Abstract
Mitochondrial transcription factor A (TFAM) plays key roles in transcription and maintenance of mtDNA. It has been reported that TFAM could promote the proliferation and tumorigenesis of cells under stressed conditions. Previous evidence showed ionizing radiation stimulated the expression of TFAM, the replication of mtDNA, and the activity of mtDNA‐encoded cytochrome C oxidase. However, little is known about the mechanism of TFAM regulation in irradiated cells. In this article, we explored the role of mRNA stability in regulating TFAM expression in irradiated cancer cells. Our results showed that radiation stimulated the levels of TFAM mRNA and protein. RNA‐binding protein HuR associated and stabilized TFAM mRNA to facilitate the expression of TFAM, which was enhanced by radiation. Furthermore, radiation‐activated ataxia‐telangiectasia mutated kinase/p38 signaling positively contributed to the nucleus to cytosol translocation of HuR, its binding and stabilization of TFAM mRNA, without affecting the transcription and the stability of TFAM. Our current work proposed a new mechanism of DNA damage response‐regulated mitochondrial function variations, and indicated that TFAM might be a potential target for increasing the sensitization of cancer cells to radiotherapy.
Keywords: ATM/p38 signaling, HuR, mRNA stability, radiosensitivity, TFAM
1. INTRODUCTION
Mitochondrial transcription factor A (TFAM) is essential for the replication, transcription, and maintenance of mtDNA,1, 2, 3, 4, 5 and is transcriptionally controlled by peroxisome proliferator‐activated receptor‐γ coactivator‐1α/nuclear respiratory factor‐1.6, 7 It was reported that suppressing the expression of TFAM increased the sensitivity of cancer cells to chemotherapeutic drugs.8, 9 Ueta et al10 found that knockdown of TFAM expression increased radiation‐induced apoptosis in a radioresistant squamous cell carcinoma cell line. These findings suggested that TFAM might be a promising target for the control of tumors. The expression of TFAM can be stimulated by the binding of c‐Myc11 and p5312 to its promoter. In MDA‐MB‐231 cells, microRNA‐199a‐3p‐mediated attenuation in TFAM expression correlates with cellular sensitivity to cisplatin treatment.8 In addition, Torrens‐Mas et al13 found that knockdown of sirtuin 3, the major deacetylase of mitochondrial target proteins, affected TFAM protein level in breast cancer cells.
Previous studies reported that the stability of TFAM mRNA is related to striated muscle oxidative capacity and is mediated by RNA‐binding proteins (RBPs), suggesting that mRNA stability might constitute a critical rate‐limiting step in TFAM expression.14 RNA‐binding proteins interact with AU‐rich elements (AREs) within the 3′‐UTR of mRNAs.15, 16 AU‐rich elements might act as repressors of gene expression because their insertion into stable mRNAs renders them unstable.17, 18, 19 Elav‐type RNA‐binding proteins comprise the primarily neuronal proteins HuB/Hel‐N1, HuC, HuD, and the ubiquitous protein HuR.20, 21, 22 HuR can bind with high affinity and specificity to AREs to regulate the stability of mRNAs encoding stress‐response and proliferation‐related proteins such as cyclins, tumor suppressors, and oncogenes.23 Ionizing radiation increases intracellular reactive oxygen species (ROS), which is generated in large concentrations by the mitochondrial respiratory chain.24 Mitochondrial DNA is highly susceptible to damage caused by ROS.25 Previous studies indicated that radiation increased the TFAM‐mediated mitochondrial biogenesis in lung cancer cells.26 Therefore, we proposed a hypothesis that, in irradiated cancer cells, RBPs bind and stabilize TFAM mRNA, which resulted in accumulated expression of TFAM and decreased sensitivity to radiation. In this research, we examined the role of HuR in regulating TFAM expression. Our results showed that ataxia‐telangiectasia mutated kinase (ATM)/p38 DNA damage response facilitated the binding of HuR with TFAM mRNA. This led to stabilization of mRNA and increased protein expression of TFAM in irradiated cancer cells.
2. MATERIALS AND METHODS
2.1. Cell culture and irradiation
U‐2 OS, Hep G2, MCF7, and HCT 116 cells were from ATCC (Manassas, VA, USA) and cultured in DMEM/F12 supplemented with 10% FBS at 37°C in a humidified 5% CO2 incubator. Gamma irradiation was carried out in a Biobeam GM gamma irradiator (Leipzig, Germany) containing a cesium‐137 source with the dose rate of 3.27 Gy/min.
2.2. Chemicals and reagents
Cycloheximide (CHX) was from Calbiochem (Darmstadt, Germany). SB203580 was from Selleck (Houston, TX, USA). Actinomycin D was from Solarbio (Beijing, China). Ku55933 was from Santa Cruz (Dallas, TX, USA). The following primary antibodies were used: TFAM, β‐actin, proliferating cell nuclear antigen (Santa Cruz Biotechnology), α‐tubulin (Abcam, Cambridge, UK), p38 (pT180/pY182; Becton Dickinson, San Jose, CA, USA), ATM (pSer1981; Calbiochem), HuR (Millipore, Darmstadt, Germany), cleaved caspase 3 (Cell Signaling Technology, Danvers, MA, USA), B‐cell lymphoma 2, and BAX (Proteintech, Wuhan, China). The siRNA oligonucleotides were synthesized by GenePharma (Shanghai, China) and DNA primers were synthesized by General Biosystems (Chuzhou, China) (Table 1). TFAM shRNAs were purchased from OriGene (Rockville, MD, USA).
Table 1.
Oligonucleotides used in this research
| Human HuR siRNA1 | GAACGAAUUUGAUCGUCAATT |
| Human HuR siRNA2 | GCAGAUGUUUGGGCCGUUUTT |
| Human TFAM siRNA1 | GGACGAAACUCGUUAUCAUTT |
| Human TFAM siRNA2 | GGCAAGUUGUCCAAAGAAATT |
| Negative control siRNA | UUCUCCGAACGUGUCACGUTT |
| Human TFAM qRT‐PCR primers | GCGCTCCCCCTTCAGTTTTG |
| GTTTTTGCATCTGGGTTCTGAGC | |
| Human ACTIN qRT‐PCR primers | CCTGGCACCCAGCACAAT |
| GGGCCGGACTCGTCATAC | |
| Human GAPDH qRT‐PCR primers | TGCACCACCAACTGCTTAGC |
| GGCATGGACTGTGGTCATGA | |
| 12S rDNA qPCR primers | TAACCCAAG TCAATAGAAGCC |
| CTAGAGGGATATGAAGC ACC | |
| Human ACTIN qPCR primers | GAGCGGGAAATCGTGCGTGAC |
| GGAAGGAAGGCTGGAAGAGTG |
qPCR, quantitative PCR; qRT‐PCR, quantitative RT‐PCR.
