Whereas the emergence of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) clonal complex 398 (CC398) in animal husbandry and its transmission to humans are well documented, less is known about factors driving the epidemic spread of this zoonotic lineage within the human population. One factor could be the bacteriophage phi3, which is rarely detected in S. aureus isolates from animals but commonly found among isolates from humans, including those of the human-adapted methicillin-susceptible S. aureus (MSSA) CC398 clade.
KEYWORDS: Staphylococcus aureus, livestock-associated MRSA, clonal complex 398, spa-CC011, spa t034, spa t011, (re)adaptation, bacteriophage, phi3, immune evasion cluster, whole-genome sequencing, zoonosis
ABSTRACT
Whereas the emergence of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) clonal complex 398 (CC398) in animal husbandry and its transmission to humans are well documented, less is known about factors driving the epidemic spread of this zoonotic lineage within the human population. One factor could be the bacteriophage phi3, which is rarely detected in S. aureus isolates from animals but commonly found among isolates from humans, including those of the human-adapted methicillin-susceptible S. aureus (MSSA) CC398 clade. The proportion of phi3-carrying MRSA spa-CC011 isolates, which constitute presumptively LA-MRSA within the multilocus sequence type (MLST) clonal complex 398, was systematically assessed for a period of 16 years to investigate the role of phi3 in the adaptation process of LA-MRSA to the human host. For this purpose, 632 MRSA spa-CC011 isolates from patients of a university hospital located in a pig farming-dense area in Germany were analyzed. Livestock-associated acquisition of MRSA spa-CC011 was previously reported as having increased from 1.8% in 2000 to 29.4% in 2014 in MRSA-positive patients admitted to this hospital. However, in this study, the proportion of phi3-carrying isolates rose only from 1.1% (2000 to 2006) to 3.9% (2007 to 2015). Characterization of the phi3 genomes revealed 12 different phage types ranging in size from 40,712 kb up to 44,003 kb, with four hitherto unknown integration sites (genes or intergenic regions) and several modified bacterial attachment (attB) sites. In contrast to the MSSA CC398 clade, phi3 acquisition seems to be no major driver for the readaptation of MRSA spa-CC011 to the human host.
INTRODUCTION
During the past decade, attention was drawn to the emergence of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) of clonal complex 398 (CC398) (1–3). Mainly pigs, but also cattle, poultry, and other livestock act as zoonotic LA-MRSA CC398 reservoirs for humans (4–6). Currently, LA-MRSA CC398 constitutes a significant portion of MRSA detected in human and animal health care centers and substantially contributes to the burden of disease attributable to MRSA, especially in regions with a high density of livestock production (7–9).
Whereas resistance toward antibiotics and heavy metals as well as the general pathogenic potential of MRSA CC398 has been thoroughly investigated (10–13), the factors driving the epidemic of this clonal lineage in humans are only slightly understood. Interestingly, differences in the content of mobile genetic elements (MGEs) between the ancestral methicillin-susceptible CC398 clade that originated in humans, the methicillin-resistant livestock-associated CC398 clade, and an emerging methicillin-susceptible human-specific CC398 clade have been described previously (14–16). In the livestock-associated CC398 clade, the staphylococcal cassette chromosome mec (SCCmec) types IV and V, the tet(M) resistance gene, and the bacteriophages phi2 and phi6 were commonly found, whereas the bacteriophage phi3 and the genes cadDX and rep27 predominated in the human clade (17). Additionally, further studies observed that the host switch of methicillin-susceptible S. aureus (MSSA) CC398 from the ancestral human host to livestock was linked to a loss of phage phi3 and an uptake of tet(M) (15, 18). Therefore, we suggested that the adaptation of the CC398 lineage back to the human host might be linked to an uptake of bacteriophage phi3 into the genome.
MGEs such as prophages are known to serve as drivers in bacterial adaptation, enabling the pathogen to colonize broader host ranges due to the acquisition of new virulence and resistance genes (19, 20). The uptake of the Siphoviridae family member phi3, which is recognized as a genetic marker for human-associated, but not for animal-associated, isolates is of particular importance (17, 21, 22). The genomes of Siphoviridae are characterized by a mosaic structure, and they are typically organized in six functional modules containing genes for lysogeny, DNA replication, regulation of transcription, packaging, head and tail, and lysis (23). The integration of bacteriophages in the bacterial genome occurs via site-specific recombination by targeting the bacterial attachment site (attB site), which contains a 14-bp core sequence (5′-TGTATCCAAACTGG-3′) (24).
