Bacterial infections affect more than 2 million people annually. Of these, systemic infections caused by bacteria in critically ill patients may lead to life-threatening conditions such as sepsis.
KEYWORDS: gold nanoparticles, bloodstream bacterial infection, lipopolysaccharide, lipoteichoic acid, Gram stratification, point of care, bedside
ABSTRACT
Bacterial infections affect more than 2 million people annually. Of these, systemic infections caused by bacteria in critically ill patients may lead to life-threatening conditions such as sepsis. We have developed a point-of-care (POC) device called Septiflo that can detect and stratify the Gram status of bloodstream bacterial infections in less than 10 min from a drop of human plasma. It works on the principle of identifying pathogen-associated molecular patterns (PAMPs), such as lipopolysaccharides (LPS) and lipoteichoic acid (LTA) that are released into the bloodstream at the onset of Gram-negative and Gram-positive bacterial infections, respectively. The biomarkers are captured on a membrane without a receptor, and the Gram status specificity is conferred by the ligands attached to gold nanoparticles (AuNPs) used as signal amplification probes. The ultrasensitive colorimetric results are read by eye down to a 100-fg/ml detection limit without an instrument. No cross-interference between the PAMPs is seen during Gram stratification. Septiflo results also display better performance than commercial enzyme-linked immunosorbent assays (ELISAs). Tests performed on 60 clinical samples from patients showed a correlation accuracy of 70% against procalcitonin (PCT), an accepted surrogate biomarker for sepsis. A direct comparison with eubacterial PCR yielded up to 94% accuracy in 31 patients at a chosen cutoff level for LPS and LTA and area under the curve (AUC) values of 0.927 and 0.885, respectively, though blood culture was negative for most samples. The high sensitivity, low cost, and simple bedside utility of the assay may aid in better sepsis management apparently at the presymptomatic stage, lowering empirical therapy, medical costs, antimicrobial resistance, and mortality.
INTRODUCTION
Bloodstream bacterial infections (BSIs) are now the world's leading cause of premature deaths in neonates and patients admitted to intensive care units (ICUs) or emergency rooms (ER) and the third largest killer after cancer and cardiovascular diseases (1). Conservative estimates suggest that they affect around 1% to 3% of the total hospitalized population globally (2). A report based on 17 studies performed on a total of 40,644 patients from South and Southeast Asia indicated that between 1990 and 2010, the most common community-acquired bloodstream bacterial pathogen (found in 3,506 patients) was Salmonella enterica serotype Typhi, a Gram-negative strain, accounting for 55% isolates (3). In another study comprising 181 neonatal BSIs, the majority (∼59%) were diagnosed with Gram-positive bacteria (4). These data reflect urgent needs for bacterial stratification and appropriately directed treatment.
The sources of community-acquired BSI are classified on the basis of routes of pathogen entry as urinary (47.65%), gastrointestinal (25.99%), or respiratory (26.36%) (5). When bacteria disseminate into the bloodstream, they release pathogen-associated molecular patterns (PAMPs) as a result of bacterial death due to administered antibiotics or the host's immune response. PAMPs include endotoxin molecules such as lipopolysaccharides (LPS) in Gram-negative bacteria and lipoteichoic acid (LTA) in Gram-positive bacteria, bacterial DNA, peptidoglycans, lipoproteins, etc. Endotoxins, especially LPS, may also invade the blood by translocation from the human gut (6, 7). Once inside, these PAMPs can stimulate the innate immune response in the host, leading to a cytokine-chemokine storm and resulting in a massive inflammatory response. This critical medical condition is known as septicemia or in short sepsis. The condition is characterized by clinical features such as hemodynamic instability, mental confusion, tachypnea, and a rapid deterioration of systemic functions resulting in organ failure. According to the latest definition recommended at the Washington Consensus Conference 2016, sepsis 3.0 is a life-threatening organ dysfunction caused by a dysregulated host response to infection, while “suspected infection” is left undefined. The clinical criterion for organ dysfunction is a sequential organ failure assessment (SOFA) score of 2 or more. This inadequacy of current diagnostic procedures enables the infection to spread, increasing the severity which can only be combated by timely and evidence-based antimicrobial therapy (8, 9). Proper diagnosis not only improves outcomes but also reduces the administration of broad-spectrum antibiotics and, hence, antimicrobial resistance (10, 11). It is thus imperative to diagnose infection at an early stage before it becomes unmanageable. Further, stratification of bacterial Gram status at the onset of infection will add value to the antimicrobial treatment strategy.
There are currently several methods available for bacterial typing and species identification which can only be employed after bacterial growth in blood culture, staining, and isolation on solid culture media. Thus, conventional microbiology takes several days to weeks to get the final results, which makes it less useful for rapidly deteriorating conditions during sepsis (12, 13). Culture screening methods are generally limited by their poor detection sensitivities because of low bacterial loads in the blood (<100 CFU/ml) and decrease in bacterial viability due to empirical antibiotic therapy (14). Nucleic acid-based methods such as eubacterial PCR, on the other hand, are highly sensitive and less time consuming but involve a number of steps, expertise, and sophisticated instruments and are typically prone to contamination. These issues greatly limit their use in an ICU/ER space of a hospital setting (15).
In this study, we demonstrate a point-of-care (POC) approach that combines the simplicity of culture methods and the sensitivity of PCR. Such a POC test for detecting pathogen-derived molecules was reported for the first time by our group for identifying Gram-negative bacterial endotoxin in blood samples (16). This test was designed with a drug-gold nanoparticle (AuNP) bioconjugate, where polymyxin B sulfate was used as an affinity ligand for LPS detection. Here, we report the results of a pilot study involving our second-generation POC test devices called Septiflo, in which peptide-AuNP bioconjugates are used as affinity ligands for LPS and monoclonal antibodies (mAbs) for LTA detection. We demonstrate that these devices are comparable in performance to commercial enzyme-linked immunosorbent assays (ELISAs) and eubacterial PCR and, at the same time, surpass them in their simplicity, affordability, and adoptability for use in ICUs. Septiflo is an in vitro rapid card test that works on the facile principle of stratifying bacterial infections on the basis of the Gram-specific bacterial biomarkers LPS and LTA. Results can be visualized by the naked eye in less than 10 min, making it the fastest test for distinguishing Gram-positive and Gram-negative bacterial infections in plasma. This information can be of immense value in rapid decision making for the selection of bacterial Gram-specific narrow-spectrum antibiotics, thus reducing drug resistance and improving outcome. In the following sections, we discuss the preparation of nanobioconjugate probes, the developmental stages of the second-generation Septiflo assay, and its performance benchmarking against conventional methods and clinical validation.
MATERIALS AND METHODS
Materials.