2.3. Quantitative real‐time PCR
Total RNA was extracted using RNAiso (Takara, Shiga, Japan). Quantitative RT‐PCR (qRT‐PCR) was undertaken using One‐Step SYBR PrimeScript PLUS RT‐PCR Kit (Takara) according to the ΔΔCt method. Reverse transcription was carried out at 42°C for 10 minutes. The PCR conditions were: 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds, for 35 cycles. Primers are listed in Table 1. For relative mtDNA copy number quantification, the relative standard curve method was used. 12S rDNA encoded by mtDNA and β‐ACTIN encoded by nuclear DNA (Table 1) were quantified by SYBR Green quantitative PCR. The mtDNA/nDNA ratio was used to estimate the relative mtDNA copy number.
2.4. Western blot analysis
Samples were separated by SDS‐PAGE and then transferred to PVDF membranes. After blocking in PBST containing 1% BSA, the PVDF membrane was incubated with primary antibody at 4°C overnight. The membrane was then washed with PBST and incubated with a corresponding HRP‐linked secondary antibody for 2 hour at room temperature. Protein bands were visualized using chemiluminescence substrate and band density was analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.5. Messenger RNA and protein stability assays
Actinomycin D (1 μg/mL) was added to the culture medium 4 hours after irradiation. At the indicated time points, total RNA was prepared for qRT‐PCR. The TFAM mRNA half‐lives were calculated by being normalized to GAPDH mRNA levels and plotted on logarithmic scales. Time zero referred to the time point when actinomycin D was added to the medium. Cycloheximide (30 μg/mL) was added to the medium 6 hours after irradiation. At the indicated times, total cell lysates were prepared and analyzed by immunoblotting. Half‐lives of TFAM protein were calculated using a similar method as that used for mRNA.
2.6. Immunofluorescence assay
Cells were rinsed with PBS and fixed in 4% paraformaldehyde for 30 minutes. After washing with PBS, cells were blocked with PBS containing 1% BSA and 0.3% Triton X‐100 at 37°C for 1 hour. Then the cells were incubated with anti‐HuR antibody overnight at 4°C. After washing with PBS, cells were incubated for 1 hour with Alexa Fluor‐488 linked secondary antibody. Cell nuclei were stained with DAPI. Images were captured under a fluorescence microscope.
2.7. RNA immunoprecipitation
The association of endogenous HuR with TFAM mRNA was assessed by immunoprecipitation (IP) of the HuR–mRNA complex. Cytoplasmic lysates from U‐2 OS cells (3 × 107) were prepared in polysome lysis buffer: 100 mmol/L KCl, 5 mmol/L MgCl2, 10 mmol/L HEPES (pH 7.0), 0.5% NP‐40, 1 mmol/L dithiothreitol, RNase inhibitor, and protease inhibitors. After centrifugation, the supernatants were precleared for 30 minutes at 4°C using 15 μg normal rabbit IgG and 50 μL protein A‐agarose, which had been previously swollen in NT2 buffer (50 mmol/L Tris‐HCl [pH 7.4], 150 mmol/L NaCl, 1 mmol/L MgCl2, and 0.05% NP‐40). Protein A‐agarose (100 μL) was incubated with 10 μg normal rabbit IgG or rabbit anti‐HuR (Millipore) for 18 hours at 4°C, then incubated with precleared supernatants for further 1 hour at 4°C. After extensive washing and treatment with proteinase K, the RNA was extracted with RNAiso and used for qRT‐PCR analysis of TFAM mRNA. GAPDH mRNA, which does not bind to HuR, was used as the endogenous control.
2.8. Clonogenic assay
Cells were seeded into culture dishes. After irradiation, the dishes were incubated for 3 weeks. Then the dishes were washed with prewarmed PBS, fixed with methanol and acetic acid (9:1), and stained with crystal violet for 15 minutes. The colonies containing more than 50 cells were counted.
2.9. Cell proliferation assay
Cells were seeded into 24‐well plates at a density of 3 × 104 cells per well and incubated. Every 24 hours later, the growth of the cells was estimated by digesting the cells and counting the cell numbers. Cell growth within 7 days was recorded and plotted.
2.10. Cell cycle and apoptosis analysis
Analysis of the cell cycle distribution and the sub‐G1 population of cells was carried out on a cytoFLEX flow cytometer (Beckman Coulter, Brea, CA, USA). Briefly, cells were trypsinized then washed in PBS. After centrifugation, cell pellets were resuspended in 0.3 mL hypotonic buffer containing 0.1% sodium citrate, 0.1% Triton X‐100, 10 mg/mL RNase A, and 25 μg/mL propidium iodide, and then incubated for 30 minutes at 4°C. Samples were gated to exclude cell debris and analyzed by flow cytometry in linear mode by using the CytExprrt Software (Beckman Coulter).
2.11. Statistical analysis
Statistical data are represented as mean ± SD from at least three independent experiments undertaken in triplicate. Significant differences between two groups were determined by one‐way ANOVA using spss software (Armonk, NY, USA). *P < .05, **P < .01.
3. RESULTS
3.1. Ionizing radiation stimulates expression of TFAM
The protein levels of TFAM in U‐2 OS, Hep G2, and MCF7 cells exposed to 4 Gy γ‐radiation were detected. It was found that radiation stimulated the expression of TFAM (Figure 1A). We then detected the dose–response and post‐radiation time dependency of the levels of TFAM protein and mRNA in U‐2 OS cells. As shown in Figure 1B, 12 hours post exposure, the protein and mRNA levels of TFAM were upregulated with the increase of radiation dose. The results shown in Figure 1C indicate that the levels of TFAM protein and mRNA increased with the elongation of post‐radiation time within 24 hours after radiation. To determine the role of TFAM in mediating radiation sensitivity, TFAM in U‐2 OS and Hep G2 cells was knocked down by shRNA transfection (Figure 2A). Mitochondrial DNA copy number is a reflection of mitochondrial biogenesis. In the control U‐2 OS and Hep G2 cells, the mtDNA copy numbers showed an increasing tendency within 12 hours after irradiation, up to approximately 3.5‐fold compared to non‐irradiated cells. However, after TFAM was knocked down, the irradiation‐induced elevation of mtDNA copy number was suppressed (Figure 2B), confirming the role of TFAM in mitochondrial biogenesis. In addition, it was observed that knockdown of TFAM retarded cellular proliferation (Figure 2C). We further investigated whether TFAM affects irradiation‐induced cell killing effects. Both the clonogenic (Figure 2D) and apoptosis assay (Figure 3) showed that knockdown of TFAM sensitized U‐2 OS and Hep G2 cells to radiation. These results indicated that TFAM responded to irradiation and contributed to cell survival.
Figure 1.