Acquisition of phi3 in the S. aureus CC398 clade might be beneficial for the adaptation process to the human host as it confers a cluster of human-specific immune-modulatory elements, which counteract with the human innate immunity (25). The genes harbored on the immune evasion cluster (IEC) comprise the chemotaxis inhibitory protein (chp), the staphylococcal complement inhibitor (scn), and the plasminogen activator staphylokinase (sak). These proteins are known as human-specific virulence factors due to their high specificity to human immune cells and serum protein (26–28). In some cases, enterotoxin A or P (sea or sep) is additionally encoded in the IEC (25, 29). Previous studies evaluating the occurrence of phi3 in CC398 isolates derived from animals and humans with livestock contact found a comparatively low prevalence for this bacteriophage (30, 31). Therefore, systematic studies covering the complete time period since the advent of this S. aureus lineage in livestock are warranted to evaluate the uptake of bacteriophage phi3 in human LA-MRSA isolates over time.
Consequently, this study aimed to investigate the prevalence of bacteriophage phi3 in human-derived MRSA spa-CC011. This spa-CC with t011 and t034 as the most frequent spa types constitutes presumptively LA-MRSA within the multilocus sequence type (MLST) clonal complex 398. Overall, 632 MRSA spa-CC011 isolates covering the entire time period from the beginning of the MRSA CC398 epidemic in 2000 until 2015 and recovered from inpatients at the University Hospital Münster (UKM) were included. Furthermore, a whole-genome sequencing (WGS) approach was applied to detect the genetic location of phi3 within the MRSA spa-CC011 genome and to assess the genetic variability within the phi3 genomes.
MATERIALS AND METHODS
Strain collection, molecular typing, and cultivation conditions.
Human MRSA isolates were recovered from patients at the University Hospital Münster (UKM) in Germany. The hospital is located in a German region characterized by a very high density of pig production, and epidemiological investigations have demonstrated that carriage of MRSA spa-CC011 by patients in this region is predominantly associated with livestock contact (32). At the UKM, all MRSA isolates were identified by matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF MS) (Microflex-LT system, MALDI-Biotyper, version 3.0; Bruker Daltonics, Germany) as previously described (33, 34). Phenotypic and genotypic characterization of methicillin resistance was assessed for every isolate by using a Vitek-2 automated system (bioMérieux, Nürtingen, Germany), applying the antimicrobial susceptibility test card AST-P632, and via S. aureus-specific PCR detecting the genes mecA and mecC (GenoType MRSA; Hain-Lifescience, Germany) as described previously (35, 36).
Since 2006, every first MRSA isolate of each patient had been typed by S. aureus protein A (spa) sequence-based typing and stored at −80°C (37, 38). For isolates obtained before 2006, spa typing was retrospectively performed. The “based upon repeat pattern” (BURP) was used for the identification of MRSA isolates belonging to the spa clonal complex CC011 (spa-CC011) (39). Therefore, spa types clustering to spa clonal complex t011 and closely related clades, which are representative for the CC398 clade, were selected (see Fig. S1 in the supplemental materials). Additionally, one isolate of each clade that forms branches in the BURP analysis was selected, and MLST analysis was performed. In early years of the epidemic (2000 to 2006), all available MRSA spa-CC011 isolates were included (n = 92). Later (2007 to 2015), the first 15 isolates of each quarter were included in the study, resulting in 60 isolates per year. Only the first isolate of each patient was included in the study. The antibiotic resistance profile of the phi3-positive isolates (n = 21) was determined at the genetic level by the use of DNA microarray analysis (Identibac Microarray; Alere Technologies GmbH, Jena, Germany).
For all experiments, S. aureus strains were cultivated on Columbia agar (Becton Dickinson, Franklin Lakes, NJ, USA) containing 5% sheep blood (Oxoid, Wessel, Germany).
Genomic DNA isolation and PCR-based detection of IEC genes.
