033 [DOI] [PubMed] [Google Scholar]
- Schiffmann C. L., Jehmlich N., Otto W., Hansen R., Nielsen P. H., Adrian L., et al. (2014). Proteome profile and proteogenomics of the organohalide-respiring bacterium Dehalococcoides mccartyi strain CBDB1 grown on hexachlorobenzene as electron acceptor. J. Proteomics 98, 59–64. 10.1016/j.jprot.2013.12.009 [DOI] [PubMed] [Google Scholar]
- Schiffmann C. L., Otto W., Hansen R., Nielsen P. H., Adrian L., Seifert J., et al. (2016). Proteomic dataset of the organohalide-respiring bacterium Dehalococcoides mccartyi strain CBDB1 grown on hexachlorobenzene as electron acceptor. Data Brief 7, 253–256. 10.1016/j.dib.2016.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B. (1997). Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61, 262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B., Thauer R. K. (1988). Energetics of syntrophic methane formation and the influence of aggregation, in Granular Anaerobic Sludge: Microbiology and Technology, ed Lettinga G., Zehnder A. J. B., Grotenhuis J. T. C., Hulshoff Pol L. W. (Wageningen: Pudoc; ), 5–17. [Google Scholar]
- Schmidt A., Müller N., Schink B., Schleheck D. (2013). A proteomic view at the biochemistry of Syntrophic Butyrate Oxidation in Syntrophomonas wolfei. PLoS ONE 8:e56905. 10.1371/journal.pone.0056905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber J. R., Crable B. R., Sheik C. S., Hurst G. B., Rohlin L., Gunsalus R. P., et al. (2015). Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei. Front. Microbiol. 6:115. 10.3389/fmicb.2015.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits T. H. M., Devenoges C., Szynalski K., Maillard J., Holliger C. (2004). Development of a real-time PCR method for quantification of the three genera Dehalobacter, Dehalococcoides, and Desulfitobacterium in microbial communities. J. Microbiol. Methods 57, 369–378. 10.1016/j.mimet.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Stams A. J. M., Sousa D. Z., Kleerebezem R., Plugge C. M. (2012). Role of syntrophic microbial communities in high-rate methanogenic bioreactors. Water Sci. Technol. 66, 352–362. 10.2166/wst.2012.192 [DOI] [PubMed] [Google Scholar]
- Urakawa H., Martens-Habbena H. W., stahl D. A. (2010). High abundance of ammonia oxidizing Archaea in coastal waters, determined using a modified DNA extraction method. Appl. Environ. Microbiol. 76, 2129–2135. 10.1128/AEM.02692-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Agteren M. H., Keuning S., Oosterhaven J. (1998). Environmental Chemistry: Handbook on Biodegradation and Biological Treatment of Hazardous Organic Compounds. Dordrecht: Springer. [Google Scholar]
- Wang D. Z., Kong L. F., Li Y. Y., Xie Z. (2016). Environmental microbial community proteomics: status, challenges and perspectives. Int. J. Mole. Sci. 17:1275. 10.3390/ijms17081275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu H., Jiang L., Tang J. (2014). Effect of the coexistence of chlorobenzene homologue on anaerobic degradation of hexachlorobenzene. Huanjing Kexue/Environ. Sci. 35, 1358–1365. 10.13227/j.hjkx.2014.04.022 [DOI] [PubMed] [Google Scholar]
- Wei K., Grostern A., Chan W. W. M., Richardson R. E., Edwards E. A. (2016). Electron acceptor interactions between organohalide-respiring bacteria: cross-feeding, competition, and inhibition, in Organohalide-Respiring Bacteria, eds Lorenz F. E. L. (Berlin; Heidelberg: Springer; ), 283–308. [Google Scholar]
- Worm P., Koehorst J. J., Visser M., Sedano-Núñez V. T., Schaap P. J., Plugge C. M., et al. (2014). A genomic view on syntrophic versus non-syntrophic lifestyle in anaerobic fatty acid degrading communities. Biochim. Biophys. Acta 1837, 2004–2016. 10.1016/j.bbabio.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Wu J. H., Wu F. Y., Chuang H. P., Chen W. Y., Huang H. J., Chen S. H., et al. (2013). Community and proteomic analysis of methanogenic consortia degrading terephthalate. Appl. Environ. Microbiol. 79, 105–112. 10.1128/AEM.02327-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm D. B., Sun K., Winniford W. L. (2003). Analysis of low levels of oxygen, carbon monoxide, and carbon dioxide in polyolefin feed streams using a pulsed discharge detector and two PLOT columns. J. Chromatogr. Sci. 41, 545–549. 10.1093/chromsci/41.10.545 [DOI] [PubMed] [Google Scholar]
- Yan J., Im J., Yang Y., Löffler F. E. (2013). Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Philo. Trans. 368:20120320. 10.1098/rstb.2012.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Ritalahti K. M., Wagner D. D., Löffler F. E. (2012). Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to Organohalide-Respiring Dehalococcoides mccartyi Strains. Appl. Environ. Microbiol. 78, 6630–6636. 10.1128/AEM.01535-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. H., Coppi M. V., Lovley D. R., Sun J. (2010). Metabolic response of Geobacter sulfurreducens towards electron donor/acceptor variation. Microbial Cell Factories 9, 1–15. 10.1186/1475-2859-9-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Dolan M. E., Semprini L. (2005). Kinetics and inhibition of reductive dechlorination of chlorinated ethylenes by two different mixed cultures. Environ. Sci. Technol. 39, 195–205. 10.1021/es0496773 [DOI] [PubMed] [Google Scholar]
- Yu S., Semprini L. (2004). Kinetics and modeling of reductive dechlorination at high PCE and TCE concentrations. Biotechnol. Bioeng. 88, 451–464. 10.1002/bit.20260 [DOI] [PubMed] [Google Scholar]
- Zhuang W. Q., Yi S., Bill M., Brisson V. L., Feng X., Men Y., et al. (2014). Incomplete wood-ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc. Nat. Acad. Sci. U.S.A. 111, 6419–6424. 10.1073/pnas.1321542111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv-El M., Popat S. C., Cai K. (2012). Managing methanogens and homoacetogens to promote reductive dechlorination of trichloroethene with direct delivery of H2 in a membrane biofilm reactor. Biotechnol. Bioeng. 109:2200–2210. 10.1002/bit.24487 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


