Abstract
Circulating tumor cells (CTCs) are a tumor‐derived material utilized for liquid‐based biopsy; however, capturing rare CTCs for further molecular analysis remains technically challenging, especially in non‐small‐cell lung cancer. Here, we report the results of a clinical evaluation of On‐chip Sort, a disposable microfluidic chip‐based cell sorter, for capture and molecular analysis of CTCs from patients with lung adenocarcinoma. Peripheral blood was collected from 30 metastatic lung adenocarcinoma patients to enumerate CTCs using both On‐chip Sort and CellSearch in a blind manner. Captured cells by On‐chip Sort were subjected to further molecular analysis. Peripheral blood samples were also used for detection of EGFR mutations in plasma using droplet digital PCR. Significantly more CTCs were detected by On‐chip Sort (22/30; median 5; range, 0–18 cells/5 mL blood) than by CellSearch (9/30; median, 0; range, 0–12 cells/7.5 mL) (P < 0.01). Thirteen of 30 patients who had a negative CTC count by CellSearch had a positive CTC count by On‐chip Sort. EGFR mutations in CTCs captured by On‐chip Sort were observed in 40.0% (8/20) of patients with EGFR‐mutated primary tumor. EGFR mutations were often observed in 53.3% (8/15) of patients detected in plasma DNA. Expressions of EGFR and vimentin protein on CTCs were also successfully assessed using On‐chip Sort. These results suggest that On‐chip Sort is an efficient method to detect and capture rare CTCs from patients with lung adenocarcinoma that are undetectable with CellSearch. Mutation detection using isolated CTCs remains to be further tackled (UMIN000012488).
Keywords: cell sorter, circulating tumor cell, EGFR, liquid biopsy, lung cancer
1. INTRODUCTION
Lung cancer is the most common cause of cancer‐related deaths, and approximately 84% of new lung cancer cases are classified as non‐small‐cell lung carcinomas (NSCLC), and 15% as small‐cell carcinomas,1 with the majority of patients being diagnosed at an advanced stage (56%).2 Recent advances in molecularly targeted cancer therapy have offered a wide variety of therapeutic strategies for patients with NSCLC. For example, identification of EGFR‐activating mutations in patients with NSCLC is required prior to starting treatments with epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs).3 However, kinase inhibition frequently leads to the appearance of drug resistance mutations within the target kinase itself, such as the EGFR T790M mutation.4 In addition to identifying gene mutations, there is also a need for the detection of protein expression and gene amplification of targeted molecules on primary tumor cells, for further stratification of patients.5 To optimize treatment, real‐time monitoring of tumors over the course of the treatment, especially at the point of treatment failure, is necessary. However, the classic biopsy approach does not allow monitoring of primary tumor evolution over time, and sampling of metastatic sites is not always possible for practical reasons.
Through a simple blood draw, circulating tumor cells (CTCs) could potentially serve as an alternative to the tumor tissue as a source of material for the detection of genetic alterations, an approach that is termed “liquid biopsy”6 owing to its minimal invasiveness. To date, the CellSearch system (Veridex, Raritan, NJ, USA) is the only US FDA‐approved CTC enumeration system for the provision of prognostic information regarding survival.7, 8, 9, 10, 11 However, CTCs are very rare and make up a small minority of cells circulating in blood, so their molecular analysis beyond enumeration is technically very challenging.6, 12 Various methods to overcome this issue have been under development and evaluation.13, 14, 15, 16, 17
The potential of single cell sorting by FACS and whole‐genome amplification of CTCs after CellSearch was described previously.18, 19 The main limitation of both of these approaches is that only epithelial cell adhesion molecule (EpCAM)‐positive epithelial cells can be isolated and analyzed, because this is the isolation system used by CellSearch. Thus, invasive phenotypes of CTCs that undergo epithelial–mesenchymal transition (EMT) cannot be analyzed.20
Recently, we have established a protocol for rare CTC enumeration and sorting using a newly developed cell sorting system.21, 22 This cell sorting system, called On‐chip Sort (On‐chip Biotechnologies, Tokyo, Japan), is a novel benchtop cell sorter equipped with a disposable microfluidic device, allowing the detection and isolation of rare tumor cells for subsequent molecular analyses.21 This protocol also enables a detection of EpCAM‐negative/cytokeratin (CK)‐negative cells using the incorporation of an EMT marker.22 These results indicate that our system is a precise system for the detection and capture of tumor cells within whole blood. To confirm our previous findings, we compared the capacity and efficiency of our On‐chip Sort system and the current gold standard CellSearch system in undertaking CTC detection and enumeration in whole blood samples drawn from a cohort of patients with lung adenocarcinoma. Also, we carried out molecular characterization of the sorted CTCs and analysis of circulating tumor DNA using droplet digital PCR in paired blood samples.
2. MATERIALS AND METHODS
2.1. Study design and ethics statement
This prospective study was carried out to evaluate CTC analysis using the CellSearch system and the On‐chip Sort system in patients with advanced lung cancer in a blinded experiment (UMIN clinical trial registry no. UMIN000012488). The presence of CTCs was assessed individually according to their criteria before other results were known. The study inclusion criteria were age 20 years or older when giving informed consent, histologically or cytologically confirmed advanced NSCLC, and enrollment at the Shizuoka Cancer Center (Shizuoka, Japan). The institutional review boards of the Shizuoka Cancer Center approved the study protocol, and all patients and healthy volunteers provided written informed consent. Blood was collected from each of the 30 patients and 10 healthy volunteers (5–15 mL) in EDTA tubes for CTC capture by the On‐chip Sort system in our laboratory (Shizuoka Cancer Center), 5 mL was collected in EDTA tubes for genotyping plasma DNA using digital PCR, and 10 mL was collected in CellSave collection tubes (Menarini Silicon Biosystems Inc, PA) (San Jose, CA, USA) for CTC enumeration by the CellSearch system in the laboratory of SRL (Tokyo, Japan).