Genomic DNA was extracted from S. aureus using a QIAamp DNA minikit according to the manufacturer's instructions (Qiagen, Hilden, Germany) with the exception that lysostaphin (20 μg/ml) (Wakchemie, Steinbach, Germany) was used for bacterial cell lysis. To detect the bacteriophage phi3, PCRs specific for genes located on the IEC (chp, scn, sak, sea, and sep) using the oligonucleotides chp-for, chp-rev, sak-for, sak-rev, scn-for.2, scn-rev.2, sea-for, sea-rev, sep-for, and sep-rev were performed as follows: 95°C for 5 min, followed by 35 cycles of 30 s at 95°C, 30 s at 50°C, 30 s at 72°C, and a final elongation of 7 min at 72°C. To investigate whether phi3 was integrated within the hlb gene, PCRs using the oligonucleotides hlb-1-for, hlb-2-rev, hlb-3-for, and hlb-4-rev were performed using the following conditions for all oligonucleotide combinations: 95°C 5 min, followed by 35 cycles of 30 s 95°C, 30 s 59°C, 60 s 72°C, and a final elongation of 7 min at 72°C. A truncated hlb gene indicative of phage integration was detected using the oligonucleotide pair hlb-2-rev/hlb-3-for, giving no PCR product after phage integration. The DNA sequences of PCR oligonucleotides are given in Table S1.
WGS and bioinformatics.
Whole-genome sequencing (WGS) was performed using the PacBio RS II platform. Staphylococcal DNA was extracted using a Genomic-tip 20/G kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions with lysostaphin incubation (20 μg/ml) (Wakchemie, Steinbach, Germany) for 1 h at 37°C. Five micrograms of extracted, high-quality, double-stranded DNA was sequenced using P6-C4 chemistry and Pacific Biosciences RSII instrumentation using a movie collection time of 4-h and 110 pmol/liter of complexed 20-kb SMRTbell library. Initial de novo assembly was performed using the HGAP3, version 2.3.0, pipeline (Icahn Institute for Genomics and Multiscale Biology at the Icahn School of Medicine at Mount Sinai, New York, NY, USA). The sequenced samples presented coverages between 37× and 323× and 9,473 to 84,090 mapped reads, with a mean read length of 4,581 bp to 13,804 bp (N50 of 6,271 bp to 19,453 bp). The assembled genomes were annotated using the GenDB pipeline (40). For classification of the functional phage modules, an in silico PCR using oligonucleotides characteristic for the specific phage module types was applied using SnapGene (version 4.0.6; GSL Biotech, LLC, Chicago, IL, USA) (41). Structural comparison of the phi3 phages was performed using Easyfig software (42). Detection of genes carried by MGEs and indicative for human or animal origin of the isolates was performed by an in silico PCR using oligonucleotides for the genes int of phi6, int of phi7, cadDX, rep27, rep7 (43), and int of phi2 (20).
Statistical analysis.
A two-tailed Fisher's exact test (GraphPad Prism, version 5; GraphPad Software, Inc.) was used to compare the proportion of phi3-positive isolates among all MRSA spa-CC011 isolates detected in early and late years of the epidemic. P values of <0.05 were considered significant.
Accession number(s).
The assembled genome sequences of the phi3-positive strains were deposited in the European Nucleotide Archive (ENA) under the following accession numbers: LT992456 to LT992458, LT992460 to LT992477, and OVTT01000001 to OVTT01000003.
RESULTS
Prevalence of the bacteriophage phi3 in MRSA spa-CC011 over 16 years.
In total, 632 MRSA spa-CC011 isolates were screened for the presence of bacteriophage phi3. At the study hospital, the MRSA spa-CC011 epidemic started in 2000 with first one isolate; in 2013 MRSA spa-CC011 accounted for 35% of all MRSA isolates detected among local patients, as published elsewhere (8). Based on BURP analysis, the complete set of isolates comprised 26 different MRSA spa-CC011-associated spa types, with t011 (50.3%, n = 318), t034 (38%, n = 240), t108 (2.8%, n = 18), t1451 (2.1%, n = 13), t2011 (1.1%, n = 7), t1255 (0.9%; n = 6), t2582 (0.6%, n = 4), t2576 (0.5%, n = 3), t571 (0.5%, n = 3), t1580 (0.3%; n = 2), t1793 (0.3%; n = 2), and t2346 (0.3%, n = 2) being predominant. BURP analysis revealed four closely related clades belonging to spa-CC011 that form distinct branches in the clustering (CC034, CC108, CC1451, and CC1580). From each group, one isolate was selected for MLST analysis, which confirmed that these isolates belong to ST398.