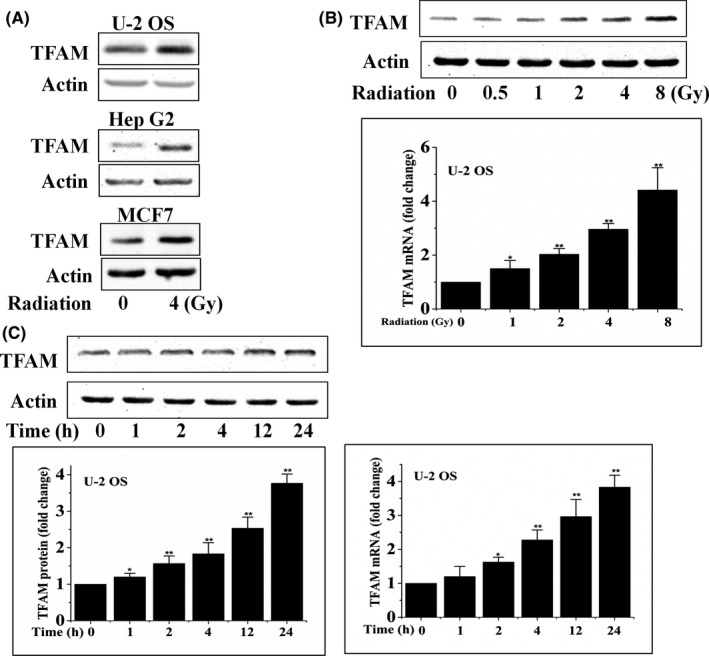
Mitochondrial transcription factor A (TFAM) expression was activated by radiation. A, Various cell lines were irradiated with 4 Gy γ‐rays. Twelve hours later, TFAM protein expression was analyzed by immunoblotting. B, U‐2 OS cells were irradiated with different doses of γ‐rays, and the protein and mRNA levels of TFAM were analyzed by immunoblotting and quantitative PCR at the indicated time points. C, U‐2 OS cells were irradiated with 4 Gy γ‐rays. At different time points, the protein and mRNA levels of TFAM were analyzed by immunoblotting and quantitative PCR, respectively. *P < .05; **P < .01
Figure 2.
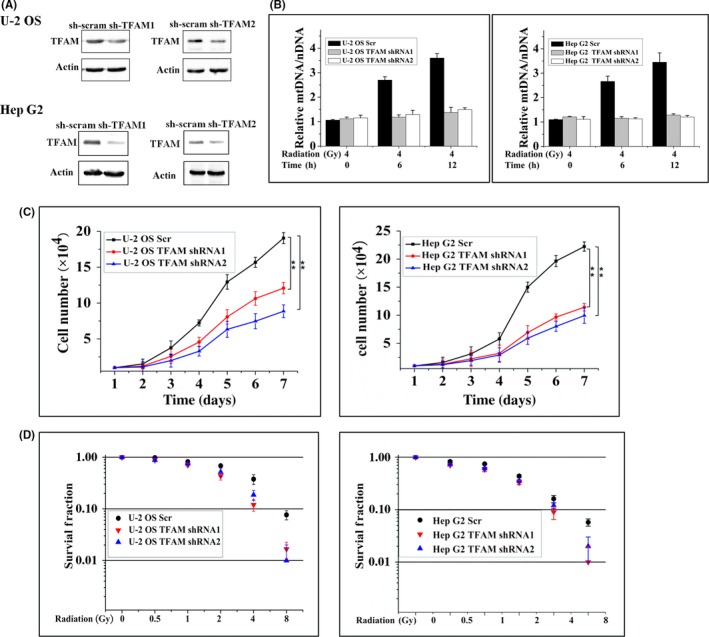
TFAM knockdown inhibits tumor cell proliferation and mtDNA copy number. A, Construction and verification of TFAM‐knockdown U‐2 OS and Hep G2 cell lines. scram, scramble control. B, Relative mtDNA copy number in irradiated control (Scr) or TFAM‐knockdown cells. C, Growth curves of control and TFAM knockdown cells. D, Colony formation results of control and TFAM‐knockdown cells treated with different doses of γ‐rays. **P < .01
Figure 3.
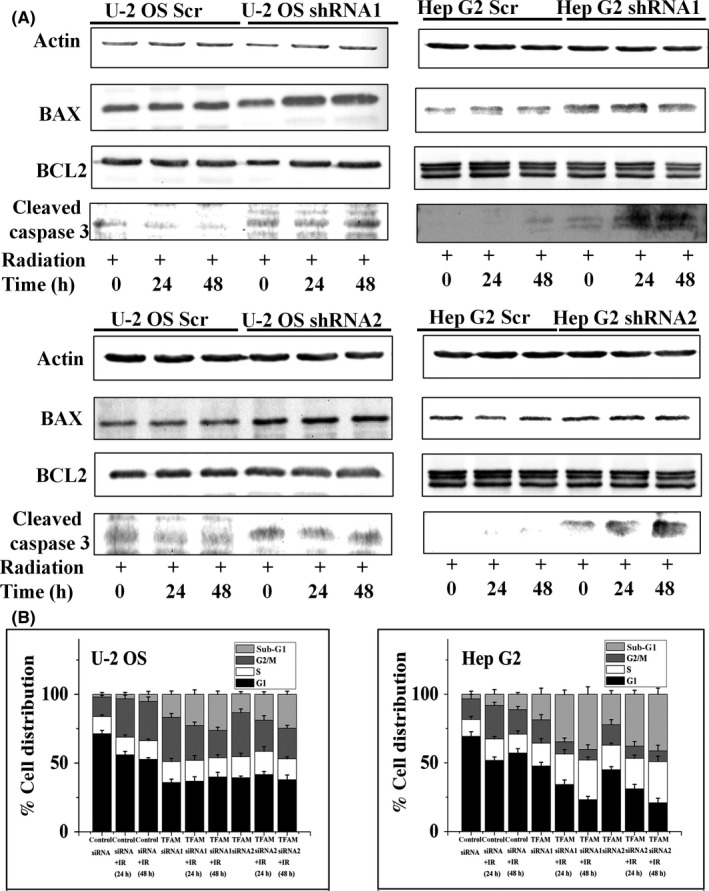
TFAM knockdown promotes apoptosis of tumor cells. A, B‐cell lymphoma 2 (BCL2), BAX, and cleavage of caspase 3 were analyzed in lysates from control and TFAM knockdown U‐2 OS and Hep G2 cells by immunoblotting at the indicated time points. B, U‐2 OS and Hep G2 cells were transfected with mitochondrial transcription factor A (TFAM) or scramble siRNA for 48 h. Cells were irradiated with 4 Gy γ‐rays. After incubation for 24 or 48 h, cells were harvested and cell cycle progression was analyzed on a flow cytometer. The percentage of sub‐G1 phase cells represents the ratio of apoptotic cells
3.2. Activation of ATM/p38 upregulates TFAM in irradiated cells
We next investigated whether the ATM/p38 pathway is involved in radiation‐induced upregulation of TFAM. Enhanced phosphorylation of p38 was observed in U‐2 OS cells 1 hour after different doses of radiation (Figure 4A). Application of p38 inhibitor SB203580 suppressed the elevation of TFAM in irradiated cells (Figure 4B). Additionally, as shown in Figure 4C, SB203580 treatment inhibited the upregulation of TFAM mRNA in irradiated U‐2 OS cells. We further explored the role of ATM in radiation‐stimulated phosphorylated p38/TFAM upregulation. As observed in Figures 5 and 6, in U‐2 OS and HCT 116 cell lines, phosphorylation of ATM was enhanced within 2 hour of irradiation. The ATM inhibitor ku55933 completely blocked the phosphorylation of ATM and p38. However, SB203580 did not inhibit the phosphorylation of ATM in irradiated cells. Both ku55933 and SB203580 suppressed the upregulation of TFAM. Taken together, activation of ATM/p38 signaling in irradiated cells promoted the expression of TFAM.