Gold(III) chloride trihydrate (HAuCl4·3H2O), sodium citrate dihydrate (C6H5Na3O7·2H2O), bovine serum albumin (BSA), purified LPS (molecular weight [MW] 10 to 20 kDa) extracted from Escherichia coli 055:B5, purified LTA extracted from Staphylococcus aureus, phosphate-buffered saline (PBS), Purpald, and HEPES were from Sigma-Aldrich (India). This study also used mouse anti-LTA mAbs (IBT Biosciences, USA), sushi peptides of purity >95% (HAEHKVKIGVEQKYGQFPQGTEVTYTCSGNYFLMC; MW, 4,083 Da; Genemed Synthesis, USA), ortho-phthaldehyde ([OPA] SRL, India), pluronic acid ([PF68] Ranbaxy, India), tannic acid, sodium phosphate salts, sodium periodate, Tween 20, and carboxythiol-polyethylene glycol (CT-PEG) linker (MW, 634.77 Da) from Fisher Scientific (India), PEG6-hydrazide aromatic dialkane thiol linker (MW, 708.19 Da; SensoPath Technologies, USA), 30-kDa MW cutoff (MWCO) centrifuge filters (Millipore India Pvt. Ltd.), absolute ethanol (EMSURE; Merck, Germany), and membranes (CX-1, SCNM-I) and absorbent pads (type-AP080) (Advanced Micro Devices Pvt. Ltd., India). ELISA kits for LPS (MBS725451; MyBiosource, USA) and LTA (E1405Ge; Wuhan EIAab Science, Antibodies Online Inc., USA) were used, as well as a QIAamp DNA minikit (catalog no. 51304; Qiagen, India), deoxynucleoside triphosphates ([dNTPs] Fermentas), Taq DNA polymerase and PCR buffer mix (New England BioLabs Inc.), agarose, and BRAHMS procalcitonin (PCT) sensitive kit and reagents (Thermo Fisher Inc.). All other standard reagents used were of analytical grade. All solutions were prepared using ultrapure deionized Milli-Q water (∼18.8 mΩ · cm resistivity, CDUFBI001; Millipore, USA). Due to the known pyrogenicity and toxicity of LPS and LTA, strict safety guidelines were followed in the laboratory while handling these molecules (17).
Preparation and characterization of AuNPs.
Citrate-capped AuNPs of approximately 15-nm diameter were synthesized using the Turkevich method (18) and characterized by UV-visible spectroscopy (UV-2600; Shimadzu), dynamic light scattering ([DLS] Zetasizer nano ZS90; Malvern Instruments Ltd.), and transmission electron microscopy ([TEM] Tecnai G2). For TEM, a 4× diluted AuNP suspension was dried on a carbon-coated copper TEM grid (CF200 CU; Electron Microscopy Sciences, USA) and imaged at 200 kV. The micrographs obtained at ×44,000 magnification were analyzed using ImageJ software to determine the average particle size from 100 particles. The procedure was repeated thrice, and the data were plotted as the mean ± 1 standard deviation (SD).
Preparation of AuNP-bioconjugate probes.
LPS-specific sushi peptides were conjugated to the AuNPs at a 1,000:1 molar ratio using thiol chemistry and then characterized via Fourier-transform infrared (FTIR) spectroscopy (Nicolet, Protege 460) and OPA analysis as described earlier (19). The LTA-specific antibodies were conjugated to the AuNPs using a protocol reported in the literature with slight modifications to the procedure (20, 21). Briefly, 50 μl of 100 mM sodium phosphate buffer at pH 7.4 and 10 μl of 100 mM sodium periodate were added to 100 μl of a 1-mg/ml antibody solution to oxidize the hydroxyls present in the Fc regions of the antibodies. At room temperature, the mixture was kept in a rotary shaker (45 rpm) and incubated for 1 h, protected from light. The reaction was quenched by adding 500 μl of 10 mM PBS at pH 7.4, and the completion of the reaction was confirmed by mixing 10 μl of the above antibody solution with 20 μl of freshly prepared 20-mg/ml Purpald solution prepared in 1 M sodium hydroxide. The change in color to purple confirmed the successful oxidation of the hydroxyls to aldehydes. The aldehyde-activated antibody was then mixed with 10 μl of a 10 mM hydrazide linker solution in absolute ethanol (99% purity). The mixture was incubated for 1 h followed by the addition of 1 ml of 40 mM HEPES buffer at pH 8.5. The entire volume was filtered using a 30-kDa MWCO centrifuge filter at 14,490 rcf for 15 min to remove the excess linker. The filtered linker-conjugated antibodies were finally resuspended in 40 mM HEPES buffer at pH 7.4 and kept at 4°C until further use.
The antibody-linker conjugates were mixed with 2 nM AuNPs at a 1:10 molar ratio and incubated for 1 h to assist in Au-thiol bond formation. Finally, the antibody-conjugated gold suspension was centrifuged at 7,245 rcf to remove any unconjugated antibodies. The pellet was resuspended in 40 mM HEPES buffer at pH 7.4, and the conjugation was confirmed by UV-Vis spectroscopy, DLS, and Bradford assay (22).
Preparation of spiked plasma samples.
Master stocks of 1 mg/ml LTA and LPS were prepared in Milli-Q (MQ) water. The solutions were vortexed for 30 min followed by a 10-min sonication and preparation of aliquots of different concentrations in MQ water. A fixed 10% (vol/vol) LPS or LTA solution was then spiked in human sera to obtain concentrations ranging from 100 fg/ml to 10 ng/ml. The spiked solutions were stored at 4°C until further use. Before testing, the samples were again vortexed for 15 min at room temperature.
Selection criteria for clinical samples.
Blood plasma samples (n = 60) for Septiflo validation were obtained from Global Hospitals Hyderabad, India, after obtaining the requisite approval from the institutional ethical committee (protocol no. MDXBM:001, version 1, 29 October 2009) and informed consent from the patients. The patients included (i) people who visited the outpatient department for consultation and routine health check-up, (ii) patients from the inpatient wards who were suspected to have infections, and (iii) patients who were admitted in the ICU following various surgical procedures and monitored postsurgery. No other exclusion/inclusion criteria for sample collection were imposed. The blood samples were collected in EDTA and tested for various hematological parameters such as differential cell count (Sysmax-XN1000/100) and procalcitonin (Kryptor Compact). The toxic granules were observed under Leishman's stain of peripheral blood smears and blood culture using the Bactec-9120/9050 system. Approximately half of these samples were also randomly chosen to perform eubacterial PCR (procedure described below).
Septiflo procedure.
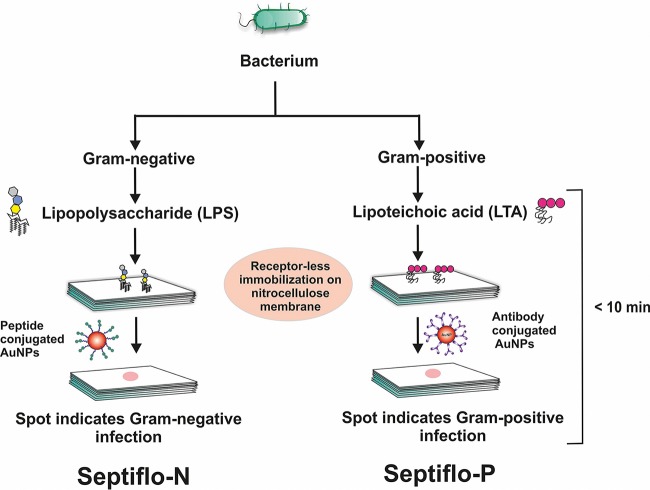
A typical Septiflo assay comprised a membrane kept on top of a set of absorbent pads enclosed in custom-fitted plastic cassettes. The absorbent pads served as liquid reservoirs and mechanical support for the thin membrane. The surface of the membrane was first treated with 50 μl of surfactant (PF68 for the Septiflo-N assay and Tween 20 for the Septiflo-P assay). After 2 min, 2 μl of test sample containing LPS or LTA was drop casted in the middle of the membrane followed by the addition of 50 μl of 1% (wt/vol) BSA. After 2 min, 50 μl of 1 nM AuNP suspension preconjugated to peptides/antibodies was applied to the membrane followed by a second injection after 10 s. The appearance of a pink spot against a white background indicated the presence of LPS/LTA in plasma. Finally, the membrane was washed with 100 μl of MQ water to remove any unbound NPs. Cross-reactivity experiments were performed with mixtures cospiked with various ratios of LPS and LTA. The entire assay was performed under ambient conditions (relative humidity, 30% to 70%; temperature, 30 ± 5°C). On the basis of the Gram status, the assay for LPS detection was termed Septiflo-N and that for LTA was termed Septiflo-P (Fig. 1).