Within this group, 21/632 (3.3%) isolates tested positive for IEC genes and were thus classified as phi3 positive (Table 1). Over time, the rate of phi3-positive MRSA spa-CC011 isolates increased nonsignificantly from 1.1% (n = 1/92 in 2000 to 2006) to 3.9% (n = 20/540 in 2007 to 2015) (P > 0.05). The majority of phi3-positive isolates were found in 2011 (n = 7/60; 11.7%), followed by 2010 (n = 4/60, 6.7%), 2012 (n = 3/60, 5%), 2008 and 2013 (each, n = 2/60; 3.3%), and 2006, 2014, and 2015 (each, n = 1/60; 1.7%) (Table 1).
TABLE 1.
phi3-positive isolates belonging to spa-CC011-related spa types from 2000 to 2015a
| Year(s) of isolation | Frequency of phi3-positive isolates by spa type (no. of positive isolates/no. tested [%]) |
Total no. (%) | ||||
|---|---|---|---|---|---|---|
| t011 | t034 | t1451 | t1793 | Other | ||
| 2000 to 2006 | 1/50 | 0/37 | 0/2 | 0/0 | 0/3 | 1/92 (1.1) |
| 2007 | 0/34 | 0/24 | 0/0 | 0/0 | 0/2 | 0/60 (0) |
| 2008 | 0/32 | 2/21 | 0/0 | 0/0 | 0/7 | 2/60 (3.3) |
| 2009 | 0/31 | 0/18 | 0/1 | 0/0 | 0/10 | 0/60 (0) |
| 2010 | 1/28 | 2/21 | 1/3 | 0/0 | 0/8 | 4/60 (6.7) |
| 2011 | 2/30 | 5/20 | 0/2 | 0/1 | 0/7 | 7/60 (11.7) |
| 2012 | 3/30 | 0/29 | 0/0 | 0/0 | 0/1 | 3/60 (5) |
| 2013 | 0/27 | 2/23 | 0/1 | 0/0 | 0/9 | 2/60 (3.3) |
| 2014 | 0/30 | 0/25 | 0/2 | 1/1 | 0/2 | 1/60 (1.7) |
| 2015 | 1/26 | 0/22 | 0/2 | 0/0 | 0/10 | 1/60 (1.7) |
| Total | 8/318 (2.5) | 11/240 (4.6) | 1/13 (7.7) | 1/2 (50) | 0/59 (0) | 21/632 (3.3) |
As detected by PCR targeting immune evasion cluster (IEC)-associated genes chp, scn, sak, sea, and sep.
All MRSA spa-CC011 isolates positive for IEC genes were characterized as methicillin and tetracycline resistant by the possession of the mecA and tet(M) genes, respectively (see Table S3 in the supplemental material). Furthermore, livestock origin was underlined by the presence of the phages phi2 and phi6 in 14.2% (n = 3/21) and 61.9% (n = 13/21) as well as the replication protein 7 in 95.2% (n = 20/21) of the isolates, respectively (Table S4). However, human markers such as the cadmium resistance gene cadX were found in 9.5% (n = 2/21). Furthermore, MLST analysis of the phi3-positive isolates revealed that all isolates belonged to ST398.
Most of the IEC-positive isolates (18/21) were associated with colonization and were obtained from classical screening specimens, including nasal, pharyngeal, or superficial skin swabs. The remaining three isolates were derived from clinical specimens (wound swabs and blood culture) potentially associated with infections. Among the phi3-positive isolates, four different spa types were observed, with t034 being the most prevalent (n = 11), followed by t011 (n = 8) and t1451 and t1793 (each, n = 1) (Table 1).
The bacteriophages belonged to three different IEC types. The most prevalent IEC type was type B carrying the genes sak, chp, and scn in 66.7% (n = 14/21) of phi3-positive S. aureus isolates, followed by IEC E carrying the genes sak and scn in 23.8% (n = 5/21) and IEC type A (sea, sak, chp, scn) in 9.5% (n = 2/21) of these isolates (Table 2).
TABLE 2.
Diversity of immune evasion cluster types among the phi3-positive isolates
| IEC typea | Immune-modulatory gene profile | % of phi3-positive isolatesb | |||
|---|---|---|---|---|---|
| A | sea | sak | chp | scn | 9.5 (2/21) |
| B | sak | chp | scn | 66.7 (14/21) | |
| C | chp | scn | |||
| D | sea | sak | scn | ||
| E | sak | scn | 23.8 (5/21) | ||
| F | sep | sak | chp | scn | |
| G | sep | sak | scn | ||
Immune evasion cluster (IEC) type classification according to van Wamel et al. (25).
