Bacterial vaginosis (BV) is the most common cause of vaginal discharge in reproductive-age women. BV has been associated with poor reproductive outcomes such as preterm delivery, the acquisition of sexually transmitted infections, including HIV, and pelvic inflammatory disease.
KEYWORDS: bacterial vaginosis, real-time PCR, vaginal flora
ABSTRACT
Bacterial vaginosis (BV) is the most common cause of vaginal discharge in reproductive-age women. BV has been associated with poor reproductive outcomes such as preterm delivery, the acquisition of sexually transmitted infections, including HIV, and pelvic inflammatory disease. BV represents the acquisition of a diverse community of anaerobic and facultative bacteria and a reduction in lactobacilli. It can be diagnosed using several tests ranging from clinical indicators, point-of-care tests, and molecular assays. Molecular technologies are objective, are able to detect fastidious bacteria, enable quantitation, and are ideal for self-collected vaginal swabs. This paper reviews the currently available BV diagnostic tests in the United States.
INTRODUCTION
Bacterial vaginosis (BV) is a condition characterized by an alteration in vaginal flora and is the most common cause of vaginal discharge in reproductive-age women (1). In the United States, it is estimated that nearly 30% of the general population of women has had BV, and the prevalence varies with race and ethnicity (2). Black/African-American women (odds ratio [OR], 3.13; 95% confidence interval [CI], 2.58 to 3.80) and Mexican-American women (OR, 1.29; 95% CI, 0.99 to 1.69) have higher odds of BV than white non-Hispanic women. BV has been associated with poor reproductive outcomes such as preterm delivery, the acquisition of sexually transmitted infections, including HIV, and pelvic inflammatory disease (3–6). Many women report a fishy odor and vaginal discharge; however, up to 50% may be asymptomatic (7, 8). BV can be diagnosed using several different tests ranging from clinical indicators, point-of-care tests, and molecular assays. However, studies show that symptomatic patients may be misdiagnosed, as clinicians might not have access to specific diagnostic tools or may not adhere to the strict diagnostic criteria of clinical indicator tests (9).
MICROBIOLOGY
BV is not caused by a single organism, instead, it represents the acquisition of a diverse community of anaerobic and facultative bacteria and a reduction in lactobacilli (10, 11). The most common types of lactobacilli in the vagina include Lactobacillus crispatus, L. jensenii, L. gasseri, and L. iners (11, 12). Most of these lactobacilli act as important host-defense mechanisms against BV by secreting substances that inhibit the growth of microbial pathogens and indigenous anaerobes (13). For example, L. crispatus produces lactic acid and hydrogen peroxide, which help to maintain a low vaginal pH (14, 15). Recent data suggest that lactic acid has greater bactericidal activity against BV-associated bacteria than hydrogen peroxide (14, 16). L. iners, in contrast, has been found in women with and without BV (17). In a study that used a combination of broad-range PCR, cloning, and sequencing, women with BV had greater bacterial diversity, with 9 to 17 bacterial species (mean, 12.6), while women without BV had 1 to 6 bacterial species (mean, 3.3) (18). This study also reported bacteria that were frequently detected in women with BV that included Gardnerella vaginalis, Atopobium vaginae, Megasphaera types, Leptotrichia amnionii, Sneathia sanguinegens, Porphyromonas asaccharolytica, a bacterium related to Eggerthella hongkongensis, and bacteria related to Prevotella species. Less frequently detected bacteria included Peptostreptococcus spp., Aerococcus, Anaerococcus, Gemella, and Veillonella genera. Thirty-five unique bacterial species were identified in women with BV, 16 of which were newly characterized, including fastidious bacteria termed BV-associated bacterium 1 (BVAB1), BVAB2, and BVAB3. BVAB1, -2, and -3 were highly specific indicators for BV.
A substantial body of literature exists on G. vaginalis, as it was originally thought to be the single bacterium responsible for BV. In 1955, before informed consent was required for clinical investigation, Gardner and Dukes inoculated the vaginas of 15 healthy women with secretions from women with BV, and 11 of them developed BV symptoms (19). Since their early work, numerous studies have shown that G. vaginalis can be recovered from 98% to 100% of women with BV; however, it can also be recovered from up to 55% of women without BV (20).
Despite this lack of specificity, G. vaginalis is still a key bacterium in the etiology of BV. G. vaginalis is the main bacterium that produces a biofilm matrix, whereas other bacterial species such as A. vaginae and Fusobacterium spp. stimulate and enhance the biofilm formed by G. vaginalis (21). These biofilms are communities of microorganisms attached to the surface of epithelial cells and are encased in a polymeric matrix of polysaccharides, proteins, and nucleic acids (22) Biofilms are important clinically, because they are able to adhere to epithelial cells even in the presence of lactic acid-producing bacteria such as lactobacilli, which may inhibit their elimination by the immune system or prevent microbiologic cure after antibiotic usage (23–26). Some in vitro studies have shown that G. vaginalis biofilms are more tolerant to lactic acid and hydrogen peroxide than G. vaginalis in broth culture (24).