FIG 1.
Schematic of the Septiflo bioassay developed for the rapid bedside detection and stratification of bacterial infections in blood.
Development of the color chart.
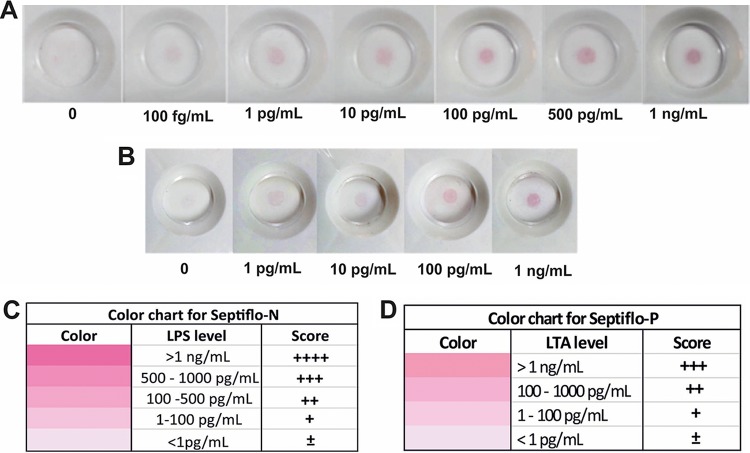
Septiflo assays were performed in the range of 1 pg/ml to 10 ng/ml LPS/LTA concentrations. The color intensities of the spots were visualized by the naked eye and calibrated by comparing with the colors generated in Excel (version 2013) by adjusting the red, green and blue (RGB) values in a particular cell (see Fig. S1 in the supplemental material). The motivation was to make the signal semiquantitative, such that a human eye was sufficient to assess the concentration gradient. The concentrations were represented in terms of a score for Septiflo-N as “±” for <1 pg/ml, “+” for 1 to 100 pg/ml, “++” for 100 to 500 pg/ml, “+++” for 500 to 1,000 pg/ml, and “++++” for >1 ng/ml and for Septiflo-P as “±” for <1 pg/ml, “+” for 1 to 100 pg/ml, “++” for 100 to 1,000 pg/ml, and “+++” for >1 ng/ml. The assays were performed on clinical plasma samples and given an appropriate score by comparing against the color chart by at least four independent reviewers. Each sample was tested thrice, and the final results were reported as an average of all the data points (i.e., at least 12) ± 1 standard deviation (SD).
ELISAs.
Sandwich ELISAs for LPS were performed by adding 50 μl of spiked plasma samples (50 to 1,200 pg/ml) and standards (provided in the kit) to each microtiter plate well, precoated with mAbs specific to LPS. Next, 100 μl of horseradish peroxidase (HRP)-conjugated polyclonal antibody was added to each well to sandwich the LPS immobilized on the plate. After incubating for 30 min at 37°C, all the unbound components were removed by washing with 200 μl of buffer. Next, 100 μl of substrate was added to each well and incubated for 30 min at 37°C for the enzyme-substrate reaction to take place. The reaction was terminated by adding 50 μl sulfuric acid, and the LPS concentration-dependent color change was measured spectrophotometrically at a wavelength of 450 nm (SpectraMax i3x multimode plate reader; Molecular Devices). A standard curve was plotted with ELISA standards, and the unknown concentrations were interpolated from the curve.
LTA ELISA was also performed on spiked plasma samples (0.31 to 20 ng/ml) and ELISA standards. For this, 50 μl of sample was added to each microtiter well precoated with LTA-specific antibodies followed by the addition of 50 μl of biotin-labeled LTA. The plate was incubated for 1 h at 37°C. The unbound entities were washed thrice using 400 μl of wash buffer each. Next, 100 μl of avidin-conjugated HRP was added to each well and incubated for 45 min at 37°C. The washing step was repeated five times followed by the addition of 90 μl of substrate for 15 to 30 min (protected from light). The enzyme substrate reaction was terminated by adding 50 μl of the stop solution, and the color change was measured spectrophotometrically at 450 nm. The concentration of LTA in samples was then determined by comparing the optical density of samples to the standard curve.
Bacterial DNA isolation from blood samples.
A QIAamp DNA Minikit was used to isolate bacterial DNA from peripheral blood samples collected in EDTA. The manufacturer's instructions were followed with some changes in the duration of lysis (56°C) and final elution buffer volume to account for the unusually low number of bacteria in blood. Briefly, 200 μl of whole blood sample, 200 μl of lysis buffer AL, and 25 μl of proteinase K were placed in a 1.5-ml microcentrifuge tube. The mixture was pulse-vortexed for 15 s and incubated at 56°C for 20 min. Next, 200 μl of 100% ethanol was added, and the mixture was again pulse-vortexed for 15 s followed by 5 min of incubation at room temperature. The tube was briefly centrifuged to spin down the droplets from inside the lid, and the contents were carefully transferred to the QIAamp Minispin column (without wetting the rim) and centrifuged at 6,000 rcf for 1 min. The spin column was washed with the wash buffers AW1 and AW2 and finally with 200 μl ethanol. The alcohol was drained out, and the column was air-dried by spinning at 6,000 rcf for 6 min. Sixty microliters of the elution buffer AE was then added to the column and incubated at room temperature (15 to 25°C) for 1 min. The eluted DNA after a final centrifugation step of 6,000 rcf for 1 min was stored at −20°C for PCR analysis.
PCR for detecting eubacteria.
A known protocol for Gram type-specific broad-range PCR identification of 62 pathogenic bacteria was used to screen the blood samples (23). The PCR mixture contained one universal 16S rRNA primer DG74 (AGGAGGTGATCCAACCGCA) for all bacteria, a reverse primer 143 (GAYGACGTCAARTCMTCATGC) for Gram-positive bacteria, and 68d (AYGACGTCAAGTCMTCATGG) for Gram-negative bacteria. The PCR reagents were mixed in a 20-μl volume in individual 0.2-ml PCR tubes and amplified (thermocycling conditions mentioned in Table S1). The amplified products were then analyzed in 2% (wt/vol) agarose gel (see Fig. 7).
FIG 7.
