Infections due to colistin-resistant (Colr) Gram-negative rods (GNRs) and colistin-resistant Klebsiella pneumoniae isolates in particular result in high associated mortality and poor treatment options. To determine the risk factors for recovery on culture of Colr GNRs and Colr K. pneumoniae, analyses were chosen to aid decisions at two separate time points: the first when only Gram stain results are available without any bacterial species information (corresponding to the Colr GNR model) and the second when organism identification is performed but prior to reporting of antimicrobial susceptibility testing results (corresponding to the Colr K. pneumoniae model).
KEYWORDS: antibiotic resistance, antibiotic stewardship, antimicrobial agents, clinical decision making, colistin, Gram-negative bacteria, antimicrobial testing, Gram-negative rods
ABSTRACT
Infections due to colistin-resistant (Colr) Gram-negative rods (GNRs) and colistin-resistant Klebsiella pneumoniae isolates in particular result in high associated mortality and poor treatment options. To determine the risk factors for recovery on culture of Colr GNRs and Colr K. pneumoniae, analyses were chosen to aid decisions at two separate time points: the first when only Gram stain results are available without any bacterial species information (corresponding to the Colr GNR model) and the second when organism identification is performed but prior to reporting of antimicrobial susceptibility testing results (corresponding to the Colr K. pneumoniae model). Cases were retrospectively analyzed at a major academic hospital system from 2011 to 2016. After excluding bacteria that were intrinsically resistant to colistin, a total of 28,512 GNR isolates (4,557 K. pneumoniae isolates) were analyzed, 128 of which were Colr (i.e., MIC > 2 μg/ml), including 68 of which that were Colr K. pneumoniae. In multivariate analysis, risk factors for Colr GNRs were neurologic disease, residence in a skilled nursing facility prior to admission, receipt of carbapenems in the last 90 days, prior infection with a carbapenem-resistant organism, and receipt of ventilatory support (c-statistic = 0.81). Risk factors for Colr K. pneumoniae specifically were neurologic disease, residence in a skilled nursing facility prior to admission, receipt of carbapenems in the last 90 days, receipt of an anti-methicillin-resistant Staphylococcus aureus antimicrobial in the last 90 days, and prior infection with a carbapenem-resistant organism (c-statistic = 0.89). A scoring system derived from these models can be applied by providers to guide empirical antimicrobial therapy in patients with infections with suspected Colr GNR and Colr K. pneumoniae isolates.
INTRODUCTION
The rising prevalence of infection caused by multidrug-resistant organisms (MDROs) is a worldwide problem with increasing costs and associated morbidity and mortality (http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf). In the United States, there are currently approximately 2 million annual cases of infections due to MDROs, with ∼23,000 attributable deaths and $50 million in directly attributable costs (https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf). Initial antibiotic selection remains a challenge. A delayed start of microbiologically active antibiotic therapy has been shown to increase mortality and hospital length of stay (1, 2), while overuse of broad-spectrum antibiotics has been linked with an increased prevalence of MDROs (3–7).
Colistin (polymyxin E) has been considered an antibiotic of last resort for the treatment of infections caused by multidrug-resistant (MDR) Gram-negative bacteria, due to neurotoxicity and nephrotoxicity, but it has become an increasingly important therapy when MDR pathogens are suspected and is, at times, the sole antimicrobial with activity against these organisms (8). Early, appropriate treatment of patients infected with colistin-resistant (Colr) Gram-negative rods (GNRs) and colistin-resistant Klebsiella pneumoniae isolates has been associated with reduced mortality (9–11). The prevalence of colistin resistance among GNRs has been increasing over the past 2 decades, particularly among isolates of Klebsiella pneumoniae (8, 10, 12–16), limiting the potential utility of colistin. Infection with Colr GNRs is associated with a higher mortality than infection with colistin-susceptible (Cols) isolates (9, 10, 17–20), making the rapid identification of resistance important. However, the early identification of Colr GNRs is challenging.
Prior literature has identified multiple risk factors for colistin resistance in GNRs, including recent prior hospitalization (17, 21, 22), prior carbapenem resistance (10, 13–15, 21, 23), prior treatment with colistin (18, 21, 23, 24), exposure to chlorhexidine (25), the presence of multiple comorbidities in the patients (21, 22), and increasing age, male sex, length of hospitalization, and the presence of indwelling urinary catheters (22). Other risk factors for the development of MDROs in general included prior residence in a nursing home, hemodialysis, intensive care unit (ICU) admission (26), the presence of a medical comorbidity (19, 27), and prior beta-lactam usage and invasive surgery (27).
We hypothesized that a large, adequately powered study would provide sufficient observations to identify easily obtainable clinical factors that could serve as a prediction tool for identifying patients at high risk for infection with colistin-resistant organisms, specifically, Colr K. pneumoniae.
MATERIALS AND METHODS
We conducted a retrospective study of all patients with positive cultures of specimens from any source over a 6-year period to develop a comprehensive model for the risk of infection with Colr GNRs, with a specific focus on Colr K. pneumoniae. The study was performed at two hospitals in metropolitan Los Angeles, CA. Ronald Reagan UCLA Medical Center is a 520-bed tertiary care center with five adult intensive care units totaling 109 beds. Santa Monica UCLA Hospital has 266 beds in total with 22 mixed intensive care beds in a single unit. Both are part of UCLA Health and serve patients with solid organ and bone marrow transplants, cancer, and various medical and surgical conditions. The Integrated Clinical and Research Data Repository (xDR) serves as a warehouse for all clinical data throughout UCLA Health since the implementation of electronic health records in 2006. The initial data set contained information from all admissions with start dates from January 2006 through November 2016 to either hospital for patients ≥18 years of age and at least one positive culture of a specimen from any source (blood, urine, sputum, wounds, or other fluids). Approval for this study was granted through the UCLA Institutional Review Board.
Since the endpoint of this analysis was prediction of development of the first colistin-resistant isolate, once the culture of a specimen from a patient grew a Colr GNR organism (defined as an organism for which the colistin MIC was >2 μg/ml), results for all cultured specimens collected from that patient at a time later than the time of collection of the original cultured specimen were removed from the data set. In order to focus on the clinically relevant event of transition from colistin susceptibility to resistance, isolates with intrinsic colistin resistance (Burkholderia, Morganella, Proteus, Providencia, and Serratia spp.) were excluded from the analysis.
Routine susceptibility testing was performed by the Clinical and Laboratory Standards Institute (CLSI) reference broth microdilution (BMD) method, using panels prepared in-house. Only data from 2011 and onwards were used in this study, as routine colistin susceptibility testing was not performed prior to 2011. After 2011, colistin susceptibility testing was routinely performed on all nonurinary specimens and was routinely performed for urinary specimens demonstrating carbapenem resistance. All antimicrobial susceptibility data were interpreted using CLSI breakpoints current to the year of testing. Because no CLSI breakpoints exist for the Enterobacteriaceae and colistin, the EUCAST breakpoints of ≤2 μg/ml to define susceptible and >2 μg/ml to define resistance were applied. These breakpoints are specific to the intravenous route of administration.