Figure 4.
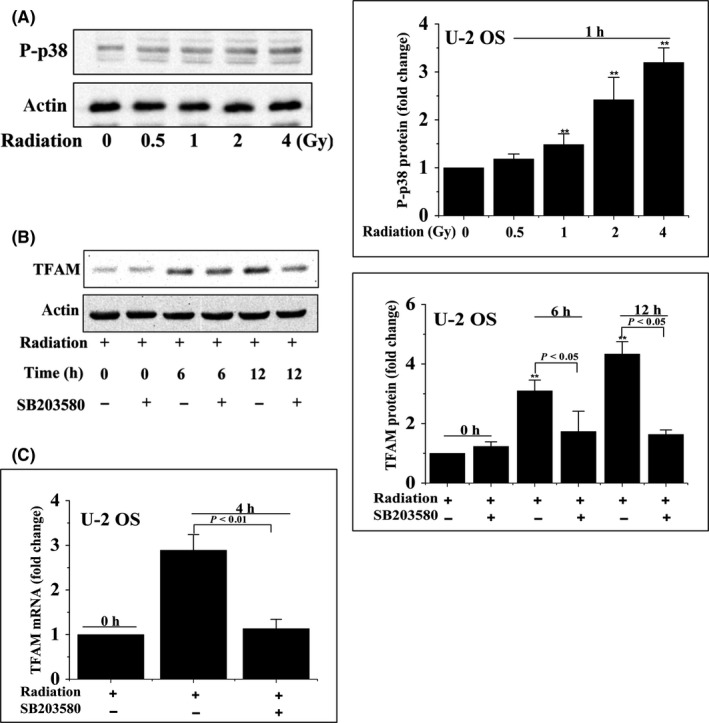
Activation of p38 upregulated mitochondrial transcription factor A (TFAM) in irradiated cells. A, U‐2 OS cells were irradiated and incubated for 1 h. The levels of phosphorylated p38 (P‐p38) were analyzed by immunoblotting. B, C, U‐2 OS cells were pretreated with SB203580 for 1 h and then irradiated with 4 Gy γ‐rays. Total cell lysates were analyzed by immunoblotting and quantitative PCR. **P < .01
Figure 5.
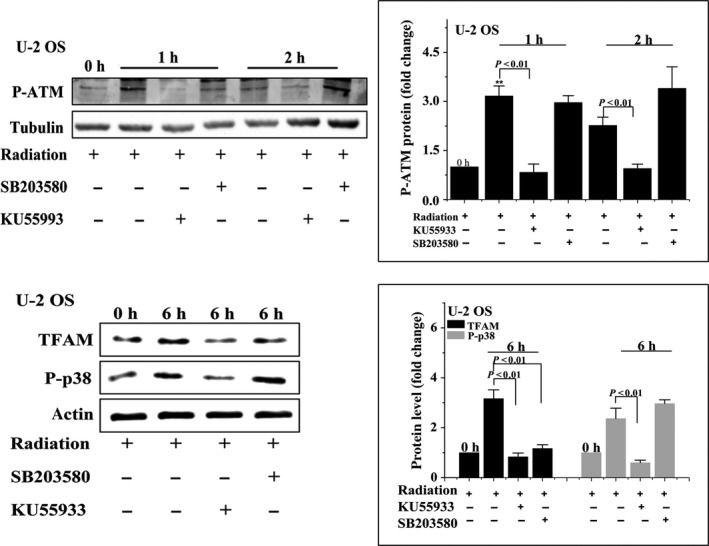
U‐2 OS cells were pretreated with ku55933 or SB203580 and then irradiated with 4 Gy γ‐rays. Phosphorylated ataxia‐telangiectasia mutated kinase (P‐ATM), phosphorylated p38 (P‐p38), and mitochondrial transcription factor A (TFAM) were analyzed by immunoblotting at the indicated time points. **P < .01
Figure 6.
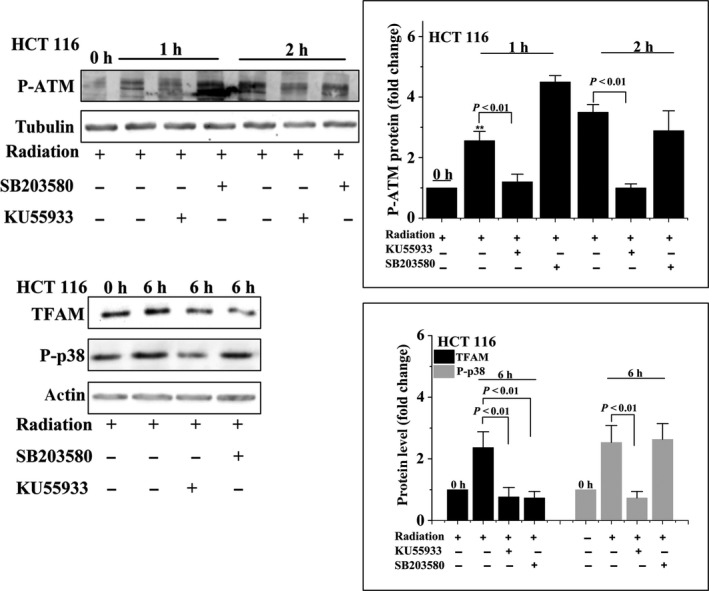
HCT 116 cells were pretreated with ku55933 or SB203580 and then irradiated with 4 Gy γ‐rays. Phosphorylated ataxia‐telangiectasia mutated kinase (P‐ATM), phosphorylated p38 (P‐p38), and mitochondrial transcription factor A (TFAM) were analyzed by immunoblotting at the indicated time points. **P < .01
3.3. HuR contributes to TFAM expression in irradiated cells
To explore the role of HuR in radiation‐induced upregulation of TFAM, HuR was knocked down by siRNA in U‐2 OS cells. The expression of TFAM in irradiated HuR knockdown cells was analyzed. As shown in Figure 7A, two different siRNAs targeted HuR‐blocked 4 Gy radiation‐induced upregulation of TFAM at 6 and 12 hours time points, respectively, indicating that HuR plays a positive role in TFAM expression in irradiated cells.