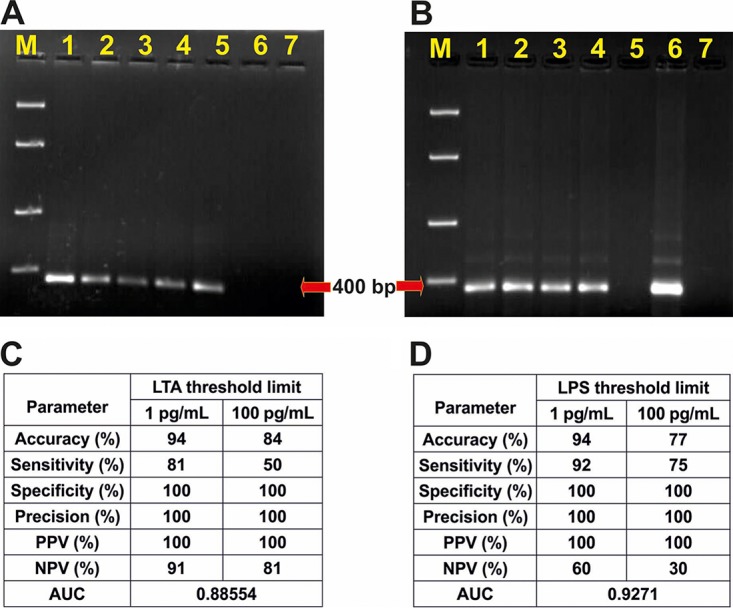
Typical PCR gel images for Gram-positive (A) and Gram-negative (B) blood samples. Lanes 1 (A) and 6 (B), PCR positive controls; 2 to 5 (A), Gram-positive blood samples; 1 to 4 (B), Gram-negative blood samples; 6 (A) and 5 (B), negative clinical blood samples; 7 (A and B), PCR negative controls; M (A and B), molecular size ladder (100, 400, 850, 2,000, and 5,000 bp). Septiflo performance correlations with PCR for two different threshold limits of LTA (C) and LPS (D) in blood.
RESULTS AND DISCUSSION
Membranes make a superior choice for POC affinity scaffolds, as their surface properties can be precisely engineered for receptorless immobilization of target molecules and their porosity can be modulated to control fluid flow. By receptorless, we mean that the analytes can be trapped/bound on the membrane surface without the aid of any ligands or receptor molecules such as antibodies. With some prior knowledge of the membrane from our previous study (16), the second-generation Septiflo assay was further optimized to enable both LPS and LTA (PAMPs released from the cell wall of the two bacteria types) molecules to be captured on the same type of membrane without the use of any specific receptors. The specificity for Gram status was conferred by the ligand moieties attached to the colloidal probes used in the subsequent step; sushi peptide was used against LPS (in Septiflo-N assay) and anti-LTA mAb was used to target LTA (in Septiflow-P assay). The two assays were carried out simultaneously in separate cassettes by injecting a drop of the same sample into each followed by the addition of the respective AuNP suspension (Fig. 1). The result was a pink spot that was easily visualized by the naked eye only if LPS/LTA was present in the sample above a certain threshold. The pink color was imparted by the AuNPs that display a distinct absorbance in the visible range (at 521 nm for 15-nm-diameter particles).
A variety of other parameters were also extensively optimized to identify the operating conditions for best assay performance (see Table S2 in the supplemental material). For instance, surfactants were seen to play a crucial role during the immobilization process. Since both LPS and LTA molecules contain large lipophilic tails favoring their hydrophobic immobilization on the membrane, the addition of surfactant helped in the proper wettability of the injected solution, providing the optimum hydrophilic-lipophilic balance for uniform adsorption of the endotoxins. Uneven patchy spots were obtained in the absence of surfactant (see Fig. S2). Minimizing the nonspecific interaction of the AuNPs with the membrane or plasma protein interface was also an important design criterion for the successful execution of the bioassay. To address this issue, the unoccupied sites on the nanobioconjugate surface were blocked with a PEG-thiol linker which not only nullified any nonspecific signal but also provided more stability to the NPs (see Fig. 2, Fig. S3, and reference 19 for a detailed characterization of the AuNP probes). Any surface-based nonspecific binding was eliminated by blocking the membrane with BSA solution (24).
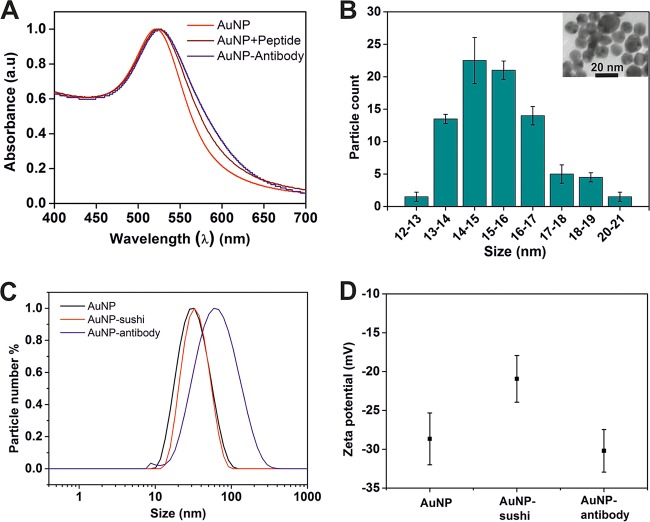
FIG 2.
Detailed characterization of the AuNPs after synthesis and bioconjugation. (A) Typical UV-Vis graph showing a peak at 521 nm characteristic of AuNPs. The subsequent red shifts in absorbance obtained after sushi peptide and mAb conjugation indicated successful binding. (B) TEM illustrated spherical particles with a narrow size distribution of 15 ± 4 nm. (C) DLS showed an expected average particle size of 30 ± 4 nm (hydrodynamic diameter), which increased systematically upon conjugation with the smaller sushi peptide and a larger mAb. (D) Changes in the zeta-potential values were also suggestive of successful conjugation.
The performance of Septiflo using well-characterized AuNP probes was tested with plasma/sera of critically ill subjects/patients suspected of carrying bacterial infection. Among these, noninfectious cases formed our true control besides the healthy people (without any medical condition) whose samples were used during the actual design and optimization of the assay. The threshold values of LPS in peripheral blood are reported to be around 100 to 120 pg/ml (16). Patients with endotoxin contents higher than these levels are potentially at higher risk of developing sepsis or septic shock (16, 25). On the other hand, while the crucial role of LTA in sepsis is well established (26), not much data exist on its blood levels and cutoff concentrations in cases of Gram-positive infections (27). In reality, both Gram-positive and Gram-negative bacterial infections may coexist (mixed infections) and are, in fact, quite common due to excess amounts of gut-derived LPS released into the bloodstream in several clinical conditions that alter the mucosal permeability of the intestines or due to sepsis-associated intestinal hypoperfusion (28). It thus becomes imperative to develop a test that is not only sensitive in the clinically relevant range but also selective toward LPS/LTA without any cross-interference from other molecules and, more importantly, each other.