Predictor variables were chosen on the basis of prior studies, as were those with biologic plausibility that could readily be obtained from the medical record. Data collected for each patient included admission hospital, the number of days since admission, location prior to admission (home versus long-term-care facility or other hospital), demographic information, comorbidities (grouped into categories based on Elixhauser score designations) (28), laboratory results from the date of the culture, vital signs on the date that the specimen for culture was collected (maximum temperature, heart rate, and respiratory rate and minimum blood pressure), vital signs from initial hospital presentation, oxygen/ventilation method, presence of a tracheostomy, presence of a urinary catheter, administration of antibiotics and other selected medications (vasopressors, probiotics, blood products, immunosuppressants, and acid suppressants), the source of the specimen cultured, and prior culture positivity for carbapenem-resistant GNRs. Administration of antibiotics and the medications listed above was coded as the number of days since the last receipt of the medication, Winsorized to a maximum value of 100 (received within 24 h of the time of culture was coded as 0; never received was coded as 100 days since receipt). The variable “antipseudomonal carbapenem” refers to the receipt of meropenem, imipenem, or doripenem. The variable “anti-methicillin-resistant Staphylococcus aureus (MRSA)” agents refer to vancomycin, linezolid, and daptomycin, as these were used at the two institutions in cases of suspected hospital-acquired MRSA infection. Receipt of colistin was by any route, including the intravenous or inhalation route. An infection was coded as “hospital acquired” if the specimen for culture was submitted to the laboratory >48 h after the time of first presentation to the hospital.
The Elixhauser category of neurologic disease includes cerebral degeneration, movement disorders, degenerating neuropathies, seizure disorders, and anoxic brain injury (28). The construct of advanced ventilatory support includes patients receiving either noninvasive or invasive mechanical ventilation.
In cases where laboratory tests were not performed before specimens for culture were sent (typically at the beginning of a patient's admission), the first set of laboratory test results was used for that patient, provided that the tests were performed on specimens collected within 24 h of culture positivity. For laboratory tests not typically performed daily (e.g., liver function tests, measures of coagulation, and protein/prealbumin concentrations), the most recent result within a 48-hour period prior to culture positivity was used.
Statistical analysis.
Two separate analyses were performed: one compared all Cols GNRs against all Colr GNRs, and one compared only colistin-susceptible K. pneumoniae isolates against Colr K. pneumoniae isolates. These two analyses were chosen to aid decisions at two separate time points: the first when only Gram stain results were available without any bacterial species information (corresponding to the Colr GNR model) and the second when organism identification was performed but was performed prior to reporting of antimicrobial susceptibility testing results (corresponding to the Colr K. pneumoniae model). The measured variables in each case were compared between the cases and controls by a two-sided Mann-Whitney U test, Student's t test, or chi-squared test, as appropriate. In each case, after bivariate associations were examined, variables with P values of <0.10 or strong biologic plausibility were included in a stepwise forward model selection procedure to create a logistic regression model for each case/control pair; a P value of <0.10 was chosen as a threshold to include variables in the model to ensure that marginally significant variables with strong predictive effects were not inappropriately excluded. Only complete cases were included in the model selection. Model discrimination was assessed with the area under the receiver operating characteristic curve (c-statistic), and models were compared by the chi-squared test if they were nested or by use of the Akaike information criterion if they were not. In the original analysis, an MIC of >2 μg/ml was used for all organisms, but for Pseudomonas aeruginosa, the CLSI breakpoint is defined as an MIC of >4 μg/ml. As a sensitivity analysis, models were rerun by defining colistin resistance in Pseudomonas aeruginosa as an MIC of >4 μg/ml; this did not substantially change the results. All analyses were performed using the Stata statistical software package, version 14.2 (29).
RESULTS
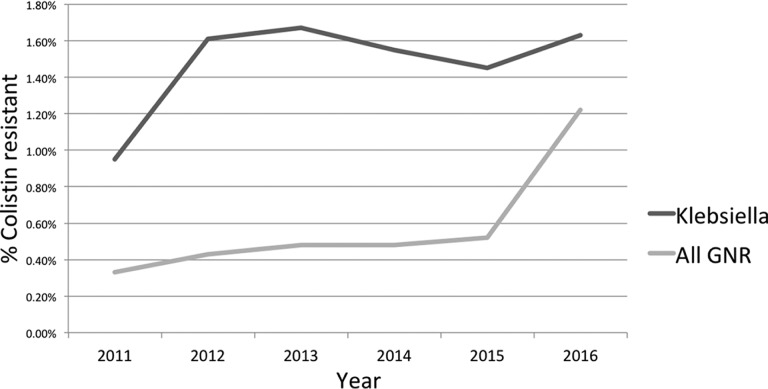
The overall data set included 28,512 GNR isolates from 12,388 patients, and 128 of these isolates were Colr. A total of 4,557 K. pneumoniae isolates were in the data set, and 68 of these were Colr. Since only complete cases were analyzed for the multivariate model, the final model for all GNRs comprised 15,372 cultures, 80 of which were Colr GNRs, and the final model for K. pneumoniae comprised 2,234 cultures, 34 of which were Colr K. pneumoniae. The incidence of development of Colr showed an upward trend for both GNRs and K. pneumoniae over the study period (Fig. 1). Among the Colr organisms, K. pneumoniae and Acinetobacter baumannii were overrepresented compared to colistin-susceptible organisms, while Escherichia and Pseudomonas spp. were underrepresented (Table 1). Among both all GNR and K. pneumoniae isolates, a respiratory source for the culture was predictive of Colr (Table 2). Since urinary isolates were not consistently tested for colistin resistance, they were excluded from Table 2 and the accompanying chi-squared test.
FIG 1.
Rates of new cases of colistin resistance by year for K. pneumoniae and all Gram-negative rods, excluding isolates with intrinsic colistin resistance.
TABLE 1.
Distribution of organisms for Cols and Colr culturesa
| Organism | % of isolates |
|
|---|---|---|
| Cols | Colr | |
| A. baumannii | 2.4 | 11.1 |
| Enterobacter spp. | 8.5 | 6.2 |
| Escherichia spp. | 37.6 | 0.4 |
| K. pneumoniae | 16.0 | 68.4 |
| Pseudomonas spp. | 22.3 | 7.6 |
| Stenotrophomonas spp. | 3.7 | 2.7 |
| Other | 9.5 | 3.6 |
P < 0.001 by the chi-squared test. Colr was defined as an MIC of >2 μg/ml.
TABLE 2.
Distribution of specimen source for Cols and Colr cultures among both all GNRs and K. pneumoniaea
| Specimen | % of isolates |
|||
|---|---|---|---|---|
| Cols GNRs | Colr GNRs | Cols K. pneumoniae | Colr K. pneumoniae | |
| Blood | 20.0 | 10.9 | 29.0 | 17.0 |
| Respiratory | 47.4 | 67.3 | 40.2 | 55.3 |
| Externalb | 11.5 | 10.9 | 8.8 | 17.0 |
| Other | 21.1 | 10.9 | 22.1 | 10.7 |
P < 0.001 for GNRs and P = 0.011 for K. pneumoniae by the chi-squared test.
External includes skin or a wound specimen source.
Bivariate analyses.