Furthermore, G. vaginalis has four different clades that differ in genome sizes and core gene contents. Each of these clades possesses unique genetic markers, which may lead to different metabolic by-products and virulence (27). For example, sialidase, which enhances biofilm formation through its mucinase activity, has been detected in all clades, but enzymatic activity has been detected only in two of the four (28, 29). Additionally, certain G. vaginalis clades, e.g., clades 1 and 3, are found more commonly among women with BV than among women without BV (30). Research is ongoing to determine whether quantitative assessments of G. vaginalis might be more diagnostic than qualitative assessments and whether there are differences in virulence among the different clades.
Several bacterial species present in women with BV produce metabolic by-products that can be detected in vaginal fluid. First, anaerobic bacteria produce proteolytic carboxylase enzymes that metabolize peptides into volatile amines (putrescine, cadaverine, and trimethylamine). Amines, such as trimethylamine, lead to the classic fishy odor characteristic of BV, which is one of the clinical indicators for a BV diagnosis. Second, organic acids such as succinate, acetate, and other short-chain acids can be detected in women with BV. Third, the enzymes proline aminopeptidase and sialidase produced by several bacteria in women with BV are used in some diagnostic assays.
POINT-OF-CARE TESTS
Saline microscopy or wet mount.
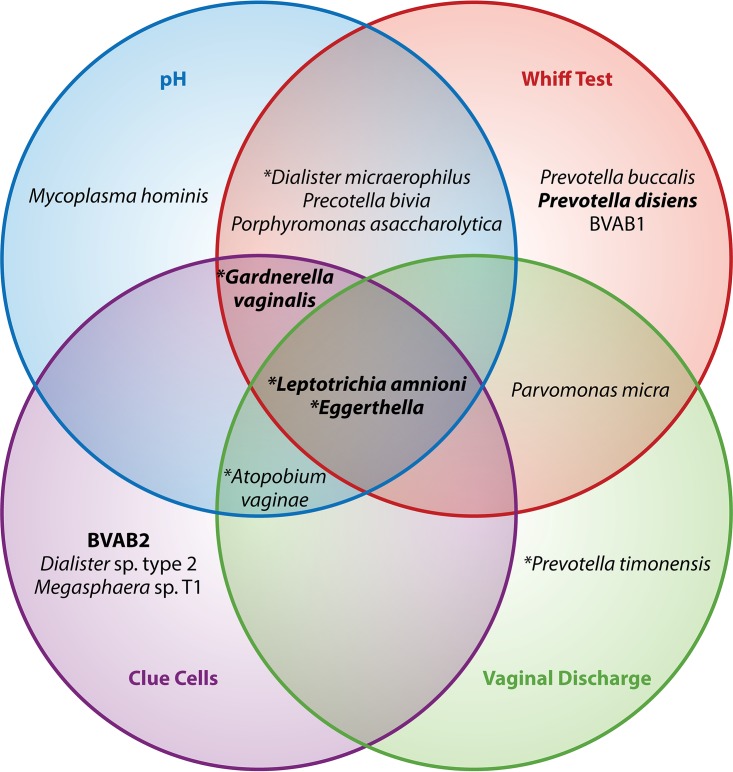
Amsel's criteria are generally used to make the diagnosis of BV using saline microscopy. A diagnosis requires the presence of at least three of the four criteria (i.e., thin, watery homogenous discharge; elevated vaginal pH [>4.5]; 20% of clue cells on saline microscopy; and a fishy odor after the addition of 10% potassium hydroxide to a slide of secretions [whiff test]) (7). The rise in pH that occurs in BV facilitates the adherence of G. vaginalis, which then produces cytotoxins and a biofilm that is exfoliated, leading to appearance of clue cells. Figure 1 presents a model showing bacteria that are associated with Amsel's criteria for the diagnosis of BV. It attempts to demonstrate how some of the newer molecular 16S rRNA sequencing research on noncultivable organisms associated with BV may relate to some of the more standard Amsel's methods with which BV has traditionally been diagnosed by clinicians. The sensitivity and specificity for Amsel's criteria have ranged from 37% to 70% and 94% to 99%, respectively, compared to the gold-standard Gram stain Nugent score, and the reproducibility is moderate (31, 32). Microscopy is desired by some clinicians, because it is relatively fast and a diagnosis can be made during the office visit. However, data suggest that clinicians do not routinely assess all criteria or may inaccurately diagnose BV on the basis of a lack of time or skills (33).
FIG 1.
Relationship between Amsel's criteria and bacterial communities in women with bacterial vaginosis. Diagram showing the bacterial taxa that are associated with the four factors used to diagnose BV using Amsel's criteria (50). For example, G. vaginalis is associated with elevated pH, clue cells, and a positive whiff test. Asterisks represent bacteria present in >75% of women with BV, and taxa in bold are those associated with Amsel's criteria as a composite unit.
OSOM BV Blue.