Figure 7.
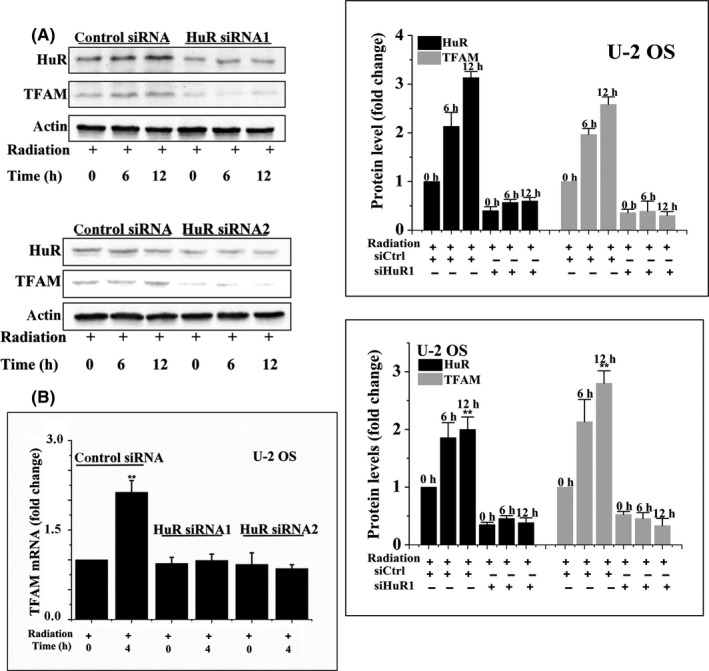
U‐2 OS cells were transfected with HuR siRNA1, HuR siRNA2, or control siRNA (siCtrl) for 48 h and then treated with 4 Gy γ‐rays. At the indicated time points, the levels of mitochondrial transcription factor A (TFAM) protein (A) and mRNA (B) were analyzed by immunoblotting and quantitative PCR. **P < .01
3.4. HuR binds and stabilizes TFAM mRNA in irradiated cells
As HuR is a well‐known RNA‐binding protein, we next examined whether HuR binds TFAM mRNA. Through a RNA‐IP followed by qRT‐PCR assay, it was confirmed that HuR bound the mRNA of TFAM in U‐2 OS and Hep G2 cells. The level of TFAM mRNA in the immunoprecipitate collected with HuR antibody increased by more than 20‐fold compared with that collected with control IgG. Furthermore, radiation significantly enhanced the binding of HuR with TFAM mRNA (Figure 8). However, knockdown of HuR by siRNAs in U‐2 OS cells prevented the induction of TFAM mRNA by radiation (Figure 7B). These results indicated that HuR binds and stabilizes the mRNA of TFAM in irradiated cells.
Figure 8.
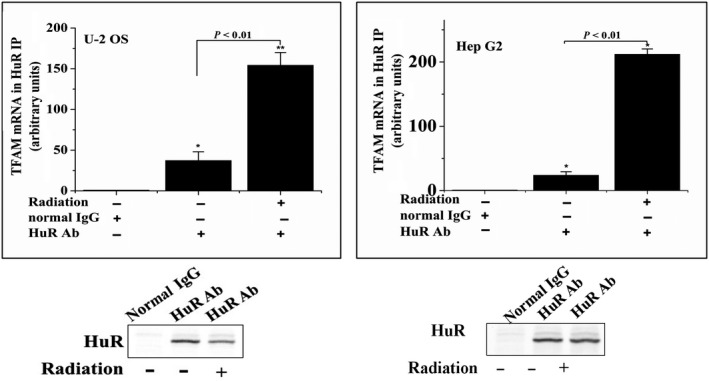
U‐2 OS and Hep G2 cells were irradiated with 4 Gy γ‐rays. Total cell lysates were immunoprecipitated (IP) with either HuR antibody or control normal IgG. RNA was collected from the immunoprecipitates and subjected for qRT‐PCR analysis. The levels of HuR in the immunoprecipitates were analyzed by immunoblotting. TFAM, mitochondrial transcription factor A. *P < .05
3.5. p38 regulates HuR‐dependent TFAM mRNA stabilization
Next, we investigated whether p38 regulated the binding of HuR to TFAM mRNA. Through RNA‐IP followed by qRT‐PCR assay, it was confirmed that application of SB203580 in irradiated U‐2 OS and Hep G2 cells evidently decreased the binding of HuR with TFAM mRNA (Figure 9A). The stabilities of TFAM protein or mRNA were then detected using U‐2 OS cells. Protein synthesis inhibitor CHX was added to the culture medium 6 hours post irradiation. At the time points 0, 30, 60, 90, or 120 minutes, whole cell lysate was collected for Western blot analysis. RNA synthesis inhibitor actinomycin D was added to the culture medium 4 hours post radiation. At the time points 0, 1, 2, 4, or 8 hours, total RNA was extracted for qRT‐PCR analysis (Figure 9B). The results showed that the presence or absence of SB203580 did not affect the stability of TFAM protein, reflected by the fact that neither radiation nor SB203580 significantly changed the half‐lives of TFAM protein. In terms of TFAM mRNA, it was observed that, in non‐irradiated cells, the half‐life of TFAM mRNA was approximately 60 minutes; in irradiated cells, it was approximately 265 minutes. Furthermore, SB203580 suppressed this tendency. These results indicated that p38 promoted radiation‐induced TFAM upregulation through stabilizing TFAM mRNA.
Figure 9.