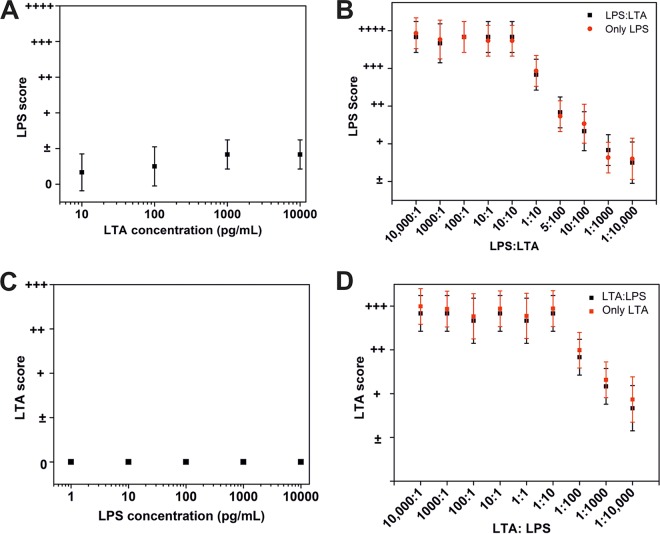
One of the first tasks required for determining the sensitivity of the assay was to estimate the extent of endotoxin levels in the analyte. To do this in a semiquantitative manner, we prepared color charts to map the spot intensity gradients obtained against various LPS/LTA concentrations in spiked plasma samples (Fig. 3). The concentration ranges for LPS and LTA in these color charts were selected on the basis of the severity of clinical conditions imposed by them during sepsis (16). Although, in the literature, these correlations are available only for LPS, we set the LTA levels to be similar to that of LPS, considering that the two molecules are structurally similar and share the inflammatory immune response pathways (29, 30). The antigenicity of LTA is, however, expected to be lower (than LPS) (31) and may have important implications as discussed later in this paper. The wide dynamic range of both Septiflo-N and Septiflo-P assays varied over three orders of magnitude, but the Septiflo-N had a 10× lower detection limit (LOD) of 100 fg/ml. These LODs are 10 to 100× better compared to those reported in the literature. The two assay platforms were also independent and highly specific to their respective bacterial component, as they showed minimal cross-reactivity across a range of endotoxin concentrations (Fig. 4A and C). A “0” score here indicates a blank membrane with no spot. To establish the robustness of our assays for handling mixed infections, a further cross-interference study was carried out in which LPS and LTA were mixed together in different ratios and tested on both platforms. The outcomes of this study again showed no change in signal due to competitive binding between the molecules (Fig. 4B and D), establishing the full utility of our assay for stratifying bacterial infections on the basis of Gram status.
FIG 3.
Actual representative images of the Septiflo assay and their corresponding color chart for LPS (Septiflo-N) (A and C) and for LTA (Septiflo-P) (B and D).
FIG 4.
Interference study of Septiflo-N and Septiflo-P with structurally similar molecules. (A and B) Interference of different LTA concentrations present in spiked samples analyzed using Septiflo-N. (C and D) Interference of LPS presence during measurement of LTA-spiked samples (zero score indicates no spot).
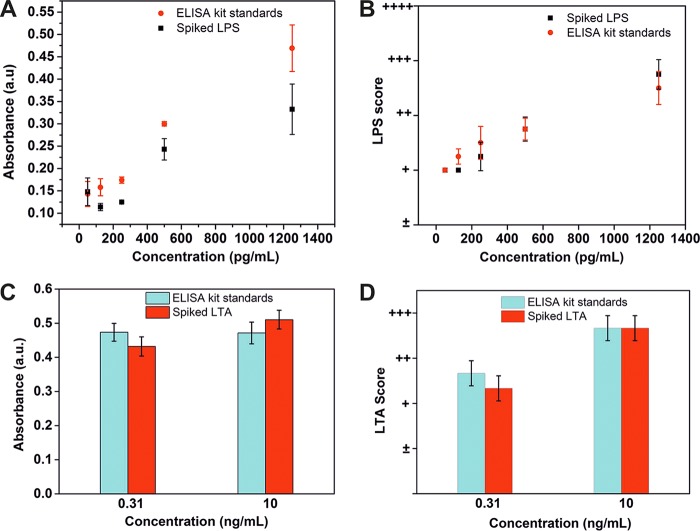
Another unique aspect of our bioassay was its potential use right by a patient's bedside inside ICUs. To highlight this feature, we compared the performance of our Septiflo devices with standard ELISAs available on the market. These ELISA kits are sold only for research and development purposes, and there are no approved in vitro diagnostic ELISA kits available for either LPS or LTA. The study comprised testing both spiked LPS/LTA plasma samples and the standard solutions provided in the kit on either platform, i.e., on Septiflo and ELISA. The LPS results in ELISAs showed overall increasing trends for both solution types, but the absorbance values obtained for spiked plasma samples were consistently lower (Fig. 5A). The Septiflo-N assay, on the other hand, exceeded our expectations and showed remarkable overlap between the two sets of data points (Fig. 5B). In the case of LTA, the performance of the ELISA kit was quite inconsistent with the manufacturer's claim. For instance, the ELISA results yielded approximately the same values for concentrations that were 30-fold apart (Fig. 5C). On the contrary, the results for Septiflo-P showed clear classification of LTA concentrations in both solution types (Fig. 5D). These results were truly encouraging and reflected the promising position of our device as an affordable and portable POC test for both urban and rural health care settings alike. Although there are other methods such as the endotoxin activity assay (EAA) or limulus amebocyte lysate (LAL) assay for detecting endotoxin, they are either not direct or limited in their application toward complex biological fluids, such as serum, plasma, and tissue homogenates. While EAA works on the principle of chemiluminescence by measuring the host's neutrophil respiratory burst activity in response to endotoxin (32), LAL is a chromogenic LPS detection assay, widely used in parenteral pharmaceutical preparations (33).
FIG 5.
Comparison of Septiflo performance with commercial ELISAs. The spiked LPS samples and the LPS ELISA standards analyzed using ELISA (A) and Septiflo-N (B). The spiked LTA samples and the LTA ELISA standards analyzed using ELISA (C) and Septiflo-P (D).
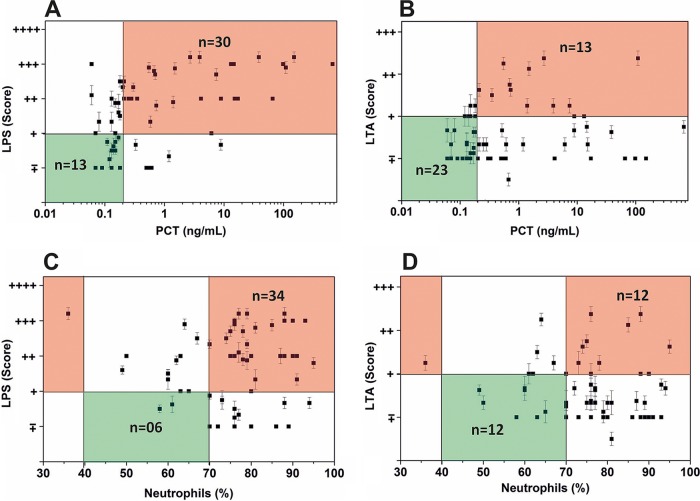
Next, a preliminary clinical validation of our assay was performed on 60 plasma samples collected from a mixed set of critical care patients and controls. For this, Septiflo results for LPS/LTA levels were plotted against two hematological parameters, PCT and neutrophils. PCT is a U.S. FDA-approved surrogate biomarker for sepsis diagnosis, which is used worldwide either on automated instruments or in a rapid card test format (34, 35). Its levels in normal individuals are reported to be less than 0.25 ng/ml but tend to elevate when inflammation and/or infection is involved (36). The circulating LPS and LTA levels, on the other hand, can vary greatly in individuals depending on demographics and the extent of physical activity undertaken by the person leading to altered intestinal permeability (37–39). Deciding the cutoff levels for LPS and LTA in the blood (to designate a blood sample as positive for infection) can thus prove tricky. While a few studies claim the peripheral blood LPS threshold levels to be around 100 pg/ml, others have found them to be lower than this (25, 40). In a study conducted by Opal et al. on 253 volunteers, septic patients were examined within 24 h of the onset of sepsis against 33 healthy volunteers. It was found that the mean average LPS value in healthy controls was 5 ± 7 pg/ml (25). No such data exist for LTA to date. Considering such variability in the literature and the absence of any concrete and definitive knowledge of threshold levels for LPS/LTA in blood, we benchmarked the performance of our Septiflo assay against PCT using two different baseline limits for the analytes as 100 and 1 pg/ml. By doing this, we hoped to get additional insight into the approximate range of concentrations of these biomarkers in nonseptic individuals and establish our own cutoff levels.