Selected bivariate associations are reported in Table 3. Risk factors were similar for all GNRs and for K. pneumoniae, although P values were generally smaller for GNRs due to the larger sample size. Consistent with information previously presented in the literature, risk factors included admission from a nursing home and location in an ICU (26), comorbidity (as measured by the Elixhauser score) (19, 22, 27), a prior specimen culture positive for a carbapenem-resistant organism (10, 13–15, 21, 23), and prior treatment with colistin (18, 21, 23, 24). Increasing age was associated with Colr K. pneumoniae but not Colr GNRs. The most prominent comorbidity associated with Colr was the neurologic disease category of the Elixhauser score. Several measures of chronic or acute respiratory failure were predictive of Colr, including whether the patient was currently receiving advanced ventilatory support, whether the patient had been on a ventilator during that hospitalization, and whether the patient had a tracheostomy at the time of specimen collection for culture or the time of admission. Laboratory values associated with ColR were higher white blood cell (WBC) counts, lower hemoglobin/hematocrit levels, and higher blood urea nitrogen (BUN) and alkaline phosphatase concentrations. The recent receipt of probiotics, blood products, and gastric acid suppressants prior to the day of culture was associated with Colr GNRs (and, occasionally, Colr K. pneumoniae; Table 3). A shorter time since receipt of the following antibiotics was associated with Colr GNRs (and, occasionally, Colr K. pneumoniae; Table 3): antipseudomonal carbapenems, ertapenem, penicillins, anti-MRSA agents, and colistin. Of note, steroids, chemotherapy (defined as cytotoxic drugs for the purposes of treating cancer), and immunosuppressants (a category including both steroids and chemotherapeutic agents) were not associated with an increased risk for Colr infections.
TABLE 3.
Selected bivariate associationsa
| Characteristic | Value(s) for GNRs |
Value(s) for K. pneumoniae |
||||
|---|---|---|---|---|---|---|
| Cols | Colr | P value | Cols | Colr | P value | |
| No. of patients | 28, 512 | 128 | 4, 557 | 68 | ||
| Mean (SD) age (yr) | 63.1 (19.3) | 65.4 (18.5) | 0.166 | 62.7 (18.4) | 69.1 (16.9) | 0.004 |
| % of male patients | 47.4 | 57 | 0.03 | 49.7 | 58.8 | 0.134 |
| % of patients by race | 0.08 | 0.054 | ||||
| White | 52.3 | 51.6 | 49.7 | 45.6 | ||
| Asian | 8.8 | 9.4 | 9.8 | 7.4 | ||
| Black | 10.9 | 17.2 | 12.0 | 23.5 | ||
| Latino | 21.5 | 14.1 | 22.4 | 16.2 | ||
| Other | 6.4 | 7.8 | 6.1 | 7.4 | ||
| Mean (SD) BMI (kg/m2) | 25.8 (6.7) | 25.6 (7.4) | 0.946 | 26.4 (7.4) | 25.5 (8.7) | 0.564 |
| % of patients admitted from health care facility | 14.3 | 45.7 | <0.001 | 13.2 | 55.9 | <0.001 |
| % of patients in hospital (RRMC vs SMH) | 65.7 | 56.1 | 0.198 | 64 | 53.8 | 0.283 |
| Median (IQR) log no. of days to culture | 0.69 (−1.35, 2.31) | 1.57 (−0.44, 3) | <0.001 | 0.86 (−1.27, 2.38) | 1.06 (−0.77, 2.82) | 0.029 |
| Proportion of cases hospital acquired | 0.463 | 0.516 | 0.237 | 0.487 | 0.515 | 0.647 |
| % of patients: | ||||||
| In ICU at time of culture | 20.3 | 33.6 | <0.001 | 21.5 | 32.4 | 0.014 |
| With any ICU stay during index hosp. | 40.3 | 61.2 | <0.001 | 43.1 | 68.3 | <0.001 |
| For whom current isolate is carbapenem resistant | 7.8 | 71.1 | <0.001 | 6.3 | 88.2 | <0.001 |
| With prior isolation of carbapenem-resistant GNR | 13.2 | 47.7 | <0.001 | 12.7 | 57.4 | <0.001 |
| With an indwelling urinary catheter | 43.8 | 76 | 0.002 | 44 | 75 | 0.01 |
| Ventilated during index hosp. | 32.4 | 65.1 | <0.001 | 33.4 | 68.3 | <0.001 |
| With tracheostomy present on day of culture | 12.1 | 30.2 | <0.001 | 8.5 | 24.4 | <0.001 |
| With tracheostomy present on admission | 5 | 16.3 | <0.001 | 3.3 | 19.5 | <0.001 |
| With advanced ventilation on day of culture | 24 | 51.2 | <0.001 | 25.1 | 51.2 | <0.001 |
| Median (IQR) Elixhauser score | 16 (7, 27) | 21 (11, 28) | 0.005 | 19 (9, 29) | 21.5 (11, 31) | 0.021 |
| % of patients with the following: | ||||||
| Congestive heart failure | 20.5 | 18 | 0.477 | 20.8 | 20.6 | 0.969 |
| Arrhythmia | 42.7 | 56.3 | 0.002 | 43.2 | 63.2 | <0.001 |
| Neurologic disease | 29.3 | 56.3 | <0.001 | 29.8 | 64.7 | <0.001 |
| Chronic pulmonary disease | 25.4 | 29.7 | 0.27 | 24 | 27.9 | 0.449 |
| Liver disease | 26.5 | 25 | 0.704 | 33 | 29.4 | 0.527 |
| Lymphoma | 4.4 | 4.7 | 0.862 | 3.8 | 2.9 | 0.726 |
| Metastatic cancer | 10.6 | 5.5 | 0.06 | 11.3 | 5.9 | 0.161 |
| Nonmetastatic cancer | 23.4 | 12.5 | 0.004 | 26.6 | 13.2 | 0.013 |
| Wt loss | 19.9 | 32 | <0.001 | 22.3 | 35.3 | 0.011 |
| Electrolyte disorder | 61.6 | 72.7 | 0.011 | 66.6 | 76.5 | 0.088 |
| Deficiency anemia | 13.2 | 14.1 | 0.777 | 14.5 | 14.7 | 0.963 |