2.2. Circulating tumor cell enumeration and capture using the On‐chip Sort system
Human blood samples were collected in a collection tube with EDTA to prevent coagulation and used within 2 h. Blood from each lung cancer patient (5–15 mL) and healthy volunteer (5 mL) was subjected to CTC capture.
Immunomagnetic enrichment was carried out as described previously.21, 22 Briefly, 5 mL of each blood sample was lysed by lysing solution (BD Biosciences, San Jose, CA, USA), and then was negatively enriched using Dynabeads coated with anti‐CD45 mAb (Thermo Fisher Scientific Inc., Waltham, MA, USA) to remove white blood cells, followed by fixation and labeling with the FITC‐conjugated anti‐CK mAb CK3‐6H5 (1:25 dilution; Miltenyi Biotec, Bergisch‐Gladbach, Germany), the phycoerythrin‐conjugated anti‐vimentin mAb D21H3 (1:50 dilution; Cell Signaling Technology, Danvers, MA, USA), and the Alexa Fluor 700‐conjugated anti‐CD45 mAb F10‐89‐4 (1:20 dilution; AbD Serotec, Oxford, UK). For some patient samples from whom >10 mL of blood was obtained, additional labeling with Alexa Fluor 647‐conjugated anti‐EGFR mAb D38B1 was carried out (1:50 dilution; Cell Signaling Technology). Samples were incubated overnight at 4°C in the dark.
Enumeration and sorting of cells were carried out by On‐chip Sort according to the manufacturer's instructions and as described previously.22 Briefly, the flow path was prewashed with 1× Through Path Plus (On‐chip Biotechnologies) and the On‐chip sample buffer (1× Through Path Plus with 1.5% polyvinylpyrrolidone; On‐chip Biotechnologies). Stained samples were dissolved in 25 μL On‐chip sample buffer and then sorted with up to 400 events/s flow rate (approximately 1 μL/min = 1.8 kPa in the On‐chip Sort setting). Total events were approximately 1 × 105 to 106 events per sample. The sorting time required for all the samples was approximately 30 min to 2 h.
The cells gated into the CK‐ and/or vimentin‐positive and CD45‐negative channels (Fig. S1A) were collected into the collection reservoir and then observed under a fluorescence microscope (Biorevo BZ‐9000; Keyence, Osaka, Japan) to confirm that the cells were CK‐ and/or vimentin‐positive and CD45‐negative. All steps were carried out at room temperature.
In clinical trials, an entire FACS dataset had been obtained using an On‐chip Sort. Subsequently, FACS data analysis had been obtained using FlowJo software version 7.6.5 (BD Biosciences) and objects that satisfied the predetermined criteria as described below had been counted. A typical example for the full gating strategy is shown in Figure S1(B). Gating of the CTCs by On‐chip Sort was carried out using the CK‐FITC staining versus vimentin–phycoerythrin staining density plot. The lower limit of the gate that discriminates CTC signals from WBCs autofluorescence as well as from debris was determined by several runs of non‐spike experiments using healthy donor control bloods. The CK‐ and/or vimentin‐positive events were then subjected to CD45‐negative gating to distinguish tumor cells from WBCs and/or debris. Finally, cell debris was removed using the forward scatter versus side scatter density plot.
2.3. Circulating tumor cell enumeration using the CellSearch system
Whole blood samples were maintained at room temperature, mailed overnight to the laboratory of SRL, and processed within 72 h of collection. All CTC evaluations were carried out without knowledge of patient clinical status, and the results were reported quantitatively as the number of CTCs/7.5 mL blood. Circulating tumor cells were defined as EpCAM‐isolated intact cells showing positive staining for cytokeratin and negative staining for CD45. In accordance with previous evaluations of the CellSearch system,23 a patient was considered CTC‐positive if >1 CTC/7.5 mL blood was detected in the patient's sample.
2.4. Primary tissue DNA sequencing
Tumor biopsy samples from patients before treatment were outsourced for detection of the EGFR mutation using the Scorpion‐ARMS method and EML4‐ALK fusion analysis immunohistochemistry confirmed by FISH by an independent clinical laboratory (SRL). The KRAS mutation was evaluated in another clinical study.24
2.5. Mutation analysis of CTCs sorted by On‐chip Sort
Sorted cells from all sample tubes (Tube 1, 2, and EGFR) were transferred from the collection reservoir to a 200‐μL PCR tube, and the collection reservoir was rinsed with sheath solution twice. After centrifugation (600g for 10 min), the supernatant was carefully removed to leave ~1 μL, which was the starting volume of the whole‐genome amplification (WGA) procedure. Whole‐genome amplification was undertaken using the Ampli1 WGA kit (Silicon Biosystems, Bologna, Italy) following the manufacturer's protocol and as described previously.22 Two microliters of amplified DNA product was subjected to mutation analysis using pyrosequencing to detect the EGFR L858R and T790M mutations, which have been previously developed,24 and using an Ampli1 EGFR Seq Kit (Silicon Biosystems) for EGFR exon 19 deletion followed by fragment analysis following the manufacturer's protocol.