Values in parentheses are the number of phi3-positive isolates/total number tested.
Comparing the PCR-based typing results of the bacteriophages with the WGS data revealed that the PCR results correspond with the WGS data in 86% of instances. Discrepancies were due to false-positive sep PCR results in three isolates. The IEC-carried genes chp, sak, and scn were correctly identified in all isolates by PCR-based typing.
Characterization and classification of the phi3 phage genomes.
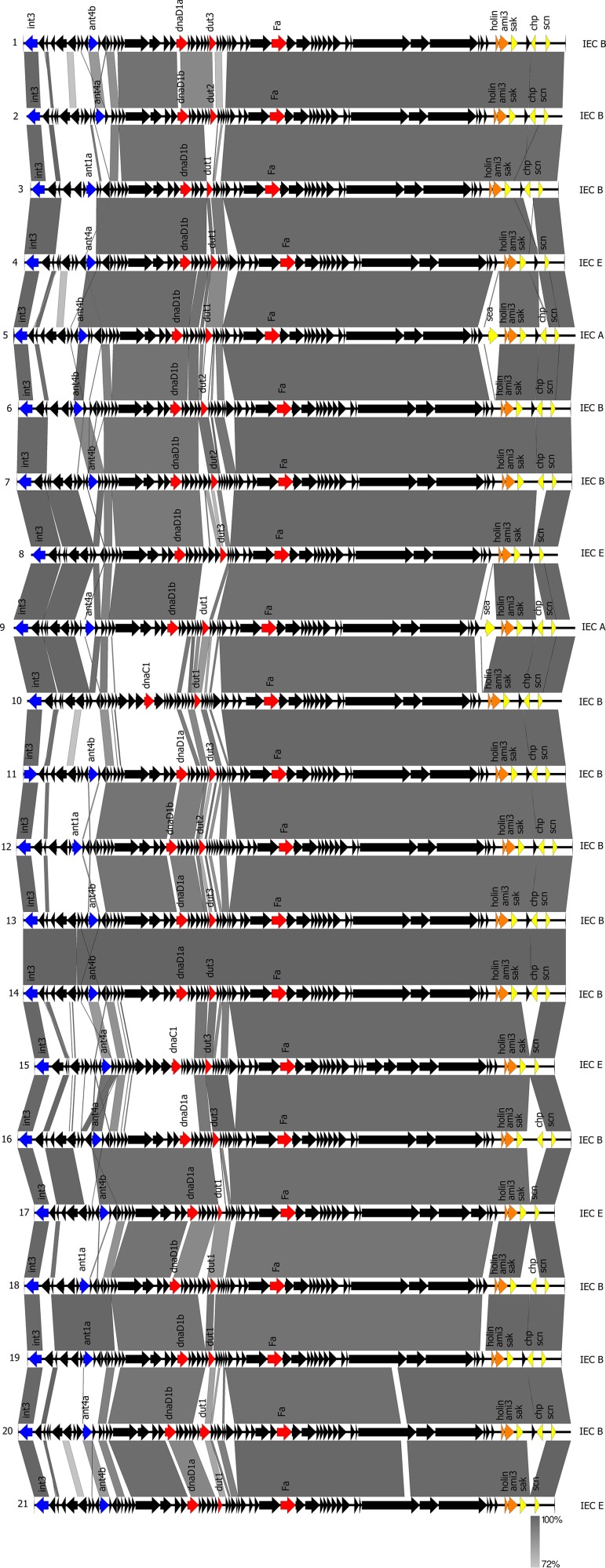
WGS of the 21 phi3-positive isolates revealed complete assembled genomes for 20/21 isolates whereas for one isolate the genome was available in several contigs. However, the phi3 phage genomes were always located within one contig and ranged between 40,712 kb and 44,003 kb, containing between 55 and 69 genes. Sequence comparison of the phi3 bacteriophages showed homologous regions for the integrase locus, the morphogenesis locus (containing genes for DNA packing), the lysis module (containing genes for host cell lysis), and the IEC (Fig. 1). Structural differences were found in modules carrying genes for lysogeny, DNA replication and packaging, and regulation of transcription.
FIG 1.