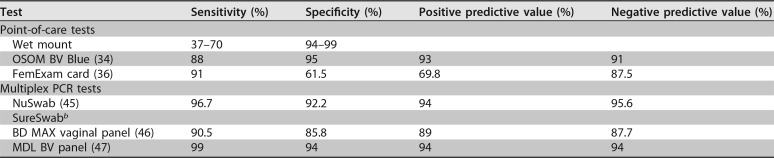
The OSOM BV Blue assay (Genzyme Diagnostics, Cambridge, MA) is a chromogenic point-of-care test (POCT) that measures sialidase levels in vaginal fluid. Sialidases, formerly known as neuraminidases, are produced by bacteria such as Gardnerella and Bacteroides species. The test is a dipstick and the results are available within 10 min; hence, it is a very useful bedside tool, especially in clinics without microscopy. The sensitivity and specificity for the OSOM BV Blue assay range from 88% to 94% and 91% to 98%, respectively, compared to Nugent and Amsel's criteria (34, 35).
FemExam card.
The FemExam (Cooper Surgical, Shelton, CT) POCT detects metabolic products of G. vaginalis, which include amines and the activity of proline aminopeptidase, and measures vaginal pH. It consists of two plastic cards; one card is for pH measurement and the presence of trimethylamine, and the second card is for proline aminopeptidase measurement. The combined sensitivity of cards 1 and 2 is 91% and the specificity is 61% compared with the Nugent score. It is very fast (2 min), objective, and easily performed (36).
LABORATORY TESTS
Gram stain.
The Gram stain is the gold-standard for the diagnosis of BV and has been used in laboratories since 1965. Gram stain results consistent with BV include Gardnerella morphotypes, including cocci, fusiforms, and curved rods, and reduced lactobacilli morphotypes. Gram stains are more specific for BV with high interobserver and intraobserver reproducibility than are Amsel's criteria (37); however, it is time consuming. The three bacterial morphotypes with the highest degree of reproducibility are Lactobacillus (large Gram-positive rods), Gardnerella and Bacteroides (small Gram-positive or Gram-variable rods), and Mobiluncus (curved Gram-negative or Gram-variable rods). A standardized scoring system for the diagnosis of BV, called the Nugent score, is most often used. With the Nugent score, the slide is examined for the quantity of Gram-positive rods and lactobacilli (i.e., normal flora) and Gram-negative or Gram-variable morphotypes (BV flora) (38). The results are given as a score of 0 to 3 (normal flora), 4 to 6 (intermediate or mixed flora), and 7 to 10 (BV). Women with intermediate or mixed flora often have elevated pH, clue cells, and a mild amine odor, which may be overdiagnosed as BV if Amsel's criteria were used alone. Prior studies found that 37% to 54% of women with intermediate flora determined by the Nugent score had BV by Amsel's criteria (34, 39). Unless arranged with a clinical laboratory, the results are typically not available at the time of an office visit. Because of the skill set needed to read slides and the volume of tests required to be performed to maintain proficiency, the Nugent score is typically used only in research settings. The scoring of morphotypes is also highly subjective and influenced by individual skill. Other disadvantages of the Nugent score include the inability to identify several of the newer bacterial morphotypes that have been associated with BV.
MOLECULAR DIAGNOSTIC ASSAYS
Given the limitations of microscopy and other POCTs for BV diagnosis, new technologies using molecular markers of BV have been developed. Molecular technologies are advantageous over POCTs and microscopy-based tests because they are objective, are able to detect fastidious bacteria, enable quantitation, and are ideal for self-collected vaginal swabs. These technologies offer higher performance and are based on the detection of specific bacterial nucleic acids. The primary types of commercial molecular assays available in the United States for BV diagnostics are direct DNA probe and nucleic acid amplification assays.
Direct probe assays.
Direct probe assays introduce a DNA probe to a vaginal fluid sample. The DNA probe binds to specific sequences from a particular bacterium and can detect the presence of different bacteria in a single sample. Some tend to have high clinical sensitivity due to the targeting of rRNA, which is present in thousands of copies in each cell in contrast to DNA, which is not as abundant.
(i) Affirm VP.
The Affirm VP assay (Becton Dickinson, Sparks, MD) is a synthetic oligonucleotide probe test that uses a rapid (30 min), semiautomated nonisotopic assay for high concentrations of G. vaginalis nucleic acids. It uses two distinct single-stranded probes, a capture probe, and a color development probe. The detection of greater than 5 × 105 CFU of G. vaginalis per milliliter of vaginal fluid is used to diagnose BV, which was similar to a BV diagnosis using clue cell detection by wet mount and the Nugent score among a population of sexually transmitted disease clinic attendees (40). The DNA probe test had a reported sensitivity of 90% and specificity of 97% compared with the detection of clue cells by microscopy and sensitivity of 94% and specificity of 81% compared with Nugent score diagnosis of BV. However, because G. vaginalis can be detected in 36% to 55% of women without clinical signs of BV (7, 41–43), this test is most useful in symptomatic patients and when used in conjunction with vaginal pH and the presence of amine odor. Using these additional clinical criteria can increase the sensitivity to 97% and specificity to 71%. Because mixed infections are common, this assay now also offers the ability to diagnose the other common causes of vaginitis, such as Candida spp. and Trichomonas vaginalis. The company markets this triple test as the Affirm VPIII microbial identification test (Becton Dickinson, Sparks, MD).