2.6. Mutation analysis of plasma DNA
Five milliliters of whole blood was centrifuged at 1500g for 10 min at 4°C. The plasma supernatant was transferred to 50‐mL conical tubes (Falcon; BD Biosciences) and stored at −80°C until use. Plasma DNA was isolated using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. DNA was eluted in AVE buffer (30 μL). DNA concentration was measured with the Qubit 2.0 Fluorometer (Thermo Fisher Scientific Inc.).
Droplet digital PCR (ddPCR) for the common EGFR mutations (L858R, exon 19 deletion, and T790M) was carried out in separate reactions for each mutation‐specific probe on the RainDrop digital PCR system (RainDance Technologies, Billerica, MA, USA) following the manufacturer's instructions (see also Doc. S1). Eight microliters of plasma DNA was used for each assay. The amplification primers and probes for mutations in EGFR are described in Table S1. Results are reported as percentage of mutant allele, as done by prior investigators.25
2.7. Statistical analysis
GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA) was used for statistical analyses. Statistical significance of difference was determined using the Wilcoxon test.
3. RESULTS
3.1. Sensitivity of the On‐chip Sort system in CTC detection
In our previous study, varying numbers of various tumor cell lines were spiked into blood, and tumor cell isolation was evaluated using our On‐chip Sort system.22 The calculated detection efficiency was constant and >70% when 5–25 tumor cells were present per 4 mL blood, and there was a 100% success rate in the detection of EGFR, KRAS, and BRAF mutations from captured tumor cells.22 The limit of blank of the On‐chip Sort system was determined as a finite number of false‐positive events of CTCs detected per assay. According to 10 healthy volunteer samples, the number of false‐positive events was two events/5 mL blood (95% confidence interval of the Poisson model fit; Fig. S2). Therefore, a patient was considered CTC‐positive if >2 CTCs/5 mL blood were detected by the On‐chip Sort system.
3.2. Patients and tumor tissue‐derived genotypes
Patient characteristics are shown in Table 1. Tumor specimens with corresponding clinical and pathological information were collected from all patients. The scorpion‐ARMS method sequencing of these 30 primary tumor specimens identified 13 EGFR exon 19 deletion mutations, six L858R mutations, and one deletion with EGFR T790M mutation yielding a frequency of 66.7% (20 of 30 tumors; Table 1). Two patients (7%) had KRAS mutation, one (3%) had EML4‐ALK gene fusion, and four (13%) had no major driver mutations. Blood samples were collected at the time of pretreatment (one patient), in the middle of first‐line treatment (eight patients), and after first‐line treatment (21 patients).
Table 1.
Characteristics of patients with non‐small‐cell lung carcinoma (n = 30)
| No. of patients | n = 30 |
|---|---|
| Gender | |
| Male | 14 |
| Female | 16 |
| Median age, years | 66 |
| Range | 38‐83 |
| Smoking | |
| Never smoker | 13 |
| Smoker | 17 |
| Mutation | |
| EGFR exon 19 deletion | 13 |
| EGFR exon 19 deletion + EGFR T790M | 1 |
| EGFR L858R | 6 |
| EGFR G719X | 3 |
| KRAS | 1 |
| EML4‐LK | 1 |
| No mutation | 4 |
| Mutation | |
| Pretreatment | 1 |
| Ongoing first line | 8 |
| After first line | 21 |
3.3. Enumeration of CTCs using the On‐chip Sort and CellSearch systems
To carry out blind comparisons of the detection sensitivity of the On‐chip Sort and CellSearch systems, blood samples were collected from 30 patients with advanced NSCLC between December 2013 and March 2014. As a result, 22 of 30 (73.3%) patients were identified as CTC‐positive using the On‐chip Sort system, but only 9 of 30 (30%) patients using the CellSearch system (Fig. 1, Table S2). Of these patients, the nine identified as CTC‐positive by CellSearch were confirmed as CTC‐positive by the On‐chip Sort system. The On‐chip Sort system identified 13 additional CTC‐positive patients. These results revealed that more CTCs were detected by the On‐chip Sort system (median cell count, 6.5; range, 0–27 cells/7.5 mL blood; Fig. 1) than by the CellSearch system (median cell count, 0; range, 0–12 cells/7.5 mL blood; Fig. 1), suggesting the statistical superiority of the On‐chip Sort system in CTC enumeration (P < 0.01, Wilcoxon test; Fig. 1A), and that CTC counts obtained with both methods were not correlated (R 2 = 0.0148) (Fig. 1B). There was no correlation between CTC counts and treatment line (R 2 = 0.0478; Fig. S3). The correlation between the measured CTC numbers in two subsequent tubes was moderately correlated with a correlation coefficient of 0.6737 (Fig. S4).
Figure 1.