| Drug abuse | 7.2 | 5.5 | 0.448 | 8 | 4.4 | 0.274 |
| Solid organ transplant | 19.1 | 21.1 | 0.559 | 20.6 | 16.2 | 0.367 |
| Bone marrow transplant | 1.6 | 1.6 | 0.991 | 1.3 | 0 | 0.345 |
| Renal failure | 15.3 | 24.2 | 0.005 | 16.8 | 29.4 | 0.006 |
| Cystic fibrosis | 1.9 | 6.3 | <0.001 | 0 | 0 | 0.863 |
| HIV infection | 0.8 | 0.8 | 0.992 | 0.5 | 1.5 | 0.292 |
| Alcohol use | 22.2 | 15.9 | 0.154 | 22.5 | 21.4 | 0.868 |
| Tobacco use | 5.6 | 5.5 | 0.972 | 5.5 | 9.4 | 0.347 |
| Vital signs on day of culture | ||||||
| Maximum temp (C) | 99.7 (1.6) | 99.5 (1.8) | 0.292 | 99.7 (1.8) | 99.6 (2.3) | 0.774 |
| Maximum pulse (beats/min) | 104.3 (23) | 108.6 (21.3) | 0.078 | 105.4 (23.5) | 111.5 (24.3) | 0.089 |
| Maximum respiratory rate (breaths/min) | 27.4 (9.8) | 28.7 (8.8) | 0.221 | 27.4 (9.7) | 28.8 (8.4) | 0.373 |
| Minimum SBP (mm Hg) | 101.2 (21.7) | 93.7 (21) | <0.001 | 100.1 (22) | 92.3 (24.3) | 0.01 |
| Minimum DBP (mm Hg) | 58.2 (12.3) | 54.6 (10.6) | 0.006 | 57.7 (12.3) | 54.5 (12.8) | 0.073 |
| Minimum MAP | 72.4 (14.3) | 67.6 (13.4) | 0.001 | 71.8 (14.3) | 67.1 (16) | 0.022 |
| % of patients with septic shock | 20.6 | 31.3 | 0.003 | 22.3 | 32.4 | 0.047 |
| % of patients with hypotension | 21.2 | 32 | 0.001 | 22.9 | 32.4 | 0.034 |
| Median (IQR) or mean (SD) lab results on day of culture | ||||||
| WBC count (109/liter) | 12.4 (8.6, 17.4) | 14.3 (9.8, 20.1) | 0.034 | 12.3 (8.3, 17.5) | 14.7 (9.8, 20.1) | 0.008 |
| Hemoglobin concn (mmol/liter) | 9.8 (8.5, 11.3) | 9.2 (8, 10.1) | <0.001 | 9.6 (8.4, 11.1) | 9.3 (8.2, 10.2) | 0.002 |
| Hematocrit (%) | 30.1 (26.3, 34.6) | 28.2 (25.2, 31.8) | <0.001 | 29.5 (25.7, 33.9) | 28.5 (25.4, 31.8) | 0.009 |
| Platelet count (109/liter) | 201 (124, 288) | 215 (123, 317) | 0.31 | 190 (106, 273) | 209 (130, 297) | 0.349 |
| Sodium concn (mmol/liter) | 137.3 (5.5) | 137.6 (5.9) | 0.55 | 137.2 (5.6) | 138.2 (6.7) | 0.159 |
| Potassium concn (mmol/liter) | 4.1 (0.6) | 4.1 (0.7) | 0.276 | 4.1 (0.6) | 4.1 (0.7) | 0.915 |
| Chloride concn (mmol/liter) | 102.4 (6.5) | 101.8 (7.2) | 0.205 | 102.7 (6.7) | 102.7 (8.7) | 0.948 |
| Bicarbonate concn (mmol/liter) | 24.5 (4.8) | 25.2 (5.4) | 0.112 | 23.7 (4.7) | 24 (4.5) | 0.612 |
| Anion gap (mmol/liter) | 10.4 (4) | 10.7 (5.2) | 0.347 | 10.8 (4.3) | 11.5 (5.6) | 0.162 |
| Creatinine concn (μmol/liter) | 1.4 (1.3) | 1.6 (1.4) | 0.048 | 1.4 (1.3) | 1.7 (1.4) | 0.042 |
| BUN concn (mmol/liter) | 28.4 (22.8) | 36.6 (26.3) | <0.001 | 29.5 (23.1) | 40.5 (27.2) | <0.001 |
| GFR (ml/min) | 71 (39, 100) | 61 (31, 100) | 0.081 | 67 (37, 100) | 62 (31.5, 100) | 0.069 |
| Glucose concn (mmol/liter) | 135.3 (57.1) | 139.5 (61) | 0.433 | 137.8 (60.9) | 141.5 (69) | 0.674 |
| Magnesium concn (mg/dl) | 1.7 (0.3) | 1.7 (0.3) | 0.399 | 1.7 (0.3) | 1.7 (0.3) | 0.859 |
| Calcium concn (mmol/liter) | 8.6 (0.9) | 8.7 (1) | 0.119 | 8.5 (0.9) | 8.6 (1) | 0.427 |
| Phosphorus concn (mmol/liter) | 3.3 (1.2) | 3.4 (1.3) | 0.171 | 3.3 (1.2) | 3.5 (1.2) | 0.187 |
| AST concn (U/liter) | 69.8 (363.5) | 51.6 (97.6) | 0.607 | 76.8 (336.1) | 65.6 (128.6) | 0.801 |
| ALT concn (U/liter) | 52 (187.7) | 47.7 (87.7) | 0.814 | 58.6 (178) | 59.4 (109.2) | 0.983 |
| Alkaline phosphatase concn (ng/ml) | 149.9 (168) | 217.5 (361.3) | <0.001 | 157.3 (178.2) | 260.7 (465.2) | <0.001 |
| aPTT (s) | 22.7 (15.3) | 20.9 (13.5) | 0.23 | 23.3 (15.9) | 22.1 (13.8) | 0.604 |
| INR | 1.3 (0.6) | 1.3 (0.3) | 0.434 | 1.4 (0.7) | 1.3 (0.4) | 0.814 |
| Lactate concn (mmol/liter) | 20.5 (22.5) | 19.6 (22) | 0.766 | 23.3 (24.4) | 22 (24.8) | 0.797 |
| D-dimer concn (ng/ml) | 3,021 (2,518) | 2,773 (2,699) | 0.686 | 2,979 (2,621) | 2,821 (2,474) | 0.85 |
| Prealbumin concn (g/liter) | 14.1 (8.1) | 14.1 (7.9) | 0.935 | 13.5 (8) | 12.6 (6.9) | 0.568 |
| Protein concn (g/liter) | 6.1 (1.1) | 6.1 (1.1) | 0.784 | 5.9 (1) | 6 (1.1) | 0.877 |
| Fibrinogen concn (g/liter) | 327.2 (180.2) | 334.6 (174.1) | 0.79 | 312.5 (177.3) | 326 (175.1) | 0.711 |
| Median (IQR) no. of days since last dose of: | ||||||
| Any antibiotic | 0 (0, 5) | 0 (0, 0) | 0.014 | 0 (0, 14) | 0 (0, 0) | 0.004 |
| An aminoglycoside | 100 (100, 100) | 100 (100, 100) | <0.001 | 100 (100, 100) | 100 (73, 100) | <0.001 |
| An antipseudomonal carbapenem | 100 (100, 100) | 100 (8, 100) | <0.001 | 100 (100, 100) | 100 (16, 100) | <0.001 |
| Ertapenem | 100 (100, 100) | 100 (100, 100) | <0.001 | 100 (100, 100) | 100 (100, 100) | <0.001 |
| A fluoroquinolone | 100 (100, 100) | 100 (29, 100) | 0.039 | 100 (100, 100) | 100 (42, 100) | 0.236 |
| A penicillin | 100 (2, 100) | 21 (0, 100) | 0.002 | 100 (0.5, 100) | 27.5 (0, 100) | 0.086 |
| An anti-MRSA agent | 60 (0, 100) | 2 (0, 42) | <0.001 | 58 (0, 100) | 1 (0, 49) | <0.001 |
| Colistin | 100 (100, 100) | 100 (63, 100) | <0.001 | 100 (100, 100) | 100 (100, 100) | <0.001 |