(ii) Bacterial vaginosis/vaginitis panel.
This DNA probe test (Quest Diagnostics, Secaucus, NJ) is FDA approved for use in symptomatic patients only. It uses nucleic acid hybridization and has two distinct single-stranded probes for G. vaginalis. The test is read as negative if there are less than 2 × 105 CFU G. vaginalis. However, as noted previously, up to 55% of women without BV may harbor G. vaginalis in low concentrations. The detection of G. vaginalis is not diagnostic of BV, and other clinical indicators (i.e., Amsel's criteria) should be used to help make the diagnosis. Additionally, this test was not studied in patients who were given treatment and cannot be used to gauge response to treatment. The expanded panel includes the detection of Candida spp. (1 × 104 CFU) and T. vaginalis (5 × 103 trichomonads).
Nucleic acid amplification tests.
Nucleic acid amplification tests (NAATs), such as PCR, are theoretically capable of detecting as little as one organism in a sample. Many research and commercially available NAAT assays have been published. They use techniques that enzymatically multiply a specific nucleic acid sequence exponentially, resulting in the production of billions of copies of the sequence in a short period of time. The amplified product is then easily detected by DNA probes or other methods. These real-time PCRs have been used by many researchers, as DNA amplification can be observed in real-time, which eliminates the need for postamplification analysis and decreases the chances for contamination.
In a follow-up from their original work, Fredricks et al. developed a panel of taxon-directed 16S rRNA gene PCR assays for the detection of 17 vaginal bacteria (44). The bacterial PCR assays were capable of detecting ≤100 molecules of cloned 16S rRNA gene per reaction. Using samples from a greater number of women with and without BV, they assessed the prevalence of each of these bacterial species. Among women with BV, there was an average of 11.1 species detected (range, 5 to 16), while women without BV had an average of 3.6 species (range, 0 to 14). The detection of either BVAB2 or Megasphaera type 1 had a sensitivity of 95.9% and specificity of 93.7% compared with the Nugent score. Given the polymicrobial nature of BV, it is desirable to amplify more than one target sequence at a time. Hence, quantitative multiplex PCR assays have been the focus of development. Multiplex PCR uses unique primer and probe sets that bind to regions of the 16S rRNA gene to provide a quick and simple alternative to Southern blot analysis for the evaluation of gene copy number or expression levels. Different bacterial species that are associated with BV have divergent positive predictive values (PPV) for diagnosis of BV if used alone. However, the combined detection of several different bacterial species might improve the test characteristics for BV diagnostics. Four quantitative or semiquantitative multiplex real-time PCR assays are commercially available in the United States. Table 1 shows the indicator organisms used in each testing algorithm.
TABLE 1.
Bacteria detected in commercially available PCR assaysa
| Organism | NuSwab | SureSwab | BD Max | MDL BV panel |
|---|---|---|---|---|
| A. vaginae | X | X | X | X |
| G. vaginalis | X | X | X | |
| Megasphaera (type 1, type 2 and/or species) | X | X | X | X |
| BVABb (type 1 and/or type 2) | X | X | X | |
| Lactobacillus species | X | X | X | X |
Commercially available PCR assays detect different combinations of indicator organisms.
BVAB, BV-associated bacteria.
(i) NuSwab.
The first quantitative multiplex PCR assay (Laboratory Corporation of America Holdings, Burlington, NC) used logistic regression to select organisms to diagnose BV. It detects three bacterial species that have been shown to be useful predictors of BV that include A. vaginae, BVAB2, and Megasphaera type 1 and one lactobacilli species, L. crispatus, which has been reported to be a negative predictor of BV (45). This assay does not include G. vaginalis to avoid the misinterpretation of results, since G. vaginalis is very common in women who do not have BV, as well as in women with BV. Using samples from symptomatic women, the assay demonstrated 96.7% sensitivity, 92.2% specificity, 94% PPV, and 95.6% negative predictive value (NPV) for BV detection compared with a combination of the Nugent score and Amsel's criteria. The assay uses a scoring system where scores of 0 to 1 are considered negative for BV, scores of 3 to 6 are considered positive for BV, and a score of 2 is considered intermediate for BV. In its validation study, 5.3% of BV samples were classified as intermediate. This assay also has an expanded version that detects Candida albicans, Candida glabrata, Chlamydia trachomatis, Neisseria gonorrhoeae, and Trichomonas vaginalis infections.
(ii) SureSwab BV DNA quantitative real-time PCR.