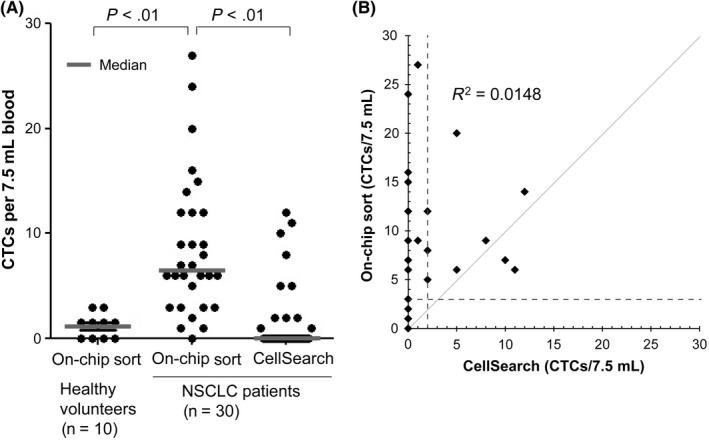
Circulating tumor cell (CTC) count using the On‐chip Sort system compared with the CellSearch system. (A) CTC count/7.5 mL blood is shown for 10 healthy donors and 30 patients with non‐small‐cell lung carcinoma (NSCLC). Paired blood samples were analyzed by CellSearch according to the manufacturer's protocol and On‐chip Sort by immunolabeling analysis. (B) Direct comparison of CTC counts between CellSearch and On‐chip Sort. Gray line represents the theoretical perfect correlation. The cut‐off levels (3 CTCs for On‐chip Sort and 2 CTCs for CellSearch per sample) are indicated by the dashed lines
3.4. Morphological features and protein expression of CTCs sorted using the On‐chip Sort
Sorted CTCs in the sorting reservoir chamber were observed under a fluorescent microscope, and then identified as being CK‐ and/or vimentin‐positive, and CD45 negative, on the basis of analysis of fluorescent images. As can be observed in Figure 2, which shows a representative gallery of CTCs identified by image analysis, CTCs expressed cytokeratin alone (CTC #1), vimentin alone (CTC #2), or both (CTC #3). No clustered CTCs (circulating tumor microembolus) were observed in this cohort.
Figure 2.
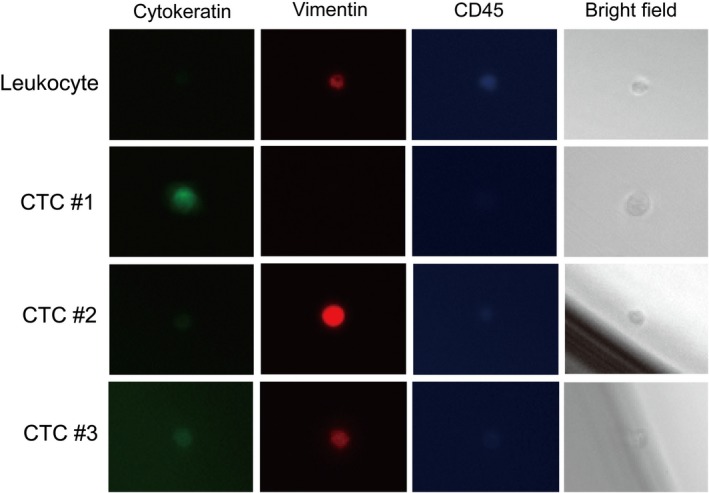
Gallery of cells captured by On‐chip Sort from patients with non‐small‐cell lung carcinoma. Cells were stained with FITC‐labeled anti‐cytokeratin antibody, phycoerythrin‐labeled anti‐vimentin, and Alexa700‐labeled anti‐CD45 antibody. CTC, circulating tumor cell
We assessed the EGFR protein expression and vimentin, EMT marker, expression on detected CTCs. We assessed CTCs from 22 patients for vimentin protein, and found them to be differently expressed between CTCs from the same patients, ranging, for example, from negative to strongly positive (Fig. 3). We also assessed CTCs from 20 patients for EGFR protein expression; EGFR‐positive CTCs (>200 intensity of EGFR‐Alexa647) could be observed in 12 of 20 (60.0%) patients, with only 3 of 20 patients (15.0%) possessing strongly EGFR‐positive CTCs (patients #10, #11, and #20) (Fig. 3A). The fluorescent signal from EGFR‐conjugated Alexa647 was observed in the peripheral region of the cell (Fig. 3B). All CTC events detected in healthy volunteers were EGFR‐negative (data not shown).
Figure 3.
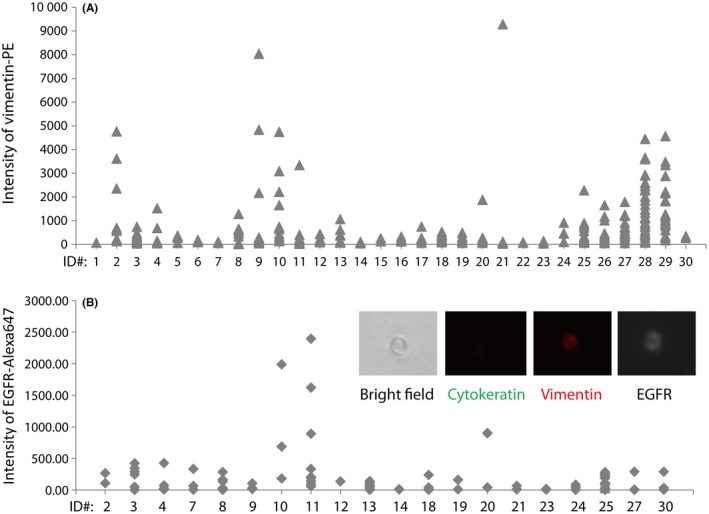
Assessment of protein expression in circulating tumor cells (CTCs). Vimentin (A) and epidermal growth factor receptor (EGFR) (B) expressions were assessed by On‐chip Sort during sorting. Intensity of fluorescent‐labeled protein expression on CTCs was analyzed by FlowJo software 7.6.5. Cell images of EGFR expression on CTCs are embedded in (B). PE, phycoerythrin
3.5. EGFR mutations detected in isolated CTCs
Given the challenge with obtaining liquid biopsy from lung cancer patients, we sought to determine therapeutically variable mutations in EGFR from isolated lung adenocarcinoma CTCs. Pooled CTCs sorted by On‐chip Sort were subjected to WGA, and WGA products were successfully amplified in all samples with enough genetic material for genotyping (data not shown). Amplified DNA samples were analyzed for EGFR L858R and T790M using pyrosequencing, and exon 19 deletion using fragment analysis for detection. We have previously shown the detection of rare alleles down to 10% frequency, even in a few target cells.22
In CTCs from patients, the common EGFR‐activating mutations were detected in 9 of 30 cases, with the remaining 21 cases sequenced as wild‐type at these nucleotide positions. Among 20 patients with tumor biopsy‐derived EGFR‐activating mutation, 8 (40%) were concordant for activation mutation status (Table 2). EGFR T790M mutation was also identified in the CTCs of two patients (#8 and #23). One of them (#23) was not detected in primary tumors; in this patient, CTCs were collected after first‐line EGFR‐TKI therapy (Table 2).