The Septiflo-N and PCT values displayed a fairly good correlation. As the PCT values increased, the clinical samples also showed higher levels of LPS (Fig. 6A). A quantitative analysis of the data using a 100-pg/ml LPS threshold limit showed that of the 60 samples, 43 lay “in range,” whereas most others were accumulated at the periphery of the PCT threshold limit of 0.25 ng/ml. Since the threshold limits of any biodiagnostic system are not intrinsically sharp, this indicated that our assay was able to pick up most of the Gram-negative bacterial infections quite effectively. In fact, there was a visibly sharper increase in the Septiflo scores compared to 0.25 ng/ml of PCT, indicating that LPS was probably a more useful biomarker to predict infection at the subclinical stage when the concentrations of host-derived biomarkers were still low (samples with PCT concentrations of 0.25 ng/ml or below are considered noninfectious, subclinical if the PCT levels range from 0.25 ng/ml to 0.5 ng/ml, and concentrations higher than >0.5 ng/ml denote the clinical range) (41). Until now, PCT is the most widely accepted and the only U.S. FDA-approved biomarker for concluding the sepsis status of a patient. Therefore, we used PCT as the rightful parameter for categorizing infectious versus noninfectious cases in our study (see Table S3). The LTA scores, on the other hand, did not correlate so well and remained in the normal range despite the high PCT values (Fig. 6B). Interestingly, more than 60% of these cases tested positive for Gram-negative infections, explaining the high levels of PCT and proving the Gram stratification potential of Septiflo (see Fig. S11). There were also several cases of mixed infection (∼25%) based on our eubacterial PCR results, where the presence of Gram-positive infection may have led to the LPS invasion via mucosal barrier permeability. The human gut flora is rich with indigenous Gram-negative bacteria (E. coli, Klebsiella pneumoniae, Enterobacter cloacae, etc.) that frequently start to translocate in cases of intestinal inflammation or altered mucosal permeability, such as with the host's immune deficiencies, immunosuppression, obstructive jaundice, stress, regional hypoperfusion, mucosal ischemia, etc. (7, 42, 43). Mixed infections have also been reported in several sepsis cases where LPS was frequently present in blood circulation irrespective of infection type (43–46).
FIG 6.
Correlation of Septiflo-N and Septiflo-P results with hematological parameters (n = 60) taking LPS/LTA threshold levels as 100 pg/ml in blood. LPS scoring against PCT (A) and neutrophil count (C). LTA scoring against PCT (B) and neutrophil count (D). The green color indicates normal range and the orange indicates out-of-range values.
To check if our hypothesis was indeed true, we replotted the PCT values against the total infection. The score for total infection was defined as the higher of the two Septiflo-N/P scores. For instance, a score of + obtained in Septiflo-P versus ++ obtained in Septiflo-N was reported as ++. By doing this analysis, we got an undeniably good correlation between infection and PCT, as shown in Fig. S4A, proving that Septiflo not only provides essential information regarding the patient's infection status, it can also give important clues about the nature of the infection. The various performance characteristics of the assay for 100 and 1 pg/ml limits are shown in Fig. S4B to S6. The receiver operating characteristic (ROC) analyses showed a fair correlation yielding area under the curve (AUC) values of 0.701 for Septiflo-N, 0.547 for Septiflo-P, and 0.695 for mixed infection cases (see Fig. S12 for detailed ROC analysis). The data obtained against neutrophil counts were more or less in agreement with our PCT data (Fig. 6C and D), though the results were slightly more correlated in the case of PCT. Most cases with high levels of PCT also tested positive for toxic granules (refer to Table S3). It is important to note that no single parameter currently in use can independently conclude or define the sepsis status, but a few biomarkers, including PCT, toxic granules, and neutrophils, can collectively lead to a provisional diagnosis. The inclusion of our assay in this list of biomarkers can add immense value even at the presymptomatic stage when the concentration of host-derived biomarkers is relatively low (Fig. 6A and Fig. S4 support this proposition).
We also compared our Septiflo results with the laboratory gold standard culture method, but most cultures yielded negative result even after 1 week of incubation (Table S3). This was most likely because the patients were already undergoing antibiotic therapy. So, we finally moved to the PCR method, which is by far the most sensitive, reliable, and direct technique for bacterial DNA confirmation. For this, we chose a protocol that was originally published in 1999 by Klausegger et al. and adopted it without any modification (23). This protocol was reported to identify 62 different human bacterial pathogens (type strains) with excellent sensitivity and specificity. This protocol was also recommended by Diane U. Leong and Kay S. Greisen in the book Diagnostic molecular microbiology-principles and applications (47) and appears to be one of the best available for eubacterial PCRs. With this knowledge in mind, fresh blood samples were analyzed for eubacteria by PCR, and the amplified DNA was visualized in an agarose gel. The appearance of bands of expected size in the gel confirmed the presence of Gram-positive (Fig. 7A) or Gram-negative (Fig. 7B) bacterial DNA. By setting the threshold limit for LPS/LTA at 100 pg/ml, a good correlation accuracy of >75% was obtained as shown in Fig. 7C and D. When the threshold limit was reduced to 1 pg/ml, the accuracy jumped to 94% and the sensitivity improved by at least 15%. The ROC curves also gave high AUC values of 0.927 for Septiflo-N and 0.885 for Septiflo-P, suggesting our assay's excellent diagnostic potential. These results also strongly pointed to the possibility of lower cutoff levels of 1 pg/ml in plasma for these biomarkers. This is also supported by the fact that unspiked plasma samples collected from healthy volunteers (Fig. 3A and B, negative controls) consistently gave a clear background. For detailed Septiflo performance classification models, refer to Fig. S7 to S10 and S13 for AUC analyses against PCR data.
Conclusions.
In summary, we have demonstrated the development of a bedside bioassay called Septiflo that semiquantitatively detects and Gram specifies bacterial infections in less than 10 min. The unique design features of the assay which make it superior to the existing endotoxin detection approaches include its simplicity, rapidness, low cost, receptorless capture of biomarkers, instrumentation-free signal output, and sub-pg/ml-level sensitivity. The performance of the Septiflo assay for concentration discrimination was found to be superior to both LPS and LTA ELISAs. While Septiflo showed consistent results with both spiked plasma samples and the calibration standards provided in the ELISA kit, the LPS ELISA showed some deviation and the LTA ELISA was completely unreliable for both solution types. The Septiflo assay was also highly specific for the pathogenic component against which it was designed, such that the presence of one biomarker did not affect the results of the other. This was particularly useful for identifying cases of mixed infection. A preliminary clinical assay validation was shown to cover ∼80% of all bloodstream infections (48). An increasing correlation was found with PCT with the exception of a sharp rise in Septiflo values around the low PCT range of 0.1 to 0.2 ng/ml PCT. This hinted that our assay could be useful for presymptomatic identification of infection from noninfectious inflammation, where PCT is not sensitive or is less discriminatory (typically after 6 h). A comparative study against PCR yielded remarkably good results for a relatively small sample population size of 31.