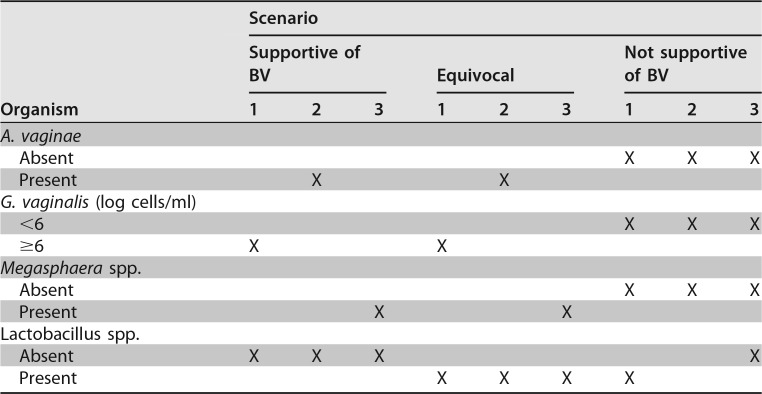
The second quantitative real-time multiplex PCR assay (Quest Diagnostics, Secaucus, NJ) detects hydrogen peroxide-producing lactobacilli (Lactobacillus acidophilus, L. crispatus, and L. jensenii) that are negative indicators of BV, as well as G. vaginalis, A. vaginae, and Megasphaera spp. Table 2 shows the complexity of a BV diagnosis based on certain indicator organisms and is an example of how the reported results can be interpreted according to data from the company's (Quest Diagnostics, Secaucus, NJ) website.
TABLE 2.
SureSwab diagnostic algorithm
(iii) BD Max vaginal panel.
The third quantitative real-time multiplex PCR assay (Becton Dickinson, Sparks, MD) that recently received FDA market authorization for the diagnosis of vaginitis in symptomatic women provides a positive or negative result for BV Candida group (C. albicans, C. tropicalis, C. parapsilosis, and C. dubliniensis), C. glabrata or C. krusei, and Trichomonas vaginalis (46). This multiplex assay uses a proprietary algorithm to diagnose BV that includes quantitative an assessment of lactobacilli (L. crispatus and L. jensenii), G. vaginalis, A. vaginae, Megasphaera type 1, and BVAB2 (46). Compared to the reference of a combined Nugent score and Amsel's criteria, the test had 90.5% sensitivity (95% confidence interval [CI], 88.3% to 92.2%), 85.8% specificity (95% CI, 83% to 88.3%), 89% PPV (95% CI, 87.1 to 90.7), and 87.7% NPV (95% CI, 85.4 to 89.8) for BV.
(iv) BV panel.
The fourth quantitative real-time multiplex PCR assay (Medical Diagnostic Laboratory, Hamilton Township, NJ) evaluated nine vaginal bacterial organisms as potential diagnostic markers of BV and, through quantitation and receiver operating curve (ROC) analyses, found that three were predictive of BV diagnosis: A. vaginae, G. vaginalis, and Megasphaera phylotypes 1 and 2 (47). The investigators found that G. vaginalis was a highly accurate predictor of BV if quantitation and ROC analysis were performed. In contrast, the inclusion of L. crispatus did not contribute to the diagnostic accuracy and was not included in the final model. Compared to the reference of a combined Nugent score and Amsel's criteria, the test had 92% sensitivity, 95% specificity, 94% PPV, and 94% NPV for BV. The company has since added BVAB2 and Lactobacillus profiling that increases the sensitivity to 99% and specificity to 94%.
Emerging assays.
Many studies have used techniques such as microarray analysis and next-generation sequencing (NGS). The use of these methods has enabled sequencing of known and unknown organisms implicated in BV, which have subsequently provided the foundation for the development of commercial molecular BV diagnostics. These assays are not commercially available and are typically performed in large reference laboratories.
(i) Microarray analysis.
Microarrays contain a unique nucleotide sequence that represents a specific gene, chromosomal location, or other nucleic acid sequence being interrogated. This technology requires knowledge of the expected organisms in the sample. Labeled nucleic acids are incubated on the surface of the microarray to enable hybridization between the microarray probes and the vaginal fluid sample-derived nucleic acids. Although the designs vary greatly, arrays may contain many thousands or millions of unique DNA probe sequences that appear as separate dots or features on a fixed surface. Gene expression arrays require the application of mRNA or cDNA to the microarray that contains all known genes expressed by a particular bacterium. The levels of gene expression can be determined by the intensity of the signals at each spot. Microarray results using mRNA isolated from two or more vaginal fluid samples can be compared to assess the up- or downregulation of each gene represented on the microarray. Oligonucleotide-based microarray analysis has been used to detect known bacterial species such as G. vaginalis, A. vaginae, Megasphaera species, Mobiluncus mulieris, Sneathia sanguinegens, and Prevotella species (48). A recent report demonstrated a microarray analysis that included probes from 17 bacterial targets associated with BV (49); however, there are not any commercially available microarray diagnostics in the United States.
(ii) Sequencing technology.
In contrast to microarrays, NGS, such as Illumina 16S rRNA and 454-sequencing, can detect sequences without prior knowledge of the bacterial species, which can help deepen the understanding of the diverse vaginal microbiota. Amplicon sequencing methods such as Illumina derive the family, genus, and species assignments from a comparison of V1 to V3 or V4 16S rRNA gene sequences with those of strains present in the ribosomal database project. These NGS technologies produce massively parallel sequencing data. Groups of genes or an entire genome with each segment of DNA being sequenced tens, hundreds, or thousands of times can be read. These provide an increased ability to detect low-level variation and make sequencing across large amounts of DNA more feasible. Assays capable of sequencing an entire gene or group of genes related to BV have been developed. The rapid development of NGS platforms have enabled costs to decrease. One limitation is that a particular species can be under- or overrepresented, since it may be difficult to discriminate the various regions of the 16S rRNA gene sequences from two separate species.