Table 2.
Analysis of EGFR mutations in primary tumors, circulating tumor cells (CTCs), and plasma from patients with non‐small‐cell lung carcinoma
| Sample ID# | CTC count | EGFR mutations in CTCs (allele frequency %) | EGFR mutation in plasma (allele frequency %) | EGFR mutations in primary tissue | Time point |
|---|---|---|---|---|---|
| 1 | 0 | Wild type | L858R (67.4%), T790M (32.8%) | L858R | Post‐gemcitabine as 4th line |
| 2 | 6 | Ex19 del | L858R (6.4%), Ex19 del (10.3%), T790M (6.4%) | Ex19 del | On palliative RT |
| 3 | 13 | Ex19 del | Ex19 del (12.4%) | Ex19 del | On erlotinib as 3rd line |
| 4 | 5 | Ex19 del | L858R (9.7%), Ex19 del (25.6%), T790M (11.5%) | Ex19 del | On gefitinib as 1st line |
| 5 | 2 | Wild type | Wild type | Wild type (EML4‐ALK) | On ALK inhibitor as 4th line |
| 6 | 2 | Wild type | L858R (9.2%) | L858R | Post‐docetaxel as 2nd line |
| 7 | 2 | Wild type | Wild type | Ex18 G719X | On amrubicin as 5th line |
| 8 | 6 | Ex19 del, T790M (21%) | Ex19 del (24.8%), T790M (22.4%) | Ex19 del, T790M | Post‐carboplatin, paclitaxel, and bevacizumab as 3rd line |
| 9 | 11 | Wild type | Wild type | L858R | On erlotinib as 4th line |
| 10 | 8 | Wild type | Wild type | Wild type (KRAS Q61H) | On gemcitabine as 3rd line |
| 11 | 9 | Wild type | Wild type | Wild type | Post‐pemetrexed as 4th line |
| 12 | 3 | Wild type | Wild type | Ex19 del | On carboplatin, paclitaxel, and bevacizumab as 3rd line |
| 13 | 4 | Wild type | Wild type | Ex19 del | On carboplatin, paclitaxel, and bevacizumab as 2nd line |
| 14 | 4 | Wild type | L858R (1.1%), Ex19 del (8.5%), T790M (1.9%) | L858R | On erlotinib as 3rd line |
| 15 | 1 | Wild type | Wild type | Ex19 del | On cisplatin and pemetrexed as 2nd line |
| 16 | 8 | Wild type | Wild type | Wild type | On carboplatin, paclitaxel, and bevacizumab as 1st line |
| 17 | 4 | Wild type | L858R (1.0%) | L858R | On carboplatin, paclitaxel, and bevacizumab as 3rd line |
| 18 | 6 | Ex19 del | Ex19 del (34.0%) | Ex19 del | On gefitinib as 1st line |
| 19 | 4 | Wild type | Wild type | Wild type (KRAS Q61H) | On gemcitabine as 7th line |
| 20 | 4 | Wild type | T790M (4.1%) | Ex18 G719X | On gefitinib as 1st line |
| 21 | 5 | Wild type | Wild type | Ex19 del | On erlotinib as 4th line |
| 22 | 1 | Ex19 del | Ex19 del (51.0%), T790M (13.0%) | Ex19 del | On gefitinib as 1st line |
| 23 | 4 | L858R (10%), T790M (17%) | L858R (21.2%), T790M (6.9%) | L858R | Post‐gefitinib as 1st line |
| 24 | 2 | Wild type | Wild type | Wild type | Post‐cisplatin and pemetrexed as 1st line |
| 25 | 16 | Ex19 del | L858R (64.2%) | Wild type | Pretreatment |
| 26 | 5 | Wild type | Wild type | Ex19 del | On carboplatin, paclitaxel, and bevacizumab as 7th line |
| 27 | 8 | Wild type | Wild type | Ex18 G719X | Post‐chemoradiotherapy |
| 28 | 18 | Wild type | Wild type | Ex19 del | Post‐cisplatin and pemetrexed as 2nd line |
| 29 | 10 | Wild type | Ex19 del (17.9%), T790M (1.4%) | Ex19 del | Post‐gefitinib as 1st line |
| 30 | 1 | Ex19 del | L858R (6.0%), Ex19 del (14.6%), T790M (5.2%) | Ex19 del | Post‐pemetrexed as 4th line |
ALK, anaplastic lymphoma kinase; Ex19 del, exon 19 deletion; RT, radiotherapy.
3.6. EGFR mutations detected in plasma DNA
Because genotyping in tumor biopsy was not undertaken concurrently with the study blood collection, matched plasma samples were collected for all patients as a reference. The amount of total plasma DNA varied among samples, ranging from 6.0 to 56.1 ng (Table S3), which is consistent with prior reports.25, 26, 27, 28 These plasma samples were analyzed for three common EGFR mutations as described above using ddPCR for detection, which is known to be ultrasensitive.29, 30 The limit of blank of the ddPCR assay was determined as a finite number of false events per assay using mutant and wild‐type plasmids. The number of false‐positive droplet events for each of the three EGFR assays is six for L858R, four for exon 19 deletion, and three for T790M (Table S4).