| Aztreonam | 100 (100, 100) | 100 (100, 100) | 0.705 | 100 (100, 100) | 100 (100, 100) | 0.876 |
| Carbapenem (any) | 100 (100, 100) | 60 (0, 100) | <0.001 | 100 (100, 100) | 100 (0, 100) | <0.001 |
| A beta-lactam | 0 (0, 83) | 0 (0, 17) | 0.041 | 3 (0, 100) | 0 (0, 19) | 0.011 |
| An acid suppressant | 0 (0, 100) | 0 (0, 0) | 0.014 | 0 (0, 88.5) | 0 (0, 0) | 0.332 |
| A probiotic | 100 (100, 100) | 100 (100, 100) | <0.001 | 100 (100, 100) | 100 (100, 100) | <0.001 |
| A steroid | 100 (20, 100) | 100 (13, 100) | 0.876 | 100 (19.5, 100) | 100 (54, 100) | 0.515 |
| Chemotherapy | 100 (100, 100) | 100 (100, 100) | 0.99 | 100 (100, 100) | 100 (100, 100) | 0.723 |
| An immunosuppressant | 100 (0, 100) | 100 (0, 100) | 0.582 | 100 (0, 100) | 100 (0, 100) | 0.811 |
| A blood product | 100 (100, 100) | 100 (11, 100) | 0.002 | 100 (40, 100) | 100 (11, 100) | 0.172 |
BMI, body mass index; RRMC, Ronald Reagan UCLA Medical Center; SMH, Santa Monica UCLA Hospital; ICU, intensive care unit; IQR, interquartile range; hosp., hospitalization; advanced ventilation, either noninvasive mask ventilation or endotracheal intubation; SBP, systolic blood pressure; DBP, diastolic blood pressure; MAP, mean arterial pressure; WBC, white blood cell; BUN, blood urea nitrogen; GFR, race-adjusted glomerular filtration rate; AST, aspartate aminotransferase; ALT, alanine aminotransferase; aPTT, activated prothrombin time; INR, international normalized ratio; antipseudomonal carbapenem, meropenem, imipenem, or doripenem.
Multivariate analyses.
Many of the variables that were significant on bivariate analysis were strongly colinear and were tested against each other in groups to determine which predictors were most representative from the various groupings of medical comorbidities, demographics, vital signs, laboratory values, indwelling devices, and recently administered medications. Next, representative predictors were added together, and the most parsimonious models were chosen. To facilitate model interpretability, the variables representing the number of days since receipt of medications were dichotomized to receipt within the prior 90 days versus not; this did not significantly affect the model fit.
For the model predicting Colr GNRs, the predictors in the final model were the presence of neurologic disease (as defined by the Elixhauser score), admission from a long-term-care facility, prior infection with a carbapenem-resistant organism, receipt of an antipseudomonal carbapenem in the prior 90 days, and receipt of advanced respiratory support at the time of culture (Table 4). The first three of these variables best represented the construct of chronic illness, while the last two represented antibiotic exposure and acute illness, respectively; this model had a c-statistic of 0.81.
TABLE 4.
Model specifications for Colr GNRs and Colr K. pneumoniae
| Organism and characteristic | Coefficient | SE | P value |
|---|---|---|---|
| Colr GNRs | |||
| Neurologic disease | 0.53 | 0.24 | 0.026 |
| Facility prior to admission | 0.96 | 0.24 | <0.001 |
| Carbapenems within 90 days | 0.75 | 0.25 | 0.002 |
| Intubation or NIVa | 0.64 | 0.25 | 0.009 |
| Prior carbapenem resistance | 0.80 | 0.26 | 0.003 |
| Colr K. pneumoniae | |||
| Neurologic disease | 0.75 | 0.37 | 0.041 |
| Facility prior to admission | 1.89 | 0.38 | <0.001 |
| Carbapenems within 90 days | 0.76 | 0.40 | 0.059 |
| Anti-MRSA within 90 days | 1.17 | 0.56 | 0.039 |
| Prior carbapenem resistance | 1.58 | 0.39 | <0.001 |
NIV, noninvasive ventilation.
For the model predicting Colr K. pneumoniae, the predictors in the final model were similar to those for the model predicting Colr GNRs: the presence of neurologic disease (as defined by the Elixhauser score), admission from a long-term-care facility, prior infection with a carbapenem-resistant organism, receipt of an antipseudomonal carbapenem in the prior 90 days, and receipt of an anti-MRSA agent in the prior 90 days. This model had a c-statistic of 0.89 (Table 4).
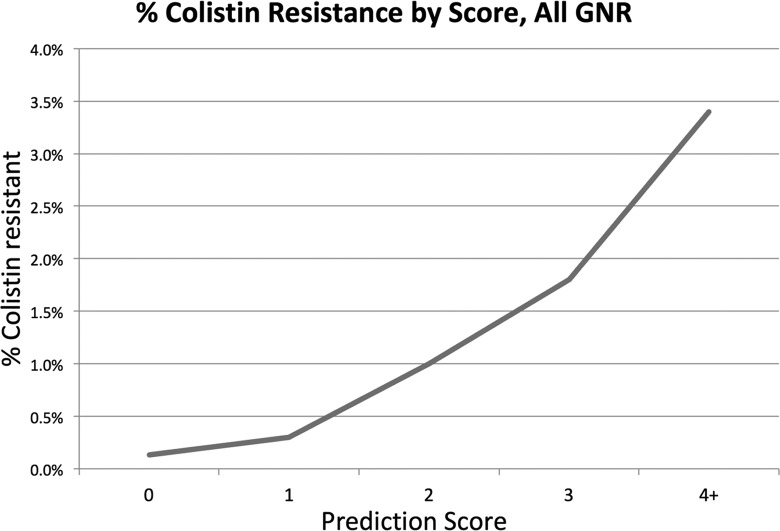
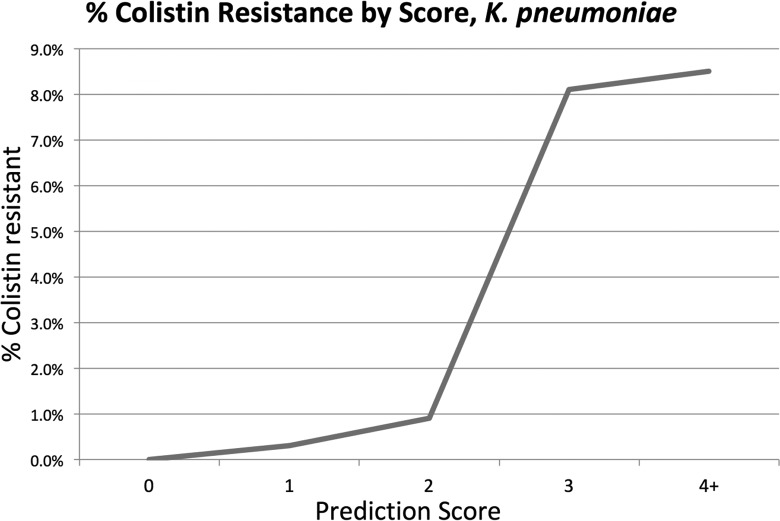
Treating each multivariate model as a score with 1 point assigned for each of the five items in the model, we created a potentially user-friendly tool to predict the probability of Colr in both situations. Figures 2 and 3 show the positive predictive value at each score total for GNRs and K. pneumoniae, respectively, and demonstrate that a higher score is associated with a higher likelihood of Colr, with scores of >3 representing a 3.5% risk of Colr GNRs and a 9.6% risk of Colr K. pneumoniae. Due to the amplified effects of random chance on a small number of cases with a score of 5, discontinuities are seen in both models at the highest score.