CONCLUSION
In summary, there are a wide variety of diagnostic assays available to diagnose BV, ranging from POCTs to molecular assays. Clinicians will need to consider costs, result time, and accuracy in their decision to select a particular assay to test for BV among symptomatic women. There are not any recommendations to screen asymptomatic women for BV. This review is intended to increase the knowledge about the different molecular assays offered and their test characteristics (Table 3), as not all of the molecular assays may be worth utilizing in clinical practice. Clinicians may want to avoid using single BV indicator organism (e.g., G. vaginalis) assays, such as the direct probe assays, in favor of multiplex PCR technology that is able to detect multiple indicator organisms. While the wet mount is advantageous for its low cost and immediate results, the multiplex PCR assays might be more useful in the diagnostic work-up of symptomatic women with recurrent vaginitis. Although these multiplex PCR assays assert an improved accuracy of diagnosing BV, a possible limitation of their performance evaluation is the reference standard to which they are compared. However, as technology continues to advance, it will enable further understanding of the complexity of BV.
TABLE 3.
Test characteristics of select clinical diagnostic tests for BVa
Reference standard is the Nugent score and/or Amsel's criteria.
b No published test characteristics found in the literature.
REFERENCES
- 1.Morris M, Nicoll A, Simms I, Wilson J, Catchpole M. 2001. Bacterial vaginosis: a public health review. BJOG 108:439–450. doi: 10.1111/j.1471-0528.2001.00124.x. [DOI] [PubMed] [Google Scholar]
- 2.Allsworth JE, Peipert JF. 2007. Prevalence of bacterial vaginosis: 2001–2004 National Health and Nutrition Examination Survey data. Obstet Gynecol 109:114–120. doi: 10.1097/01.AOG.0000247627.84791.91. [DOI] [PubMed] [Google Scholar]
- 3.Hauth JC, Macpherson C, Carey JC, Klebanoff MA, Hillier SL, Ernest JM, Leveno KJ, Wapner R, Varner M, Trout W, Moawad A, Sibai B. 2003. Early pregnancy threshold vaginal pH and Gram stain scores predictive of subsequent preterm birth in asymptomatic women. Am J Obstet Gynecol 188:831–835. doi: 10.1067/mob.2003.184. [DOI] [PubMed] [Google Scholar]
- 4.Klebanoff MA, Hillier SL, Nugent RP, MacPherson CA, Hauth JC, Carey JC, Harper M, Wapner RJ, Trout W, Moawad A, Leveno KJ, Miodovnik M, Sibai BM, Vandorsten JP, Dombrowski MP, O'Sullivan MJ, Varner M, Langer O, National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. 2005. Is bacterial vaginosis a stronger risk factor for preterm birth when it is diagnosed earlier in gestation? Am J Obstet Gynecol 192:470–477. doi: 10.1016/j.ajog.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, Mandaliya K, Ndinya-Achola JO, Bwayo J, Kreiss J. 1999. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis 180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- 6.Myer L, Denny L, Telerant R, Souza M, Wright TC Jr, Kuhn L. 2005. Bacterial vaginosis and susceptibility to HIV infection in South African women: a nested case-control study. J Infect Dis 192:1372–1380. doi: 10.1086/462427. [DOI] [PubMed] [Google Scholar]
- 7.Amsel R, Totten PA, Spiegel CA, Chen KC, Eschenbach D, Holmes KK. 1983. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am J Med 74:14–22. [DOI] [PubMed] [Google Scholar]
- 8.Koumans EH, Sternberg M, Bruce C, McQuillan G, Kendrick J, Sutton M, Markowitz LE. 2007. The prevalence of bacterial vaginosis in the United States, 2001–2004; associations with symptoms, sexual behaviors, and reproductive health. Sex Transm Dis 34:864–869. doi: 10.1097/OLQ.0b013e318074e565. [DOI] [PubMed] [Google Scholar]
- 9.Chavoustie SE, Eder SE, Koltun WD, Lemon TR, Mitchell C, Nyirjesy P, Sobel JD, Sobel R, Villanueva R. 2017. Experts explore the state of bacterial vaginosis and the unmet needs facing women and providers. Int J Gynaecol Obstet 137:107–109. doi: 10.1002/ijgo.12114. [DOI] [PubMed] [Google Scholar]
- 10.Bradshaw CS, Sobel JD. 2016. Current Treatment of Bacterial Vaginosis-Limitations and Need for Innovation. J Infect Dis 214 Suppl 1:S14–S20. doi: 10.1093/infdis/jiw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill GB. 1993. The microbiology of bacterial vaginosis. Am J Obstet Gynecol 169:450–454. doi: 10.1016/0002-9378(93)90339-K. [DOI] [PubMed] [Google Scholar]
- 12.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108 Suppl 1:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branco KM, Nardi RM, Moreira JL, Nunes AC, Farias LM, Nicoli JR, Carvalho MA. 2010. Identification and in vitro production of Lactobacillus antagonists from women with or without bacterial vaginosis. Braz J Med Biol Res 43:338–344. doi: 10.1590/S0100-879X2010007500013. [DOI] [PubMed] [Google Scholar]