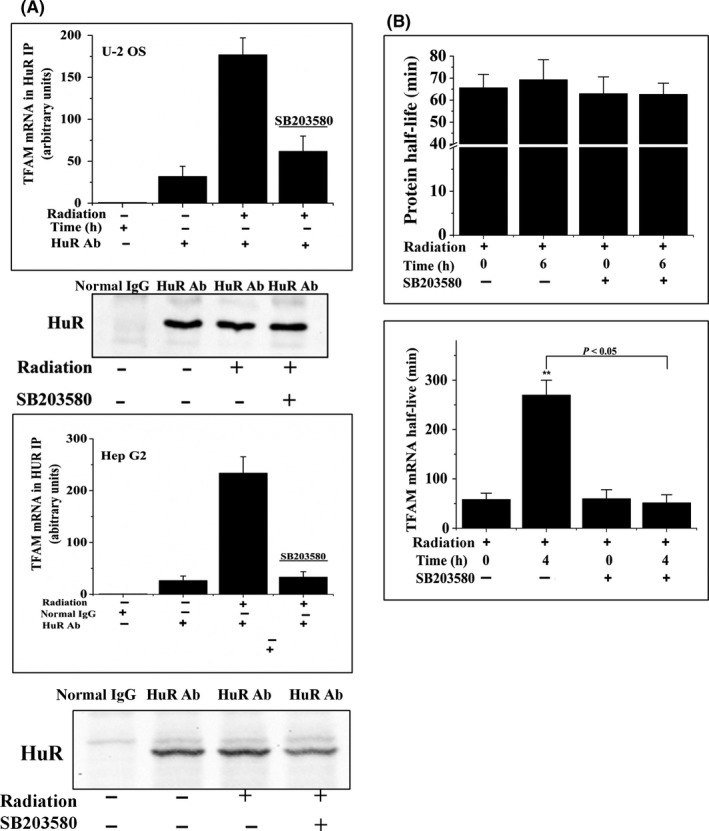
p38 regulated HuR‐dependent mitochondrial transcription factor A (TFAM) mRNA stabilization. A, U‐2 OS and Hep G2 cells were incubated in the presence or absence of SB203580 for 1 h and then irradiated with 4 Gy γ‐rays. RNA immunoprecipitation and quantitative RT‐PCR assays were carried out to analyze the levels of HuR‐bound TFAM mRNA. Levels of HuR in the immunoprecipitates were analyzed by immunoblotting. B, U‐2 OS cells were pretreated with SB203580 and then treated with radiation. Cycloheximide was added and incubated for 6 h (upper panel) or actinomycin D was added and incubated for 4 h (lower panel). Levels of TFAM protein and mRNA were analyzed by immunoblotting and quantitative RT‐PCR. **P < .01
Subcellular location of HuR in irradiated cells was also detected. As shown in Figure 10, HuR showed cytosolic translocation 1 hour after 4 Gy irradiation, reaching the maximum at 2 hour post radiation in U‐2 OS cells (Figure 10A), which was similar to Hep G2 cells (Figure 10B). However, SB203580 significantly blocked the cytosolic translocation of HuR in irradiated cells. Similar tendencies were observed in Western blotting detection of HuR in the cytosolic and nuclear fraction of irradiated cells under the conditions of mock or SB203580 treatment (Figure 11). These results showed that p38 promoted HuR cytosolic translocation in irradiated cells.
Figure 10.
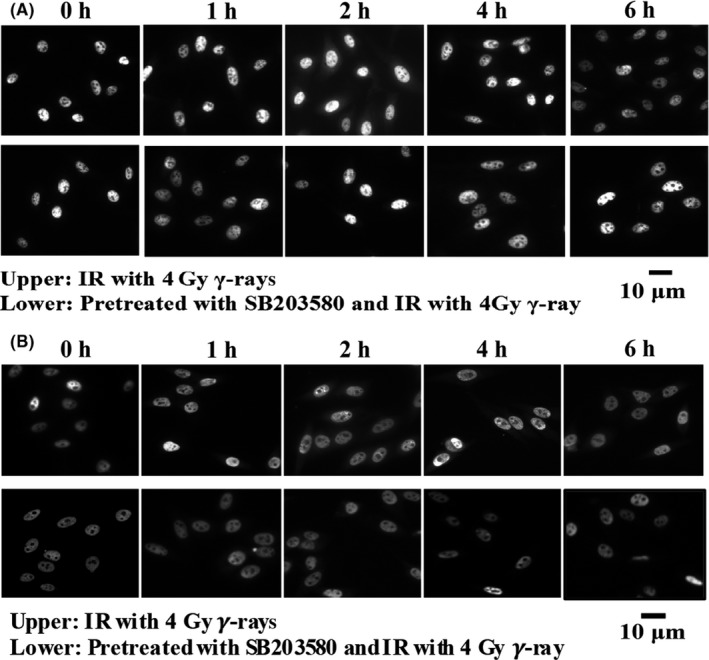
U‐2 OS (A) and Hep G2 (B) cells were irradiated (IR) with 4 Gy γ‐rays in the presence or absence of SB203580. HuR localization was visualized by fluorescence microscopy and analyzed by immunoblotting
Figure 11.
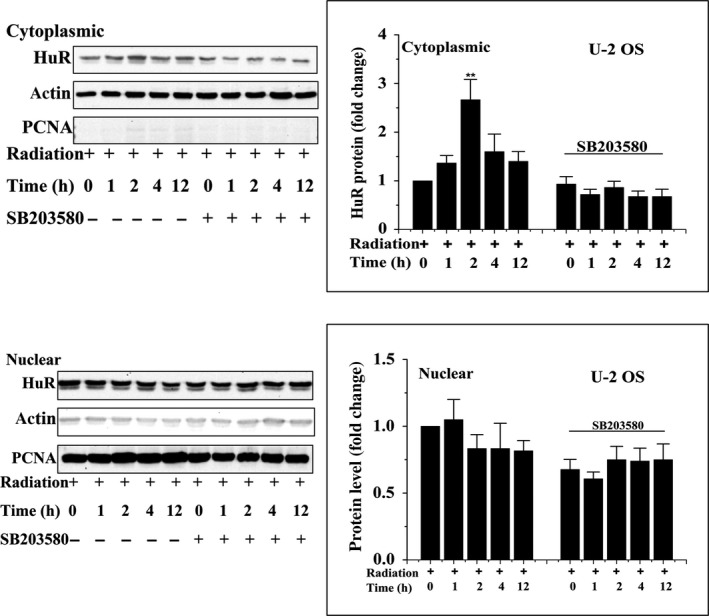
Subcellular localization of HuR in the presence or absence of SB203580 at different time points following 4 Gy γ‐ray irradiation was detected by Western blot analysis. **P < .01. PCNA, proliferating cell nuclear antigen
4. DISCUSSION
Following irradiation, mtDNA confronts elevated mitochondria‐derived active radicals and direct‐hit beams, resulting in mtDNA damage and de‐equilibrium of mitochondrial functions.27 TFAM is one of the key components involved in maintaining mtDNA integrity and mitochondrial functions. However, its roles in mediating the biological effects induced by radiation, and how it is regulated, require further exploration. Previous evidence showed that TFAM increased mtDNA copy numbers and maintained cytochrome C oxidase activity in irradiated lung cancer cells.26 Xie et al28 reported that downregulation of TFAM inhibits lung cancer cell tumorigenesis through reducing cellular bioenergetics, activating the ROS‐mediated JNK/p38 and p53/p21 signaling ways. Lee et al29 found enhanced TFAM expression in glioma. In TFAM knockout mice, massive apoptotic embryos were observed. In a heart‐specific TFAM knockout mouse model, increased apoptosis levels in heart were observed.30 Depletion of TFAM resulted in the upregulation of the levels of apoptotic proteins (BAX, caspase 3, and caspase 9 cleavage), as well as tumor suppressor p53, phosphorylated p53 (ser15), and p21 in non‐small‐cell lung cancer cells, which led to cell cycle arrest and the induction of apoptosis.28 Our study showed that radiation stimulated the levels of TFAM mRNA and protein in different cancer cells, and knockdown of TFAM reduced the colony‐forming capability and increased apoptosis rates in irradiated cells. These results were consistent with the findings that TFAM knockdown cancer cells showed increased sensitivity to cisplatin and doxorubicin treatment.9 Clinical studies have also observed that high TFAM expression was correlated with lower patient survival rate in colorectal cancers.31 Yoshida et al32 reported that in patients with metastatic colorectal cancer treated with modified 5‐fluorouracil, leucovorin, and oxaliplatin 6, immunohistochemical study of TFAM might be useful for predicting the clinical outcome. In pancreatic ductal adenocarcinoma, Yamauchi et al33 reported that TFAM expression played a pivotal role in worsening the postoperative clinical course. A univariate survival analysis showed that the survival rate of patients with TFAM‐negative endometrioid adenocarcinoma was significantly better than that for patients with TFAM‐positive endometrioid adenocarcinoma.34 These findings suggested that TFAM expression might be a useful marker for the progression of tumors and a potential target for improving tumor therapy outcomes.