Droplet digital PCR detected 14 patients with common EGFR‐activating mutations and nine patients were also identified to have an additional EGFR T790M mutation (Table 2). One patient (#20) had only the EGFR T790M mutation. Four patients (patient #2, #4, #14, and #30) had all three mutations. A comparison of EGFR mutation status of CTCs with the corresponding ddPCR plasma DNA results indicated that EGFR mutations could be identified in the CTCs of eight patients whose plasma DNA had mutated EGFR. Fifteen patients with EGFR wild‐type plasma DNA had no detectable EGFR mutation in CTC (Table 2). These data show a sensitivity of 53.3% and specificity of 100.0% of mutation detection for mutant EGFR in CTCs (Table S5).
4. DISCUSSION
In the present study, CTCs from metastatic NSCLC patients isolated on the On‐chip Sort were successfully stained with fluorescent‐labeled antibodies that target tumor cell markers. Tube‐to‐tube variation in CTC counts was confirmed by regression analysis with a correlation coefficient (R 2) of 0.6737, suggesting that our CTC counts are a reproducible and independent with any range of counts.
The On‐chip Sort system was found to possess higher detection sensitivity than the CellSearch system in lung adenocarcinoma CTC enumeration, suggesting the superiority of non‐EpCAM‐based isolation techniques compared to EpCAM‐based techniques. The low EpCAM expression in lung adenocarcinoma CTCs has been previously reported. The CTC positivity rates were recorded between 67% and 84% using the EpCAM‐independent system,31, 32, 33, 34 whereas EpCAM‐dependent systems detected between 12% and 32% of patients.32, 33, 34, 35 In line with these results, in this study, CTCs were detected in 73.3% of patients using the On‐chip Sort system and in 30% of patients using the CellSearch system. Our result confirmed that EpCAM‐independent approaches are effective to detect CTCs from patients with metastatic NSCLC.
The present study was devised to evaluate the correlation between EGFR mutation status in basal tumor biopsies and matching CTCs of patients with NSCLC. Peripheral blood specimens were subjected to CTC preparation by the On‐chip Sort system as well as investigated for EGFR mutations by pyrosequencing. Of 20 patients with EGFR mutations in the original diagnosis biopsy, 19 samples had >1 CTC and 40% (8/20) were concordant between the CTC analysis and tissue biopsy analysis. Interestingly, EGFR mutations were detected in samples that had detected only one CTC (#22 and #30), consistent with the previous study in a spike‐in model.22 This sensitivity is slightly lower than those in previous studies.36, 37 The primary reason for this lower sensitivity could be due to the timing of biopsy. Whereas tissue and liquid biopsies were obtained at the same time in previous studies,36, 37 only one sample (#25) was obtained simultaneously in our study. In fact, when matched plasma DNA was used for the reference of EGFR mutations, concordance was increased (53.3%).
The ddPCR assay on cell free DNA was much more sensitive at detecting EGFR mutations than mutation detection using CTC. This result is in line with those reported previously.38, 39 Mutation detection using isolated CTCs by the On‐Chip Sort assay might not be as sensitive as plasma assay in NSCLC, according to the results. However, CTCs offer several unique advantages over the ability to undertake cytomorphological analysis, immunocytochemistry, or FISH assay.12 Most importantly, CTCs can be used for evaluating protein expression, which can be a target of cancer therapeutics. The application of protein staining using the FL5 channel of the On‐chip Sort enables the classification of vimentin expression as surrogate of the EMT and EGFR protein expression levels on single CTCs. These applications might be utilized as a treatment choice for patients with NSCLC. The patients with NSCLC whose tumors have detectable EGFR protein could benefit from the addition of necitumumab to chemotherapy.40 The AXL protein, associated with EMT, could be a promising therapeutic target for acquired resistance to EGFR TKIs.41
Recently, immune checkpoint blockade with antiprogrammed death 1 (PD‐1)/PD‐ligand 1 (PD‐L1) antibodies was reported to be efficacious for lung cancer patients,42 and PD‐L1 expression on tumor tissues could be a predictive biomarker of clinical benefit in patients with NSCLC.43, 44 However, it is still challenging to obtain a tumor biopsy from patients with lung cancer, and tumor heterogeneity is still issue for an PD‐L1 immunohistochemistry.45 We believe that CTCs can be used as an alternative material, instead of tumor tissues, for detecting PD‐L1 expression. Preliminary data have been shown by several groups.46, 47, 48
There are some challenges to be addressed with the On‐chip Sort System. To capture the most CTCs, the sorting gate was expanded to the maximum. This might lead to the reduction in the ratio of CTCs in background normal blood cells. The optimization of this sorting gate should increase the detection rate of mutations. The prognostic and predictable value of CTCs detected by the On‐chip Sort system needs to be evaluated. Lindsay et al. recently showed the prognostic value of CTCs counted by the CellSearch system in patients with treatment‐naïve stage IIIb/IV NSCLC.49 Punnoose et al.50 reported that higher baseline CTC counts were associated with response to treatment and decreased CTC counts following treatment were associated with longer progression‐free survival. The prognostic and predictive value of CTCs counted by our system will be evaluated in future clinical studies with optimization of timing of blood collection.