- 14.O'Hanlon DE, Moench TR, Cone RA. 2011. In vaginal fluid, bacteria associated with bacterial vaginosis can be suppressed with lactic acid but not hydrogen peroxide. BMC Infect Dis 11:200. doi: 10.1186/1471-2334-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tachedjian G, Aldunate M, Bradshaw CS, Cone RA. 2017. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res Microbiol 168:782–792. doi: 10.1016/j.resmic.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 16.O'Hanlon DE, Moench TR, Cone RA. 2013. Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS One 8:e80074. doi: 10.1371/journal.pone.0080074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. 2010. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol 48:1812–1819. doi: 10.1128/JCM.00851-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredricks DN, Fiedler TL, Marrazzo JM. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 19.Gardner HL, Dukes CD. 1955. Haemophilus vaginalis vaginitis: a newly defined specific infection previously classified non-specific vaginitis. Am J Obstet Gynecol 69:962–976. doi: 10.1016/0002-9378(55)90095-8. [DOI] [PubMed] [Google Scholar]
- 20.Totten PA, Amsel R, Hale J, Piot P, Holmes KK. 1982. Selective differential human blood bilayer media for isolation of Gardnerella (Haemophilus) vaginalis. J Clin Microbiol 15:141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machado A, Jefferson KK, Cerca N. 2013. Interactions between Lactobacillus crispatus and bacterial vaginosis (BV)-associated bacterial species in initial attachment and biofilm formation. Int J Mol Sci 14:12004–12012. doi: 10.3390/ijms140612004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, Jensen PO, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T. 2011. The clinical impact of bacterial biofilms. Int J Oral Sci 3:55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerca N, Martins S, Cerca F, Jefferson KK, Pier GB, Oliveira R, Azeredo J. 2005. Comparative assessment of antibiotic susceptibility of coagulase-negative staphylococci in biofilm versus planktonic culture as assessed by bacterial enumeration or rapid XTT colorimetry. J Antimicrob Chemother 56:331–336. doi: 10.1093/jac/dki217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson JL, Girerd PH, Karjane NW, Jefferson KK. 2007. Effect of biofilm phenotype on resistance of Gardnerella vaginalis to hydrogen peroxide and lactic acid. Am J Obstet Gynecol 197:170.e1–170.e7. doi: 10.1016/j.ajog.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machado D, Castro J, Palmeira-de-Oliveira A, Martinez-de-Oliveira J, Cerca N. 2016. Bacterial vaginosis biofilms: challenges to current therapies and emerging solutions. Front Microbiol 6:1528. doi: 10.3389/fmicb.2015.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerca N, Jefferson KK, Oliveira R, Pier GB, Azeredo J. 2006. Comparative antibody-mediated phagocytosis of Staphylococcus epidermidis cells grown in a biofilm or in the planktonic state. Infect Immun 74:4849–4855. doi: 10.1128/IAI.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed A, Earl J, Retchless A, Hillier SL, Rabe LK, Cherpes TL, Powell E, Janto B, Eutsey R, Hiller NL, Boissy R, Dahlgren ME, Hall BG, Costerton JW, Post JC, Hu FZ, Ehrlich GD. 2012. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol 194:3922–3937. doi: 10.1128/JB.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santiago GL, Deschaght P, El Aila N, Kiama TN, Verstraelen H, Jefferson KK, Temmerman M, Vaneechoutte M. 2011. Gardnerella vaginalis comprises three distinct genotypes of which only two produce sialidase. Am J Obstet Gynecol 204:450.e1–450.e7. doi: 10.1016/j.ajog.2010.12.061. [DOI] [PubMed] [Google Scholar]
- 29.Schellenberg JJ, Paramel Jayaprakash T, Withana Gamage N, Patterson MH, Vaneechoutte M, Hill JE. 2016. Gardnerella vaginalis subgroups defined by cpn60 sequencing and sialidase activity in isolates from Canada, Belgium and Kenya. PLoS One 11:e0146510. doi: 10.1371/journal.pone.0146510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balashov SV, Mordechai E, Adelson ME, Gygax SE. 2014. Identification, quantification and subtyping of Gardnerella vaginalis in noncultured clinical vaginal samples by quantitative PCR. J Med Microbiol 63:162–175. doi: 10.1099/jmm.0.066407-0. [DOI] [PubMed] [Google Scholar]
- 31.Schwebke JR, Hillier SL, Sobel JD, McGregor JA, Sweet RL. 1996. Validity of the vaginal Gram stain for the diagnosis of bacterial vaginosis. Obstet Gynecol 88:573–576. doi: 10.1016/0029-7844(96)00233-5. [DOI] [PubMed] [Google Scholar]