FIG 2.
Positive predictive value for colistin resistance at each score value for Gram-negative rods.
FIG 3.
Positive predictive value for colistin resistance at each score value for K. pneumoniae.
DISCUSSION
Infection with colistin-resistant organisms is associated with a substantially increased risk for mortality, and options for treatment are limited (9–11, 17–20). Prior studies have typically been relatively small or limited in scope, either following a relatively small number of patients (9, 10, 17, 18, 24, 26, 30–33) or focusing on a single organism (16, 21, 22, 27), and none have resulted in a clinically meaningful prediction tool. Our score can be calculated by providers at the time of decision making without the help of a computer. This potentially more accurately reflects a patient's risk for Colr organisms than a hospital-wide or unit-specific antibiogram, which can provide only a flat percent expected susceptibility for a given organism and is not useful for the management of rare events. Additionally, most hospital labs do not routinely test for colistin susceptibility, and it is rarely in a hospital's antibiogram. All information used in the models was from standardized data fields that were extracted from the medical record by an automated process without any requirement for human interpretation or processing of individual patient records, allowing this score to potentially be calculated automatically.
The results of our bivariate analysis are largely in line with those of prior similar studies, demonstrating the association with prior infections with carbapenem-resistant organisms (10, 13–15, 21, 23), treatment with colistin (18, 21, 23, 24), various medical comorbidities (19, 20, 22, 27), increasing age, male sex, length of stay, and presence of indwelling urinary catheters (22), and prior residence in a nursing home and ICU admission (26). While other models have focused on prior colistin exposure as a risk factor (21, 23, 24), our multivariate models suggest that exposure to other antibiotics (carbapenems and anti-MRSA agents) may serve as better markers for disease risk. While it is improbable that exposure to these medications mechanistically leads to the development of Colr, carbapenem receipt is likely a proxy for recent infection with MDR GNRs, while anti-MRSA receipt is a proxy for a recent concern for sepsis, as nearly all patients with suspicion for sepsis receive at least one dose of vancomycin at our institutions.
Several variables indicative of acute decompensation, specifically, an increased WBC count, an increased creatinine/blood urea nitrogen concentration, a lower blood pressure, and the presence of septic shock, were associated with an increased risk for recovery of a colistin-resistant isolate on bivariate analysis. However, none of these variables remained significant in the final model, and their discriminatory power in multivariate analysis was limited. While it is possible that these variables indicated clinical decompensation in the setting of an ineffectively treated infection, their contribution to the model was better explained by other variables, implying that the effect of acute decompensation is a partial mediator for a more significant construct, such as chronic medical illness.
Some of the risk factors observed in this study, particularly neurologic disease, weight loss, and recent receipt of carbapenems, were also shown to be predictive of resistance to carbapenems (34) and aminoglycosides (S. E. Richter, unpublished data). Additionally, these risk factors did not cluster temporally, reducing the probability that an outbreak isolated to a particular population or unit was responsible for the high prevalence of these risk factors in the affected population.
Our score for ruling in Colr GNRs is of limited utility, given that the predicted probability never exceeds 4%. The model is theoretically useful for ruling out Colr GNRs at low scores; further research will focus on thresholds for clinical decision making. The model for predicting Colr K. pneumoniae is significantly more useful, as a score of >3 indicates a nearly 10% chance of Colr K. pneumoniae, at which point colistin therapy is likely risky to use as a sole therapy. Since the score is substantially more useful after species identification, it is best paired with tools that allow for early identification, such as matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry or other rapid identification technologies.
While there is no consensus at this time on the optimal treatment of infections caused by organisms expressing resistance to both colistin and carbapenems, several novel agents have shown promise in this area. Two novel beta-lactam–beta-lactamase inhibitors, ceftazidime-avibactam and meropenem-vaborbactam, should retain microbiologic activity against most colistin-resistant carbapenem-resistant Enterobacteriaceae (CRE) isolates, with improved survival and clinical outcomes for complex infections being demonstrated for ceftazidime-avibactam (35) and meropenem-vaborbactam (36) compared to those achieved with the best available treatment, including colistin. Similarly, the aminoglycoside plazomicin should also retain microbiologic activity against most colistin-resistant CRE isolates, and treatment with plazomicin has shown improved survival and clinical outcomes for complex infections compared to those achieved with colistin (37).
A significant potential limitation of this study is its external validity and generalizability. It is possible that our models are not applicable to other medical systems. The work described has several safeguards that mitigate this effect. First, the data set draws information from two hospitals that serve somewhat different populations. Ronald Reagan UCLA Medical Center has a high proportion of patients undergoing transplants of either solid organs or bone marrow, a robust neurosurgery patient population, and a large number of patients with chronic severe medical illness. Santa Monica UCLA Hospital is a community hospital with a focus on geriatrics, orthopedic procedures, and solid oncology patients. Second, the data set contains data for tens of thousands of patients and is thus 1 to 2 orders of magnitude larger than the data sets for other similar studies examining risk factors for infection with resistant organisms. Third, as shown above, the bivariate predictor variables largely match up with the predictors described in previous work. Finally, in order to prevent overfitting, the final models were restricted to a small number of predictor variables. Nevertheless, external validity is always a concern, and further work will focus on validating these prediction scores on more diverse data sets.
Another limitation of this study comes as a consequence of our institutions' testing procedures, as urinary specimens were tested for colistin resistance only as a second step if they expressed carbapenem resistance. This potentially led to the misclassification of some Colr urinary isolates as Cols, and caution should be exercised when applying the model to urinary isolates.
The final number of Colr cases was moderate, and approximately 50% could not be included in the final analysis due to a lack of complete data across the relevant domains. Additionally, we had access only to data from inpatient hospitalizations within our hospital system, potentially excluding relevant information from outpatient encounters or treatment at other facilities. These limitations reflect the real-world data that are available at the time of decision making or for eventual integration of a similar score into an electronic health record. However, it is the largest investigation to date in terms of subject number and spans a period of 6 years, allowing us to examine far more potential explanatory variables than prior investigations of risk factors for the development of Colr in GNRs or K. pneumoniae. By performing a cohort study of patients with positive cultures, we eliminate potential selection bias in choosing controls and strengthen the validity of observed associations (38).