Nuclear respiratory factor‐1 is the canonical transcription factor controlling the expression of TFAM.6, 7 It has been reported that other regulators, such as AMPK35 and nuclear factor‐κB,36 also affect the expression of TFAM. Post‐translational modification by ERK1/2 phosphorylation influenced TFAM function.37 D'souza et al14 reported that, in striated muscle, the stability of TFAM mRNA was correlated with the protein expression levels of RBPs including HuR, indicating that mRNA stability might contribute to TFAM expression. However, how this type of regulation is carried out remains unknown. HuR binds the AREs within 3′‐UTRs to stabilize mRNAs. Results from Badawi et al38 showed silencing of HuR increased the sensitivity of colorectal cancer cells to radiation. With our former observation that TFAM mRNA was significantly upregulated in irradiated cancer cells and AU‐rich motif was found in TFAM mRNA, we therefore detected whether HuR binds TFAM mRNA. Knockdown of HuR decreased the induction of TFAM protein and mRNA in irradiated cells. The RNA‐IP assay showed that HuR associated with TFAM mRNA, and the binding of HuR and TFAM mRNA was reinforced by radiation, confirming that TFAM mRNA is a new target of HuR.
It was previously reported that p38 regulated the HuR‐mediated stabilization of c‐fos mRNA in growth factor‐stimulated cells.39 Liao et al40 found in A549 cells that p38 mediated the binding of HuR with cytosolic phospholipase A2α mRNA under interleukin‐1β treatment. As p38 is an important regulator of radiation‐induced biological responses,41 we checked whether it affects the binding of HuR with TFAM mRNA. It was found that p38 inhibitor SB203580 decreased the binding of HuR with the mRNA of TFAM. In addition, SB203580 inhibited the translocation of HuR from the nucleus to cytosol, which is thought to be an indication of HuR activation.
We further confirmed that p38 affected the expression of TFAM protein through preventing mRNA decay, verifying the importance of mRNA stability in the upregulation of TFAM in irradiated cancer cells. Ataxia‐telangiectasia mutated kinase is autophosphorylated at Ser1981 following radiation to promote DNA damage responses.42 TAO kinases are intermediates in the activation of p38 by ATM under stress conditions, including radiation.43 Our work confirmed that in irradiated cells, suppression of ATM resulted in decreased phosphorylation of p38 and attenuated induction of TFAM expression. Fu et al35 found that ATM‐dependent AMPK activation promoted the expression of TFAM in etoposide‐treated cells. Our work showed that ATM could regulate TFAM through p38‐dependent regulation of mRNA stability. This finding brought new insight into the regulation of mitochondrial functions by DNA damage responses.
In conclusion, the current work provided evidence to show suppression of TFAM, a regulator of mitochondrial biogenesis, could decrease the survival of cancer cells after irradiation, which might be a potential technique to increase cancer cells’ sensitivity to radiotherapy. We also clarified an ATM/p38‐dependent and HuR‐mediated mRNA stability pathway in the regulation of radiation‐induced TFAM in cancer cells, providing new information in understanding how DNA damage responses affect mitochondrial functions in stressed conditions.
CONFLICT OF INTEREST
The authors have no conflict of interest.
ACKNOWLEDGMENTS
This work was supported by grants from the International Thermonuclear Experimental Reactor program (2014GB112006), the National Natural Science Foundation of China projects (31370842 and 11575232), the International Partnership Program of the Chinese Academy of Sciences (116134KYSB20160084), and the Innovative Program of the Development Foundation of Hefei Center for Physical Science and Technology (2016FXCX005).