Structural comparison of the phi3 genomes of the phi3-positive LA-MRSA (strain numbers on the left) performed by Easyfig software. Gray areas represent regions with nucleotide sequence similarities ranging between 72% and 100%. The integrase and antirepressor genes (lysogeny module) are shown in blue, the replication protein (dnaD or dnaC), UTPase genes, and the portal protein (DNA metabolism and DNA packaging modules) are shown in red, holin and amidase (lysis module) are shown in orange, and IEC genes (IEC type on the right) are shown in yellow.
The functional modules of phage phi3 were classified according to the multiplex scheme of Kahánková et al. (41), and 12 different phage types were identified (Table 3). For the lysogeny control module, all phages were found to belong to the same integrase group (Sa3int), and three different antirepressor types (ant1a, ant4a, and ant4b) were found. Furthermore, most phages contained a DNA replication module harboring the dnaD gene (n = 19/21) (subtypes dnaD1a [n = 7/21] and dnaD1b [n = 12/21]), and two phages were found to carry the dnaC gene (subtype dnaC1). The phages belonged to three different transcription regulation modules with the dUTPase types dut1 (n = 10/21), dut2 (n = 4/21), and dut3 (n = 7/21). All phages carried the portal protein Fa and the endolysin gene ami3.
TABLE 3.
Integration sites of phi3 phages and bacteriophage classification
| Isolate no. | Integration site | GenBank accession no.a | attB site (5′– 3′)b | Bacteriophage classificationc |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phage genome size (bp) | Integrase | Genetic control | Replication module | Transcription module | Morphogenesis subtype | Amidase | Phage type | ||||
| 1439 | hlb | TGTATCCGAATTGG | 42,460 | Sa3 | ant4b | dnaD1a | dut3 | Fa | ami3 | Sa3int-ant4b-dnaD1a-dut3-Fa-ami3 | |
| LA86 | Pyruvate oxidase (SAPIG2589) | AM990992.1f | TGTATCCAAAATGG | 41,927 | Sa3 | ant4a | dnaD1b | dut2 | Fa | ami3 | Sa3int-ant4a-dnaD1b-dut2-Fa-ami3 |
| LA115 | hlb | TGTATCCAAACTGG | 41,335 | Sa3 | ant1a | dnaD1b | dut1 | Fa | ami3 | Sa3int-ant1a-dnaD1b-dut1-Fa-ami3 | |
| LA208 | hlb | TGTATCCAAACTGG | 42,292 | Sa3 | ant4a | dnaD1b | dut1 | Fa | ami3 | Sa3int-ant4a-dnaD1b-dut1-Fa-ami3 | |
| 3949 | Phage StauST398-5 | KC595279.1f | TGTATCCAACCAGG | 44,003 | Sa3 | ant4b | dnaD1b | dut1 | Fa | ami3 | Sa3int-ant4b-dnaD1b-dut1-Fa-ami3 |
| LA232 | hlb | TGTATCCAAACTGG | 43,254 | Sa3 | ant4b | dnaD1b | dut2 | Fa | ami3 | Sa3int-ant4b-dnaD1b-dut2-Fa-ami3 | |
| 4623 | hlb | TGTATCCAAACTGG | 43,456 | Sa3 | ant4b | dnaD1b | dut2 | Fa | ami3 | Sa3int-ant4b-dnaD1b-dut2-Fa-ami3 | |
| LA272 | ilvB (SAPIG2093) | AM990992.1e | TGTATCCAAACTGG | 41,228 | Sa3 | NT | dnaD1b | dut3 | Fa | ami3 | Sa3int-dnaD1b-dut3-Fa-ami3 |
| LA281 | Between SAPIG0723 and SAPIG0724d | AM990992.1e | TGTATCCAAACTGT | 43,946 | Sa3 | ant4a | dnaD1b | dut1 | Fa | ami3 | Sa3int-ant4a-dnaD1b-dut1-Fa-ami3 |
| 5235 | Between SAPIG0723 and SAPIG0724d | AM990992.1e | TGTATCCTTACTGT | 41,849 | Sa3 | NT | dnaC1 | dut1 | Fa | ami3 | Sa3int-dnaC1-dut1-Fa-ami3 |
| LA290 | hlb | TGTATCCGAATTGG | 43,453 | Sa3 | ant4b | dnaD1a | dut3 | Fa | ami3 | Sa3int-ant4b-dnaD1a-dut3-Fa-ami3 | |