We are aware of the drawbacks of endpoint PCRs, as they can also pick up dead bacteria and circulating DNA. However, this is not really a true limitation in our opinion, as the detection of bacteria (dead or alive) or their components (above a certain threshold) does indicate bloodstream infection, as blood is normally sterile. Also, the genomic targets used in our PCR are located in rRNA, not far from a true bacterial viability PCR, but in the future, quantitative PCR (qPCR) of genes involved in the replication or metabolic activity of bacteria may also be performed to draw more quantitative comparisons. The biggest limitation of the Septiflo platform presently is that it can only pick up bacteria and not other pathogens, such as viruses, fungi, and parasites that are also known to trigger infection/sepsis, though their collective impact remains less than 25% (46). This is currently being addressed in our lab. Also, the low-cost semiquantitative mode of colorimetric detection may in the future be upgraded to an automated quantitative format by integrating the assay strips with an optical reader to make the outcomes more objective.
Despite these limitations, we believe that Septiflo has tremendous potential as a POC diagnostic device for early detection of BSIs/septicemia and for ruling out noninfectious inflammations at the preclinical stage. So far, our preliminary results suggest that 1 pg/ml is a more realistic cutoff value than 100 pg/ml to qualify for bacterial infection (subclinical stage), but more data are required to fully confirm this claim. While the sushi peptide ligand used in Septiflo-N is already a part of the protein sequence used in LAL assay, the Septiflo-P assay was limited to mAb-mediated detection due to the lack of availability of affinity ligands in the literature specific for LTA. In the future, Septiflo may be used to screen molecules specific to LTA that may serve as alternative ligands for specific targeting.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by DST-Nanomission (SR/NM/NT-1049/2016) and a GYTI award from the Society of Research and Initiatives for Sustainable Technologies and Institutions (SRISTI) and the Bitechnology Industry Research Assistance Council (BIRAC).
We thank CRF-IIT Delhi for TEM, Sudip Pattanayek for DLS and zeta measurements, and MDI for the Septiflo membrane assembly and useful discussions. We also thank Aditya Khandelwal for help with the ROC data analysis and all the lab members for blind peer reviewing of the clinical data.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00408-18.
REFERENCES
- 1.Lukaszewski RA, Yates AM, Jackson MC, Swingler K, Scherer JM, Simpson AJ, Sadler P, McQuillan P, Titball RW, Brooks TJG, Pearce MJ. 2008. Presymptomatic prediction of sepsis in intensive care unit patients. Clin Vaccine Immunol 15:1089–1094. doi: 10.1128/CVI.00486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuben AG, Musher DM, Hamill RJ, Broucke I. 1989. Polymicrobial bacteremia: clinical and microbiologic patterns. Rev Infect Dis 11:161–183. doi: 10.1093/clinids/11.2.161. [DOI] [PubMed] [Google Scholar]
- 3.Deen J, von Seidlein L, Andersen F, Elle N, White NJ, Lubell Y. 2012. Community-acquired bacterial bloodstream infections in developing countries in south and southeast Asia: a systematic review. Lancet Infect Dis 1 2:480–487. doi: 10.1016/S1473-3099(12)70028-2. [DOI] [PubMed] [Google Scholar]
- 4.Ballot DE, Nana T, Sriruttan C, Cooper PA. 2012. Bacterial Bloodstream Infections in Neonates in a Developing Country. ISRN Pediatr 2012:508512. doi: 10.5402/2012/508512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cisterna R, Cabezas V, Gomez E, Busto C, Atutxa I, Ezpeleta C. 2001. Community-acquired bacteremia. Rev Esp Quimioter 14:369–382. (In Spanish.) [PubMed] [Google Scholar]
- 6.Sheldon IM. 2016. Detection of pathogens in blood for diagnosis of sepsis and beyond. EBioMedicine 9:13–14. doi: 10.1016/j.ebiom.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaishnavi C. 2013. Translocation of gut flora and its role in sepsis. Indian J Med Microbiol 31:334–342. doi: 10.4103/0255-0857.118870. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhry H, Zhou J, Zhong Y, Ali MM, Mcguire F, Nagarkatti PS, Nagarkatti M. 2013. Role of cytokines as a double-edged sword in sepsis. In Vivo 27:669–684. [PMC free article] [PubMed] [Google Scholar]
- 9.Gotts JE, Matthay MA. 2016. Sepsis: pathophysiology and clinical management. BMJ 353:i1585. doi: 10.1136/bmj.i1585. [DOI] [PubMed] [Google Scholar]
- 10.Lee CR, Cho IH, Jeong BC, Lee SH. 2013. Strategies to minimize antibiotic resistance. Int J Environ Res Public Health 10:4274–4305. doi: 10.3390/ijerph10094274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marik PE. 2014. Early management of severe sepsis: Concepts and controversies. Chest 145:1407–1418. doi: 10.1378/chest.13-2104. [DOI] [PubMed] [Google Scholar]
- 12.Özenci V, Tegmark-Wisell K, Lundberg C, Wretlind B. 2008. Rapid culture and identification: a practical method for early preliminary laboratory diagnosis of sepsis. Clin Microbiol Infect 14:177–180. doi: 10.1111/j.1469-0691.2007.01897.x. [DOI] [PubMed] [Google Scholar]
- 13.Jagtap P, Sritharan V, Gupta S. 2017. Nanotheranostic approaches for management of bloodstream bacterial infections. Nanomedicine 13:329–341. doi: 10.1016/j.nano.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Opota O, Croxatto A, Prod'hom G, Greub G. 2015. Blood culture-based diagnosis of bacteraemia: state of the art. Clin Microbiol Infect 21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Chiquet C, Cornut PL, Benito Y, Thuret G, Maurin M, Lafontaine PO, Pechinot A, Palombi K, Lina G, Bron A, Denis P, Carricajo A, Creuzot C, Romanet JP, Vandenesch F. 2008. Eubacterial PCR for bacterial detection and identification in 100 acute postcataract surgery endophthalmitis. Invest Ophthalmol Vis Sci 49:1971–1978. doi: 10.1167/iovs.07-1377. [DOI] [PubMed] [Google Scholar]
- 16.Kalita P, Chaturvedula LM, Sritharan V, Gupta S. 2017. In vitro flow-through assay for rapid detection of endotoxin in human sera: a proof-of-concept. Nanomedicine 13:1483–1490. doi: 10.1016/j.nano.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Gorbet MB, Sefton MV. 2005. Endotoxin: the uninvited guest. Biomaterials 26:6811–6817. doi: 10.1016/j.biomaterials.2005.04.063. [DOI] [PubMed] [Google Scholar]
- 18.Slot JW, Geuze HJ. 1985. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur J Cell Biol 38:87–93. [PubMed] [Google Scholar]