Zhang R, Wang J. HuR stabilizes TFAM mRNA in an ATM/p38‐dependent manner in ionizing irradiated cancer cells. Cancer Sci. 2018;109:2446–2457. 10.1111/cas.13657
Funding information
International Thermonuclear Experimental Reactor program; National Natural Science Foundation of China; Chinese Academy of Sciences; Development Foundation of Hefei Center for Physical Science and Technology
REFERENCES
- 1. Acin‐Perez R, Salazar E, Kamenetsky M, Buck J, Levin L, Manfredi G. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab. 2009;9:265‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ekstrand M, Falkenberg M, Rantanen A, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2014;13:935‐944. [DOI] [PubMed] [Google Scholar]
- 3. Larsson N, Wang J, Wilhelmsson H, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231‐236. [DOI] [PubMed] [Google Scholar]
- 4. Lu B, Lee J, Nie X, et al. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell. 2013;49:121‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaufman B, Durisic N, Mativetsky J, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid‐like structures. Mol Biol Cell. 2007;18:3225‐3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morishita M, Kawamoto T, Hara H, et al. AICAR induces mitochondrial apoptosis in human osteosarcoma cells through an AMPK‐dependent pathway. Int J Oncol. 2017;50:23‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoshino M, Naka A, Sakamoto Y, et al. Dietary isoflavone daidzein promotes Tfam expression that increases mitochondrial biogenesis in C2C12 muscle cells. J Nutr Biochem. 2015;26:1193‐1199. [DOI] [PubMed] [Google Scholar]
- 8. Fan X, Zhou S, Zheng M, Deng X, Yi Y, Huang T. MiR‐199a‐3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM (TFAM). Biomed Pharmacother. 2017;88:507‐514. [DOI] [PubMed] [Google Scholar]
- 9. Mei H, Sun S, Bai Y, Chen Y, Chai R, Li H. Reduced mtDNA copy number increases the sensitivity of tumor cells to chemotherapeutic drugs. Cell Death Dis. 2015;6:e1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ueta E, Sasabe E, Yang Z, Osaki T, Yamamoto T. Enhancement of apoptotic damage of squamous cell carcinoma cells by inhibition of the mitochondrial DNA repairing system. Cancer Sci. 2008;99:2230‐2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin X, Tang L, Yang J, Xu W. HIF‐1 regulates insect lifespan extension by inhibiting c‐Myc‐TFAM signaling and mitochondrial biogenesis. Biochim Biophys Acta. 2016;1863:2594‐2603. [DOI] [PubMed] [Google Scholar]
- 12. Wen S, Gao J, Zhang L, Zhou H, Fang D, Feng S. p53 increase mitochondrial copy number via up‐regulation of mitochondrial transcription factor A in colorectal cancer. Oncotarget. 2016;7:75981‐75995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Torrens‐Mas M, Pons D, Sastre‐Serra J, Oliver J, Roca P. SIRT3 silencing sensitizes breast cancer cells to cytotoxic treatments through an increment in ROS production. J Cell Biochem. 2017;118:397‐406. [DOI] [PubMed] [Google Scholar]
- 14. D'souza D, Lai R, Shuen M, Hood D. mRNA stability as a function of striated muscle oxidative capacity. Am J Physiol Regul Integr Comp Physiol. 2012;303:R408‐R417. [DOI] [PubMed] [Google Scholar]
- 15. Chen C, Shyu A. AU‐rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465‐470. [DOI] [PubMed] [Google Scholar]
- 16. Kenan D, Query C, Keene J. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214‐220. [DOI] [PubMed] [Google Scholar]
- 17. Gorospe M, Wang X, Holbrook N. p53‐dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate‐sensitive regulatory system. Mol Cell Biol. 1998;18:1400‐1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shyu A, Greenberg M, Belasco J. The c‐fos transcript is targetedfor rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60‐72. [DOI] [PubMed] [Google Scholar]
- 19. Treisman R. Transient accumulation of c‐fos RNA following serum stimulation requires a conserved 5′ element and c‐fos 3′ sequences. Cell. 1985;42:889‐902. [DOI] [PubMed] [Google Scholar]
- 20. Antic D, Lu N, Keene J. ELAV tumor antigen, Hel‐N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarci‐ noma cells. Genes Dev. 1999;13:449‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brennan C, Steitz J. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma W, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav‐like protein. J Biol Chem. 1996;271:8144‐8151. [DOI] [PubMed] [Google Scholar]
- 23. Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple functions of the RNA‐binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14:10015‐10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Azzam E, Jay‐Gerin J, Pain D. Ionizing radiation‐induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012;327:48‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yakes F, VanHouten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514‐519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu J, Wang Q, Chen N, et al. Mitochondrial transcription factor A regulated ionizing radiation‐induced mitochondrial biogenesis in human lung adenocarcinoma A549 cells. J Radiat Res. 2013;54:998‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang B, Davidson M, Zhou H, Wang C, Walker W, Hei T. Cytoplasmic irradiation results in mitochondrial dysfunction and DRP1‐dependent mitochondrial fission. Cancer Res. 2013;73:6700‐6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie D, Wu X, Lan L, et al. Downregulation of TFAM inhibits the tumorigenesis of non‐small cell lung cancer by activating ROS‐mediated JNK/p38MAPK signaling and reducing cellular bioenergetics. Oncotarget. 2016;7:11609‐11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee H, Park J, Tran Q, et al. Mitochondrial transcription factor A (TFAM) is upregulated in glioma. Mol Med Rep. 2017;15:3781‐3786. [DOI] [PubMed] [Google Scholar]
- 30. Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson NG. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci USA. 2001;98:4038‐4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katsuki T, Nakayama Y, Akiyama M, Sawatsubashi Y, Nagata J, Minagawa N. Prognostic significance of mitochondrial transcription factor A expression in patients with right‐ or left‐sided colorectal cancer. Anticancer Res. 2018;38:569‐575. [DOI] [PubMed] [Google Scholar]
- 32. Yoshida Y, Hasegawa J, Nezu R, et al. Clinical usefulness of mitochondrial transcription factor A expression as a predictive marker in colorectal cancer patients treated with FOLFOX. Cancer Sci. 2011;102:578‐582. [DOI] [PubMed] [Google Scholar]
- 33. Yamauchi M, Nakayama Y, Minagawa N, Torigoe T, Shibao K, Yamaguchi K. Mitochondrial transcription factor A worsen the clinical course of patients with pancreatic cancer through inhibition of apoptosis of cancer cells. Pancreas. 2014;43:405‐410. [DOI] [PubMed] [Google Scholar]
- 34. Toki N, Kagami S, Kurita T, Kawagoe T, Matsuura Y, Hachisuga T. Expression of mitochondrial transcription factor A in endometrial carcinomas: clinicopathologic correlations and prognostic significance. Virchows Arch. 2010;456:387‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu X, Wan S, Lyu Y, Liu L, Qi H. Etoposide induces ATM‐dependent mitochondrial biogenesis through AMPK activation. PLoS ONE. 2008;3:e2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suliman H, Sweeney T, Withers C, Piantadosi C. Co‐regulation of nuclear respiratory factor‐1 by NFkappaB and CREB links LPS‐induced inflammation to mitochondrial biogenesis. J Cell Sci. 2010;123:2565‐2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang K, Zhu J, Dagda R, et al. ERK‐mediated phosphorylation of TFAM downregulates mitochondrial transcription: implications for Parkinson's disease. Mitochondrion. 2014;17:132‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Badawi A, Hehlgans S, Pfeilschifter J, Rödel F, Eberhardt W. Silencing of the mRNA‐binding protein HuR increases the sensitivity of colorectal cancer cells to ionizing radiation through upregulation of caspase‐2. Cancer Lett. 2017;393:103‐112. [DOI] [PubMed] [Google Scholar]
- 39. Degese M, Tanos T, Naipauer J, et al. An interplay between the p38 MAPK pathway and AUBPs regulates c‐fos mRNA stability during mitogenic stimulation. Biochem J. 2015;467:77‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liao W, Wang W, Chang W, Tseng J. The RNA‐binding protein HuR stabilizes cytosolic phospholipase A2α mRNA under interleukin‐1β treatment in non‐small cell lung cancer A549 cells. J Biol Chem. 2011;286:35499‐35508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Munshi A, Ramesh R. Mitogen‐activated protein kinases and their role in radiation response. Genes Cancer. 2013;4:401‐408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blackford A, Jackson S. ATM, ATR, and DNA‐PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801‐817. [DOI] [PubMed] [Google Scholar]
- 43. Raman M, Earnest S, Zhang K, Zhao Y, Cobb M. TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 2007;26:2005‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]