| LA293 | hlb | TGTATCCGAATTGG | 43,543 | Sa3 | ant1a | dnaD1b | dut2 | Fa | ami3 | Sa3int-ant1a-dnaD1b-dut2-Fa-ami3 | |
| LA301 | hlb | TGTATCCGAATTGG | 42,458 | Sa3 | ant4b | dnaD1a | dut3 | Fa | ami3 | Sa3int-ant4b-dnaD1a-dut3-Fa-ami3 | |
| 5418 | hlb | TGTATCCGAATTGG | 42,458 | Sa3 | ant4b | dnaD1a | dut3 | Fa | ami3 | Sa3int-ant4b-dnaD1a-dut3-Fa-ami3 | |
| LA305 | hlb | TGTATCCAAACTGG | 40,712 | Sa3 | ant4a | dnaC1 | dut3 | Fa | ami3 | Sa3int-ant4a-dnaC1-dut3-Fa-ami3 | |
| LA309 | hlb | TGTATCCGAATTGG | 43,331 | Sa3 | ant4a | dnaD1a | dut3 | Fa | ami3 | Sa3int-ant4a-dnaD1a-dut3-Fa-ami3 | |
| LA343 | hlb | TGTATCCGAATTGG | 40,746 | Sa3 | ant4b | dnaD1a | dut1 | Fa | ami3 | Sa3int-ant4b-dnaD1a-dut1-Fa-ami3 | |
| LA388 | hlb | TGTATCCGAATTGG | 42,310 | Sa3 | ant1a | dnaD1b | dut1 | Fa | ami3 | Sa3int-ant1a-dnaD1b-dut1-Fa-ami3 | |
| LA415 | hlb | TGTATCCGAATTGG | 41,810 | Sa3 | ant1a | dnaD1b | dut1 | Fa | ami3 | Sa3int-ant1a-dnaD1b-dut1-Fa-ami3 | |
| LA436 | Alanine racemase (SAPIG2238) | AM990992.1e | TGTATCCAATCTGG | 43,263 | Sa3 | ant4a | dnaD1b | dut1 | Fa | ami3 | Sa3int-ant4a-dnaD1b-dut1-Fa-ami3 |
| LA562 | hlb | TGTATCCGAATTGG | 40,747 | Sa3 | ant4b | dnaD1a | dut1 | Fa | ami3 | Sa3int-ant4b-dnaD1a-dut1-Fa-ami3 | |
GenBank accession numbers correspond to genes used for alignment in WGS data.
Nucleotides differing from the original attB site of phage phi3 are underlined (24).
Classification of bacteriophage modules according to Kahánková et al. (41). NT, nontypeable.
SAPIG0723 encodes a nucleoside transporter protein; SAPIG0724 is the yxkD gene.
S. aureus ST398 (GenBank accession no. NC_017333.1) (20).
S. aureus phage StauST398-5pro (GenBank accession no. KC595279.1) (16).
Alternative integration sites (genes or intergenic regions) of phage phi3 in the MRSA spa-CC011 genome.
In addition to two already described integration sites (hlb gene and alanine racemase), WGS revealed four alternative integration sites for bacteriophage phi3 in the MRSA spa-CC011 genome (Table 3). The phi3 phage was integrated within metabolic genes (genes of the pyruvate oxidase [SAPIG2589], the acetolactate synthase large subunit ilvB [SAPIG2093], and the alanine racemase [SAPIG2238] [GenBank accession no. AM990992.1]), in between a gene for a nucleoside transporter protein (SAPIG0723) and yxkD (SAPIG0724) (GenBank accession no. AM990992.1), or within another phage genome (GenBank accession no. KC595279.1). Moreover, modified bacterial attachment (attB) sites were identified (Table 3). Six of the 21 phages integrated within hlb or ilvB harbored the attB site (5′-TGTATCCAAACTGG-3′) published by Coleman et al. (24). The majority of bacteriophages contained an attB site with two nucleotide exchanges (5′-TGTATCCGAATTGG-3′; changes are underlined). This attB site was found only for phages integrated within the hlb gene. Further attB sites with one to three nucleotide exchanges (underlined) were found in the remaining bacteriophages integrated within other genes in the spa-CC011 genome (5′-TGTATCCAAAATGG-3′; 5′-TGTATCCAAACTGT-3′; 5′-TGTATCCAATCTGG-3′; 5′-TGTATCCAACCAGG-3′; 5′-TGTATCCTTACTGT-3′).