In conclusion, we have evaluated the performance of our On‐chip Sort system in detecting CTCs in whole blood from patients with lung cancer. We have also evaluated whether CTC preparations obtained by the On‐Chip Sort system represent a suitable source of tumor DNA for efficient detection of EGFR mutations. The results were promising and suggest that the On‐Chip Sort system has clinical potential for CTC diagnosis in lung cancer. Further prospective investigation with scheduled biopsies on a larger scale is warranted.
DISCLOSURE STATEMENT
The authors have no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
The authors thank Mrs. Junko Suzuki and Ms. Akane Naruoka (Shizuoka Cancer Center) for technical assistance and Drs. Kazuo Takeda, Masayuki Ishige, Yuu Fujimura, and Namiko Yamashita (On‐chip Biotechnologies) for technical support. The authors also thank Ms. Mie Yamada (Division of Thoracic Oncology, Shizuoka Cancer Center) for study management. This research was supported by the Advanced Research and Development Project on Diagnosis and Treatment for Early Stage of Cancer, Development of Automatic Testing System for Genetic Diagnosis using Peripheral Blood from the New Energy and Industrial Technology Development Organization, Japan.
Watanabe M, Kenmotsu H, Ko R, et al. Isolation and molecular analysis of circulating tumor cells from lung cancer patients using a microfluidic chip type cell sorter. Cancer Sci. 2018;109:2539–2548. 10.1111/cas.13692
Funding information
New Energy and Industrial Technology Development Organization, Japan.
Masaru Watanabe and Hirotsugu Kenmotsu contributed equally to this work.
REFERENCES
- 1. Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2012;62:220‐241. [DOI] [PubMed] [Google Scholar]
- 2. Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975‐2010. Bethesda, MD: National Cancer Institute; 2013. [Google Scholar]
- 3. Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non‐small‐cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380‐2388. [DOI] [PubMed] [Google Scholar]
- 4. Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non‐small‐cell lung cancer to gefitinib. N Engl J Med. 2005;352:786‐792. [DOI] [PubMed] [Google Scholar]
- 5. Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to anti EGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279‐286. [DOI] [PubMed] [Google Scholar]
- 6. Alix‐Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479‐491. [DOI] [PubMed] [Google Scholar]
- 7. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781‐791. [DOI] [PubMed] [Google Scholar]
- 8. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration‐resistant prostate cancer. Clin Cancer Res. 2008;14:6302‐6309. [DOI] [PubMed] [Google Scholar]
- 9. Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non‐smallcell lung cancer. J Clin Oncol. 2011;29:1556‐1563. [DOI] [PubMed] [Google Scholar]
- 10. Naito T, Tanaka F, Ono A, et al. Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J Thorac Oncol. 2012;7:512‐519. [DOI] [PubMed] [Google Scholar]
- 11. Matsusaka S, Chìn K, Ogura M, et al. Circulating tumor cells as a surrogate marker for determining response to chemotherapy in patients with advanced gastric cancer. Cancer Sci. 2010;101:1067‐1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ilie M, Hofman V, Long E, et al. Current challenges for detection of circulating tumor cells and cell‐free circulating nucleic acids, and their characterization in non‐small cell lung carcinoma patients. What is the best blood substrate for personalized medicine?. Ann Transl Med. 2014;2:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fujii T, Reuben JM, Huo L, et al. Androgen receptor expression on circulating tumor cells in metastatic breast cancer. PLoS ONE. 2017;12:e0185231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takakura M, Matsumoto T, Nakamura M, et al. Detection of circulating tumor cells in cervical cancer using a conditionally replicative adenovirus targeting telomerase‐positive cells. Cancer Sci. 2018;109:231‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Magbanua MJM, Rugo HS, Wolf DM, et al. Expanded genomic profiling of circulating tumor cells in metastatic breast cancer patients to assess biomarker status and biology over time (CALGB 40502 and CALGB 40503, Alliance). Clin Cancer Res. 2018;24:1486‐1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El‐Heliebi A, Hille C, Laxman N, et al. In situ detection and quantification of AR‐V7, AR‐FL, PSA, and KRAS point mutations in circulating tumor cells. Clin Chem. 2018;64:536‐546. [DOI] [PubMed] [Google Scholar]
- 17. Miyamoto DT, Lee RJ, Kalinich M, et al. An RNA‐based digital circulating tumor cell signature is predictive of drug response and early dissemination in prostate cancer. Cancer Discov. 2018;8:288‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swennenhuis JF, Reumers J, Thys K, Aerssens J, Terstappen LW. Efficiency of whole genome amplification of Single Circulating Tumor Cells enriched by Cell Search and sorted by FACS. Genome Med. 2013;5:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neves RPL, Raba K, Schmidt O, et al. Genomic high‐resolution profiling of single CKpos/CD45neg flow‐sorting purified circulating tumor cells from patients with metastatic breast cancer. Clin Chem 2014;60:1290‐1297. [DOI] [PubMed] [Google Scholar]
- 20. Bednarz‐Knoll N, Alix‐Panabie' res C, Pantel K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer Metastasis Rev 2012;31:673‐687. [DOI] [PubMed] [Google Scholar]
- 21. Watanabe M, Uehara Y, Yamashita N, et al. Multicolor Detection of Rare Tumor Cells in Blood Using a Novel Flow Cytometry‐based System. Cytometry Part A. 2014;85:206‐213. [DOI] [PubMed] [Google Scholar]
- 22. Watanabe M, Serizawa M, Sawada T, et al. A novel flow cytometry‐based cell capture platform for the detection, capture and molecular characterization of rare tumor cells in blood. J Transl Med. 2014;12:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of, all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897‐6904. [DOI] [PubMed] [Google Scholar]