The initial empirical antibiotic choice is dependent on the pretest probability of colistin resistance, and initial empirical treatment with the higher-intensity regimens required for Colr organisms may not always be warranted. Lower scores can theoretically rule out Colr organisms and may be helpful in allowing the empirical use of colistin therapy in cases that would otherwise be considered at higher risk for resistance development; further research will focus on specific cutoffs for clinical decision making. In particular, this score could potentially improve care while awaiting the laboratory to perform colistin susceptibility testing, which may take days to weeks to a result, due to the limited availability of tests for colistin susceptibility testing.
Our study demonstrates the potential to harness currently available information from an existing electronic medical record to better inform clinical decision makers. In the current era of data-intensive medical care, we should harness all available information to better manage our patients. Further research will focus on validation of this score in other populations with other antibiotics and other pathogens and an analysis of cost-benefit thresholds for initiating specific antibiotic regimens in cases of uncertainty.
REFERENCES
- 1.Kollef MH, Sherman G, Ward S, Fraser VJ. 1999. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 115:462–474. doi: 10.1378/chest.115.2.462. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 3.MacKenzie FM, Bruce J, Struelens MJ, Goossens H, Mollison J, Gould IM, ARPAC Steering Group. 2007. Antimicrobial drug use and infection control practices associated with the prevalence of methicillin-resistant Staphylococcus aureus in European hospitals. Clin Microbiol Infect 13:269–276. doi: 10.1111/j.1469-0691.2006.01592.x. [DOI] [PubMed] [Google Scholar]
- 4.Neuhauser MM, Weinstein RA, Rydman R, Danzinger LH, Karam G, Quinn JP. 2003. Antibiotic resistance among gram-negative bacilli in US intensive care units: implications for fluoroquinolone use. JAMA 289:885–888. doi: 10.1001/jama.289.7.885. [DOI] [PubMed] [Google Scholar]
- 5.Polk RE, Johnson CK, McClish D, Wenzel RP, Edmond MB. 2004. Predicting hospital rates of fluoroquinolone-resistant Pseudomonas aeruginosa from fluoroquinolone use in US hospitals and their surrounding communities. Clin Infect Dis 39:497–503. doi: 10.1086/422647. [DOI] [PubMed] [Google Scholar]
- 6.Torres-Gonzalez P, Cervera-Hernandez ME, Niembro-Ortega MD, Leal-Vega F, Cruz-Hervert LP, Garcia-Garcia L, Galindo-Fraga A, Martinez-Gamboa A, Bobadilla-Del Valle M, Sifuentes-Osornio J, Ponce-de-Leon A. 2015. Factors associated to prevalence and incidence of carbapenem-resistant Enterobacteriaceae fecal carriage: a cohort study in a Mexican tertiary care hospital. PLoS One 10:e0139883. doi: 10.1371/journal.pone.0139883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fortin E, Platt RW, Fontela PS, Buckeridge DL, Quach C. 2015. Predicting antimicrobial resistance prevalence and incidence from indicators of antimicrobial use: what is the most accurate indicator for surveillance in intensive care units? PLoS One 10:e0145088. doi: 10.1371/journal.pone.0145088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granata G, Petrosillo N. 2017. Resistance to colistin in Klebsiella pneumoniae: a 4.0 strain? Infect Dis Rep 9:7104. doi: 10.4081/idr.2017.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machuca I, Gutierrez-Gutierrez B, Gracia-Ahufinger I, Rivera Espinar F, Cano A, Guzman-Puche J, Perez-Nadales E, Natera C, Rodriguez M, Leon R, Caston JJ, Rodriguez-Lopez F, Rodriguez-Bano J, Torre-Cisneros J. 2017. Mortality associated with bacteremia due to colistin-resistant Klebsiella pneumoniae with high-level meropenem resistance: importance of combination therapy without colistin and carbapenems. Antimicrob Agents Chemother 61:e00406-17. doi: 10.1128/AAC.00406-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rojas LJ, Salim M, Cober E, Richter SS, Perez F, Salata RA, Kalayjian RC, Watkins RR, Marshall S, Rudin SD, Domitrovic TN, Hujer AM, Hujer KM, Doi Y, Kaye KS, Evans S, Fowler VG Jr, Bonomo RA, van Duin D, Antibacterial Resistance Leadership Group. 2017. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis 64:711–718. doi: 10.1093/cid/ciw805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutiérrez-Gutiérrez B, Salamanca E, de Cueto M, Hsueh P-R, Viale P, Paño-Pardo JR, Venditti M, Tumbarello M, Daikos G, Cantón R, Doi Y, Tuon FF, Karaiskos I, Pérez-Nadales E, Schwaber MJ, Azap ÖK, Souli M, Roilides E, Pournaras S, Akova M, Pérez F, Bermejo J, Oliver A, Almela M, Lowman W, Almirante B, Bonomo RA, Carmeli Y, Paterson DL, Pascual A, Rodríguez-Baño J, del Toro MD, Gálvez J, Falcone M, Russo A, Giamarellou H, Trecarichi EM, Losito AR, García-Vázquez E, Hernández A, Gómez J, Bou G, Iosifidis E, Prim N, Navarro F, Mirelis B, Skiada A, Origüen J, Juan RS, Fernández-Ruiz M, et al. 2017. Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae (INCREMENT): a retrospective cohort study. Lancet Infect Dis 17:726–734. doi: 10.1016/S1473-3099(17)30228-1. [DOI] [PubMed] [Google Scholar]
- 12.Saly M, Jayol A, Poirel L, Megraud F, Nordmann P, Dubois V. 2017. Prevalence of faecal carriage of colistin-resistant Gram-negative rods in a university hospital in western France, 2016. J Med Microbiol 66:842–843. doi: 10.1099/jmm.0.000497. [DOI] [PubMed] [Google Scholar]
- 13.Giamarellou H. 2016. Epidemiology of infections caused by polymyxin-resistant pathogens. Int J Antimicrob Agents 48:614–621. doi: 10.1016/j.ijantimicag.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 14.Rossi F, Girardello R, Cury AP, Di Gioia TS, Almeida JN Jr, Duarte AJ. 2017. Emergence of colistin resistance in the largest university hospital complex of Sao Paulo, Brazil, over five years. Braz J Infect Dis 21:98–101. doi: 10.1016/j.bjid.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayol A, Poirel L, Dortet L, Nordmann P. 2016. National survey of colistin resistance among carbapenemase-producing Enterobacteriaceae and outbreak caused by colistin-resistant OXA-48-producing Klebsiella pneumoniae, France, 2014. Euro Surveill 21(37):pii=30339 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=30339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parisi SG, Bartolini A, Santacatterina E, Castellani E, Ghirardo R, Berto A, Franchin E, Menegotto N, De Canale E, Tommasini T, Rinaldi R, Basso M, Stefani S, Palu G. 2015. Prevalence of Klebsiella pneumoniae strains producing carbapenemases and increase of resistance to colistin in an Italian teaching hospital from January 2012 to December 2014. BMC Infect Dis 15:244. doi: 10.1186/s12879-015-0996-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arjun R, Gopalakrishnan R, Nambi PS, Kumar DS, Madhumitha R, Ramasubramanian V. 2017. A study of 24 patients with colistin-resistant Gram-negative isolates in a tertiary care hospital in South India. Indian J Crit Care Med 21:317–321. doi: 10.4103/ijccm.IJCCM_454_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falcone M, Russo A, Iacovelli A, Restuccia G, Ceccarelli G, Giordano A, Farcomeni A, Morelli A, Venditti M. 2016. Predictors of outcome in ICU patients with septic shock caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Clin Microbiol Infect 22:444–450. doi: 10.1016/j.cmi.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Viale P, Giannella M, Lewis R, Trecarichi EM, Petrosillo N, Tumbarello M. 2013. Predictors of mortality in multidrug-resistant Klebsiella pneumoniae bloodstream infections. Expert Rev Anti Infect Ther 11:1053–1063. doi: 10.1586/14787210.2013.836057. [DOI] [PubMed] [Google Scholar]