- 19.Singh R, Patil S, Singh N, Gupta S. 2017. Dual functionality nanobioconjugates targeting intracellular bacteria in cancer cells with enhanced antimicrobial activity. Sci Rep 7:5792. doi: 10.1038/s41598-017-06014-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pal N, Sharma S, Gupta S. 2016. Sensitive and rapid detection of pathogenic bacteria in small volumes using impedance spectroscopy technique. Biosens Bioelectron 77:270–276. doi: 10.1016/j.bios.2015.09.037. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Aaron J, Sokolov K. 2008. Directional conjugation of antibodies to nanoparticles for synthesis of multiplexed optical contrast agents with both delivery and targeting moieties. Nat Protoc 3:314–320. doi: 10.1038/nprot.2008.1. [DOI] [PubMed] [Google Scholar]
- 22.Hammond JB, Kruger NJ. 1988. The Bradford method for protein quantitation. Methods Mol Biol 3:25–32. [DOI] [PubMed] [Google Scholar]
- 23.Klausegger A, Hell M, Berger A, Zinober K, Baier S, Jones N, Sperl W, Kofler B. 1999. Gram type-specific broad-range PCR amplification for rapid detection of 62 pathogenic bacteria. J Clin Microbiol 37:464–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bass JJ, Wilkinson DJ, Rankin D, Phillips BE, Szewczyk NJ, Smith K, Atherton PJ. 2017. An overview of technical considerations for Western blotting applications to physiological research. Scand J Med Sci Sports 27:4–25. doi: 10.1111/sms.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opal SM, Scannon PJ, Vincent JL, White M, Carroll SF, Palardy JE, Parejo NA, Pribble JP, Lemke JH. 1999. Relationship between plasma levels of lipopolysaccharide (LPS) and LPS-binding protein in patients with severe sepsis and septic shock. J Infect Dis 180:1584–1589. doi: 10.1086/315093. [DOI] [PubMed] [Google Scholar]
- 26.Ginsburg I. 2002. Role of lipoteichoic acid in infection and inflammation. Lancet Infect Dis 2:171–179. [DOI] [PubMed] [Google Scholar]
- 27.Weber JR, Moreillon P, Tuomanen EI. 2003. Innate sensors for Gram-positive bacteria. Curr Opin Immunol 15:408–415. [DOI] [PubMed] [Google Scholar]
- 28.Stearns-Kurosawa DJ, Osuchowski MF, Valentine C, Kurosawa S, Remick DG. 2011. The pathogenesis of sepsis. Annu Rev Pathol 6:19–48. doi: 10.1146/annurev-pathol-011110-130327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Percy MG, Gründling A. 2014. Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu Rev Microbiol 68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 30.Rosenfeld Y, Shai Y. 2006. Lipopolysaccharide (endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim Biophys Acta 1758:1513–1522. doi: 10.1016/j.bbamem.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 31.Kimbrell MR, Warshakoon H, Cromer JR, Malladi S, Hood JD, Balakrishna R, Scholdberg TA, David SA. 2008. Comparison of the immunostimulatory and proinflammatory activities of candidate Gram-positive endotoxins, lipoteichoic acid, peptidoglycan, and lipopeptides, in murine and human cells. Immunol Lett 118:132–141. doi: 10.1016/j.imlet.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishihata K, Kakihana Y, Yasuda T, Imabayashi T, Nakamura N. 2013. Newly developed endotoxin measurement method (the endotoxin activity assay) may reflect the severity of sepsis. Open J Pathol 3:27260. doi: 10.4236/ojpathology.2013.31001. [DOI] [Google Scholar]
- 33.Paulssen J, Michaelsen P. 1984. The Limulus amoebocyte lysate (LAL) assay for the detection of endotoxin in fat emulsions for total parenteral nutrition (TPN). Acta Pathol Microbiol Immunol Scand B 92:177–179. [DOI] [PubMed] [Google Scholar]
- 34.Kim H, Hur M, Moon HW, Yun YM, Di Somma S. 2017. Multi-marker approach using procalcitonin, presepsin, galectin-3, and soluble suppression of tumorigenicity 2 for the prediction of mortality in sepsis. Ann Intensive Care 7:27. doi: 10.1186/s13613-017-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsthaler F, Kettner M, Mall G, Bratzke H. 2008. The use of rapid diagnostic test of procalcitonin serum levels for the postmortem diagnosis of sepsis. Forensic Sci Int 178:139–145. doi: 10.1016/j.forsciint.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Mehanic S, Baljic R. 2013. The importance of serum procalcitonin in diagnosis and treatment of serious bacterial infections and sepsis. Mater Sociomed 25:277–281. doi: 10.5455/msm.2013.25.277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiest R, Garcia-Tsao G. 2005. Bacterial translocation (BT) in cirrhosis. Hepatology 41:422–433. doi: 10.1002/hep.20632. [DOI] [PubMed] [Google Scholar]
- 38.Balzan S, de Almeida Quadros C, de Cleva R, Zilberstein B, Cecconello I. 2007. Bacterial translocation: Overview of mechanisms and clinical impact. J Gastroenterol Hepatol 22:464–471. doi: 10.1111/j.1440-1746.2007.04933.x. [DOI] [PubMed] [Google Scholar]
- 39.Berg RD. 1999. Bacterial translocation from the gastrointestinal tract. Adv Exp Med Biol 473:11–30. doi: 10.1007/978-1-4615-4143-1_2. [DOI] [PubMed] [Google Scholar]
- 40.Cohen J. 2002. The immunopathogenesis of sepsis. Nature 420:885–891. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- 41.Lee H. 2013. Procalcitonin as a biomarker of infectious diseases. Korean J Intern Med 28:285–291. doi: 10.3904/kjim.2013.28.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rocke DA, Gaffin SL, Wells MT, Koen Y, Brock-Utine JG. 1987. Endotoxemia associated with cardiopulmonary bypass. J Thorac Cardiovasc Surg 93:832–837. [PubMed] [Google Scholar]
- 43.Hurley JC. 1995. Endotoxemia: methods of detection and clinical correlates. Clin Microbiol Rev 8:268–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danner RL, Elin RJ, Hosseini JM, Wesley RA, Reilly JM, Parillo JE. 1991. Endotoxemia in human septic shock. Chest 99:169–175. [DOI] [PubMed] [Google Scholar]
- 45.Hurley JC. 1994. Concordance of endotoxemia with gram-negative bacteremia in patients with gram-negative sepsis: a meta-analysis. J Clin Microbiol 32:2120–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schedel I, Dreikhausen U, Nentwig B, Hockenschnieder M, Rauthmann D, Balikcioglu S, Coldewey R, Deicher H. 1991. Treatment of Gram-negative septic shock with an immunoglobulin preparation: a prospective, randomized clinical trial. Crit Care Med 19:1104–1113. doi: 10.1097/00003246-199109000-00003. [DOI] [PubMed] [Google Scholar]
- 47.Leong DU, Greisen KS. 1993. In Pershing DH. (ed), Diagnostic molecular microbiology-principles and applications. ASM Press, Washington, DC. [Google Scholar]
- 48.Haddadin Y, Regunath H. 2017. Central line associated blood stream infections (CLABSI). StatPearls Publishing, Treasure Island, FL. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.