- 24. Serizawa M, Koh Y, Kenmotsu H, et al. Assessment of mutational profile of Japanese lung adenocarcinoma patients by multitarget assays: a prospective, single‐institute study. Cancer. 2014;120:1471‐1481. [DOI] [PubMed] [Google Scholar]
- 25. Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early‐stage breast cancer. Clin Cancer Res. 2014;20:2643‐2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hindson BJ, Ness KD, Masquelier DA, et al. High‐throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604‐8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hatch AC, Fisher JS, Tovar AR, et al. 1‐Million droplet array with wide‐field fluorescence imaging for digital PCR. Lab Chip. 2011;11:3838‐3845. [DOI] [PubMed] [Google Scholar]
- 28. Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR‐mutant lung cancer using quantitative next‐generation genotyping of cell‐free plasma DNA. Clin Cancer Res. 2014;20:1698‐1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe M, Kawaguchi T, Isa S, et al. Ultra‐sensitive detection of the pretreatment EGFR T790M mutation in non‐small cell lung cancer patients with an EGFR‐activating mutation using droplet digital PCR. Clin Cancer Res. 2015;21:3552‐3560. [DOI] [PubMed] [Google Scholar]
- 30. Watanabe M, Kawaguchi T, Isa SI, et al. Multiplex ultrasensitive genotyping of patients with non‐small cell lung cancer for epidermal growth factor receptor (EGFR) mutations by means of picodroplet digital PCR. EBioMedicine. 2017;21:86‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang W, Bao L, Yang S, et al. Tumor‐selective replication herpes simplex virus‐based technology significantly improves clinical detection and prognostication of viable circulating tumor cells. Oncotarget. 2016;7:39768‐39783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hosokawa M, Kenmotsu H, Koh Y, et al. Size‐based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. PLoS ONE. 2013;8:e67466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yagi S, Koh Y, Akamatsu H, et al. Development of an automated size‐based filtration system for isolation of circulating tumor cells in lung cancer patients. PLoS ONE. 2017;12:e0179744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ilie M, Szafer‐Glusman E, Hofman V, et al. Expression of MET in circulating tumor cells correlates with expression in tumor tissue from advanced‐stage lung cancer patients. Oncotarget. 2017;8:26112‐26121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang BY, Li YS, Guo WB, et al. Detection of driver and resistance mutations in leptomeningeal metastases of NSCLC by next‐generation sequencing of cerebrospinal fluid circulating tumor cells. Clin Cancer Res. 2017;23:5480‐5488. [DOI] [PubMed] [Google Scholar]
- 36. Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung‐cancer cells. N Engl J Med. 2008;359:366‐377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood‐based analyses. Clin Cancer Res. 2016;22:1103‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Freidin MB, Freydina DV, Leung M, Montero Fernandez A, Nicholson AG, Lim E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin Chem. 2015;61:1299‐1304. [DOI] [PubMed] [Google Scholar]
- 39. Guibert N, Pradines A, Farella M, et al. Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS‐mutated lung adenocarcinoma. Lung Cancer. 2016;100:1‐4. [DOI] [PubMed] [Google Scholar]
- 40. Paz‐Ares L, Socinski MA, Shahidi J, et al. Correlation of EGFR‐expression with safety and efficacy outcomes in SQUIRE: a randomized, multicenter, open‐label, phase III study of gemcitabine‐cisplatin plus necitumumab versus gemcitabine‐cisplatin alone in the first‐line treatment of patients with stage IV squamous non‐small‐cell lung cancer. Ann Oncol. 2016;27:1573‐1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR‐targeted therapy in lung cancer. Nat Genet. 2012;44:852‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non‐small‐cell lung cancer. N Engl J Med. 2015;372:2018‐2028. [DOI] [PubMed] [Google Scholar]
- 43. Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD‐L1‐positive, advanced non‐small‐cell lung cancer (KEYNOTE‐010): a randomised controlled trial. Lancet. 2016;10027:1540‐1550. [DOI] [PubMed] [Google Scholar]
- 44. Reck M, Rodríguez‐Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD‐L1‐positive non‐small‐cell lung cancer. N Engl J Med. 2016;375:1823‐1833. [DOI] [PubMed] [Google Scholar]
- 45. McLaughlin J, Han G, Schalper KA, et al. Quantitative assessment of the heterogeneity of PD‐L1 expression in non‐small‐cell lung cancer. JAMA Oncol. 2016;2:46‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nicolazzo C, Raimondi C, Mancini M, et al. Monitoring PD‐L1 positive circulating tumor cells in non‐small cell lung cancer patients treated with the PD‐1 inhibitor Nivolumab. Sci Rep. 2016;6:31726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adams DL, Adams DK, He J, et al. Sequential tracking of PD‐L1 expression and RAD50 induction in circulating tumor and stromal cells of lung cancer patients undergoing radiotherapy. Clin Cancer Res. 2017;23:5948‐5958. [DOI] [PubMed] [Google Scholar]
- 48. Ilié M, Szafer‐Glusman E, Hofman V, et al. Detection of PD‐L1 in circulating tumor cells and white blood cells from patients with advanced non‐small‐cell lung cancer. Ann Oncol. 2018;29:193‐199. [DOI] [PubMed] [Google Scholar]
- 49. Lindsay CR, Faugeroux V, Michiels S, et al. A prospective examination of circulating tumor cell profiles in non‐small‐cell lung cancer molecular subgroups. Ann Oncol. 2017;28:1523‐1531. [DOI] [PubMed] [Google Scholar]
- 50. Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non‐small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res. 2012;18:2391‐2401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