- 32.Sha BE, Chen HY, Wang QJ, Zariffard MR, Cohen MH, Spear GT. 2005. Utility of Amsel criteria, Nugent score, and quantitative PCR for Gardnerella vaginalis, Mycoplasma hominis, and Lactobacillus spp. for diagnosis of bacterial vaginosis in human immunodeficiency virus-infected women. J Clin Microbiol 43:4607–4612. doi: 10.1128/JCM.43.9.4607-4612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menard JP, Fenollar F, Henry M, Bretelle F, Raoult D. 2008. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin Infect Dis 47:33–43. doi: 10.1086/588661. [DOI] [PubMed] [Google Scholar]
- 34.Bradshaw CS, Morton AN, Garland SM, Horvath LB, Kuzevska I, Fairley CK. 2005. Evaluation of a point-of-care test, BVBlue, and clinical and laboratory criteria for diagnosis of bacterial vaginosis. J Clin Microbiol 43:1304–1308. doi: 10.1128/JCM.43.3.1304-1308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myziuk L, Romanowski B, Johnson SC. 2003. BVBlue test for diagnosis of bacterial vaginosis. J Clin Microbiol 41:1925–1928. doi: 10.1128/JCM.41.5.1925-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West B, Morison L, Schim van der Loeff M, Gooding E, Awasana AA, Demba E, Mayaud P. 2003. Evaluation of a new rapid diagnostic kit (FemExam) for bacterial vaginosis in patients with vaginal discharge syndrome in The Gambia. Sex Transm Dis 30:483–489. doi: 10.1097/00007435-200306000-00003. [DOI] [PubMed] [Google Scholar]
- 37.Forsum U, Jakobsson T, Larsson PG, Schmidt H, Beverly A, Bjornerem A, Carlsson B, Csango P, Donders G, Hay P, Ison C, Keane F, McDonald H, Moi H, Platz-Christensen JJ, Schwebke J. 2002. An international study of the interobserver variation between interpretations of vaginal smear criteria of bacterial vaginosis. APMIS 110:811–818. doi: 10.1034/j.1600-0463.2002.1101107.x. [DOI] [PubMed] [Google Scholar]
- 38.Nugent RP, Krohn MA, Hillier SL. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J Clin Microbiol 29:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor-Robinson D, Morgan DJ, Sheehan M, Rosenstein IJ, Lamont RF. 2003. Relation between Gram-stain and clinical criteria for diagnosing bacterial vaginosis with special reference to Gram grade II evaluation. Int J STD AIDS 14:6–10. [DOI] [PubMed] [Google Scholar]
- 40.Briselden AM, Hillier SL. 1994. Evaluation of affirm VP Microbial Identification Test for Gardnerella vaginalis and Trichomonas vaginalis. J Clin Microbiol 32:148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eschenbach DA, Hillier S, Critchlow C, Stevens C, DeRouen T, Holmes KK. 1988. Diagnosis and clinical manifestations of bacterial vaginosis. Am J Obstet Gynecol 158:819–828. doi: 10.1016/0002-9378(88)90078-6. [DOI] [PubMed] [Google Scholar]
- 42.Krohn MA, Hillier SL, Eschenbach DA. 1989. Comparison of methods for diagnosing bacterial vaginosis among pregnant women. J Clin Microbiol 27:1266–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spiegel CA, Amsel R, Holmes KK. 1983. Diagnosis of bacterial vaginosis by direct Gram stain of vaginal fluid. J Clin Microbiol 18:170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. 2007. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol 45:3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cartwright CP, Lembke BD, Ramachandran K, Body BA, Nye MB, Rivers CA, Schwebke JR. 2012. Development and validation of a semiquantitative, multitarget PCR assay for diagnosis of bacterial vaginosis. J Clin Microbiol 50:2321–2329. doi: 10.1128/JCM.00506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaydos CA, Beqaj S, Schwebke JR, Lebed J, Smith B, Davis TE, Fife KH, Nyirjesy P, Spurrell T, Furgerson D, Coleman J, Paradis S, Cooper CK. 2017. Clinical validation of a test for the diagnosis of vaginitis. Obstet Gynecol 130:181–189. doi: 10.1097/AOG.0000000000002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilbert DW, Smith WL, Chadwick SG, Toner G, Mordechai E, Adelson ME, Aguin TJ, Sobel JD, Gygax SE. 2016. Development and validation of a highly accurate quantitative real-time PCR assay for diagnosis of bacterial vaginosis. J Clin Microbiol 54:1017–1024. doi: 10.1128/JCM.03104-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dols JA, Smit PW, Kort R, Reid G, Schuren FH, Tempelman H, Bontekoe TR, Korporaal H, Boon ME. 2011. Microarray-based identification of clinically relevant vaginal bacteria in relation to bacterial vaginosis. Am J Obstet Gynecol 204:305.e1–305.e7. doi: 10.1016/j.ajog.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 49.Cruciani F, Biagi E, Severgnini M, Consolandi C, Calanni F, Donders G, Brigidi P, Vitali B. 2015. Development of a microarray-based tool to characterize vaginal bacterial fluctuations and application to a novel antibiotic treatment for bacterial vaginosis. Antimicrob Agents Chemother 59:2825–2834. doi: 10.1128/AAC.00225-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, Fredricks DN. 2012. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]