- 20.Capone A, Giannella M, Fortini D, Giordano A, Meledandri M, Ballardini M, Venditti M, Bordi E, Capozzi D, Balice MP, Tarasi A, Parisi G, Lappa A, Carattoli A, Petrosillo N, on behalf of the SEERBIO-GRAB Network. 2013. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect 19:E23–E30. doi: 10.1111/1469-0691.12070. [DOI] [PubMed] [Google Scholar]
- 21.Giacobbe DR, Del Bono V, Trecarichi EM, De Rosa FG, Giannella M, Bassetti M, Bartoloni A, Losito AR, Corcione S, Bartoletti M, Mantengoli E, Saffioti C, Pagani N, Tedeschi S, Spanu T, Rossolini GM, Marchese A, Ambretti S, Cauda R, Viale P, Viscoli C, Tumbarello M, Isgri S. 2015. Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case-control-control study. Clin Microbiol Infect 21:1106.e1–e8. doi: 10.1016/j.cmi.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Yang D, Xie Z, Xin X, Xue W, Zhang M. 2016. A model for predicting nosocomial carbapenem-resistant Klebsiella pneumoniae infection. Biomed Rep 5:501–505. doi: 10.3892/br.2016.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Duin D, Doi Y. 2015. Outbreak of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae: are we at the end of the road? J Clin Microbiol 53:3116–3117. doi: 10.1128/JCM.01399-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontopidou F, Plachouras D, Papadomichelakis E, Koukos G, Galani I, Poulakou G, Dimopoulos G, Antoniadou A, Armaganidis A, Giamarellou H. 2011. Colonization and infection by colistin-resistant Gram-negative bacteria in a cohort of critically ill patients. Clin Microbiol Infect 17:E9–E11. doi: 10.1111/j.1469-0691.2011.03649.x. [DOI] [PubMed] [Google Scholar]
- 25.Wand ME, Bock LJ, Bonney LC, Sutton JM. 2017. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother 61:e01162-16. doi: 10.1128/AAC.01162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shorr AF, Zilberberg MD, Micek ST, Kollef MH. 2008. Prediction of infection due to antibiotic-resistant bacteria by select risk factors for health care-associated pneumonia. Arch Intern Med 168:2205–2210. doi: 10.1001/archinte.168.20.2205. [DOI] [PubMed] [Google Scholar]
- 27.Tacconelli E, Cataldo MA, De Pascale G, Manno D, Spanu T, Cambieri A, Antonelli M, Sanguinetti M, Fadda G, Cauda R. 2008. Prediction models to identify hospitalized patients at risk of being colonized or infected with multidrug-resistant Acinetobacter baumannii calcoaceticus complex. J Antimicrob Chemother 62:1130–1137. doi: 10.1093/jac/dkn289. [DOI] [PubMed] [Google Scholar]
- 28.van Walraven C, Austin PC, Jennings A, Quan H, Forster AJ. 2009. A modification of the Elixhauser comorbidity measures into a point system for hospital death using administrative data. Med Care 47:626–633. doi: 10.1097/MLR.0b013e31819432e5. [DOI] [PubMed] [Google Scholar]
- 29.StataCorp. 2015. Stata statistical software: release 14. StataCorp LP, College Station, TX. [Google Scholar]
- 30.Vasudevan A, Mukhopadhyay A, Li J, Yuen EG, Tambyah PA. 2014. A prediction tool for nosocomial multi-drug resistant Gram-negative bacilli infections in critically ill patients—prospective observational study. BMC Infect Dis 14:615. doi: 10.1186/s12879-014-0615-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber MP, Chan CM, Shorr AF. 2010. Resistant pathogens in nonnosocomial pneumonia and respiratory failure: is it time to refine the definition of health-care-associated pneumonia? Chest 137:1283–1288. doi: 10.1378/chest.09-2434. [DOI] [PubMed] [Google Scholar]
- 32.Park SC, Kang YA, Park BH, Kim EY, Park MS, Kim YS, Kim SK, Chang J, Jung JY. 2012. Poor prediction of potentially drug-resistant pathogens using current criteria of health care-associated pneumonia. Respir Med 106:1311–1319. doi: 10.1016/j.rmed.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Depuydt PO, Blot SI, Benoit DD, Claeys GW, Verschraegen GL, Vandewoude KH, Vogelaers DP, Decruyenaere JM, Colardyn FA. 2006. Antimicrobial resistance in nosocomial bloodstream infection associated with pneumonia and the value of systematic surveillance cultures in an adult intensive care unit. Crit Care Med 34:653–659. doi: 10.1097/01.CCM.0000201405.16525.34. [DOI] [PubMed] [Google Scholar]
- 34.Richter SE, McKinnell JA, Bell D, Uslan D, Watson K, Miller LG, Humphries R. 2017. Modeling carbapenem resistance among Gram negative pathogens using commonly available clinical data, poster 373. ID Week, San Diego, CA. [Google Scholar]
- 35.van Duin D, Lok JJ, Earley M, Cober E, Richter SS, Perez F, Salata RA, Kalayjian RC, Watkins RR, Doi Y, Kaye KS, Fowler VG Jr, Paterson DL, Bonomo RA, Evans S, Antibacterial Resistance Leadership Group. 2018. Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 66:163–171. doi: 10.1093/cid/cix783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaye KS, Vazquez J, Mathers A, Daikos G, Alexander E, Loutit JS, Zhang S, Dudley MN, Cornely O. 2017. Clinical outcomes of serious infections due to carbapenem-resistant Enterobacteriaceae (CRE) in TANGO II, a phase 3, randomized, multi-national, open-label trial of meropenem-vaborbactam (M-V) vs. best available therapy (BAT), abstr 1862. Abstr OPEC Fund Int Dev Conf 2017. [Google Scholar]
- 37.McKinnell JA, Connolly LE, Pushkin R, Jubb AM, O'Keefe B, Serio AW, Smith AK, Gall J, Riddle V, Krause KM, Pogue JM. 2017. Improved outcomes with plazomicin (PLZ) compared with colistin (CST) in patients with bloodstream infections (BSI) caused by carbapenem-resistant Enterobacteriaceae (CRE): results from the CARE study, abstr 1853. Abstr OPEC Fund Int Dev Conf 2017. [Google Scholar]
- 38.Mann CJ. 2003. Observational research methods. Research design II: cohort, cross sectional, and case-control studies. Emerg Med J 20:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]