DISCUSSION
Bacterial host switches are often linked to gains and losses of MGEs. Thus, we hypothesized that the livestock phenotype (i.e., methicillin and tetracycline resistance) associated with MRSA spa-CC011, which had been transmitted to the human host, would show adaptation by taking up bacteriophage phi3 characterized by its human-specific gene cluster containing the genes chp, sak, scn, sea, and sep. The livestock origin of the phi3-positive isolates was supported by the presence on the MGEs of the genes rep7, int of phi2, and int of phi6 in the majority of isolates. These genes were shown to be present in livestock-associated clades but were absent in human-specific isolates (17). Furthermore, MGE-carried genes described for human-specific isolates (cadDX) were also present sporadically in some isolates. This might indicate an ongoing uptake of human-specific genes contained on MGEs in the livestock-associated MRSA spa-CC011 clade and thereby an expanding host range of MRSA spa-CC011. However, the percentage of human phi3-carrying MRSA spa-CC011 isolates recovered in our study was comparatively low (3.3%). The number of phi3-positive MRSA spa-CC011 isolates increased only slightly from 1.1% in 2000 to 2006 to 3.9% in 2007 to 2015. In general, it is known for S. aureus that the majority of clinical and laboratory strains from human sources carry the phi3 phage (25). In contrast, the phi3-related gene cluster is only rarely found among the livestock-associated MRSA CC398 (18, 30, 31).
Until now, human-adapted MSSA CC398 isolates harboring the phi3 phage were mainly assigned to spa types t571, t034, and t3625 and were associated with human infections (16, 18, 31, 44, 45). Furthermore, bacteriophage phi3 was found in MSSA CC398 isolates belonging to the spa types t899, t571, t1451, t5635, t6587, and t9378 obtained from bloodstream infections in France (14). In Spain, an MRSA CC398 isolate containing bacteriophage phi3 belonging to spa type t1451 was identified in a poultry farmer with abscesses and septic arthritis (46).
In our study, we found MRSA spa-CC011 from colonization and clinical specimens carrying the IEC genes in a human health care setting located in a German pig-dense area. Interestingly, the MRSA spa-CC011 isolates carrying the phage phi3 belonged to the spa types t011, t034, t1451, and t1793. This is of particular importance since isolates with spa types t011 and t034 represent the majority of MRSA spa-CC011 isolates recovered from the human population in general as well as from the hospital environment (8, 9, 47).
In a previous study by Cuny et al. (31), a slightly higher prevalence for MRSA spa-CC011 carrying the phi3 phage (10%) was found in horses, veterinary personnel from horse clinics, and humans with infections, including wound infections, furuncles, and septicemia (31). However, bacteriophage phi3 was absent in MRSA spa-CC011 isolated from nasal specimens of pig farmers and pigs. The higher percentage of phi3-positive isolates found by Cuny et al. might be due to diverse strain origins, whereas our isolates were exclusively obtained from human specimens in a clinical setting.
Recently, Kraushaar et al. identified nine alternative integration sites for a bacteriophage belonging to integrase group 3 in the CC398 genome (48). These alternative integration sites were all within genes (mostly metabolic genes), and the phage integration was found after experimental infection of CC398 strains with an int3 bacteriophage. Furthermore, phage transfer from a human-associated S. aureus donor strain to an S. aureus CC398 strain was found after treatment with sublethal concentrations of biocides, and nine alternative integration sites for prophage Φ13 were described in an LA-MRSA CC398 isolate (49). The authors speculated that mutations in attB might be the reason for the missing phage integration into hlb in their study (49). However, we found that phage phi3 was able to integrate within the hlb gene containing both the original attB site and attB sites containing two mutations. Additionally, atypical integration sites for bacteriophages encoding the sak gene were previously described for disease-related S. aureus isolates within genes or intergenically (50). In contrast, we found six naturally occurring alternative integration sites, four of which have not been previously described. The integration sites were within metabolic genes or in between metabolic and transporter protein genes. Interestingly, for one phi3 phage, we identified another bacteriophage genome as an integration site.
This study provides evidence that the PCR-based detection of the IEC genes described here is a reliable method to detect phage phi3 in MRSA spa-CC011. For the IEC-carried genes scn, sak, and chp, no discrepancies were found between WGS and PCR-based typing, and the IEC genes were found only in isolates harboring phage phi3. However, for the precise typing of isolates, especially for the genes sea and sep and the integration site of the phi3 phage, WGS is an essential tool.
Previous studies showed the presence of three separate clades of Sa3 prophages, as detected by the prophage integrase gene Sa3int (18). The PCR-based detection of IEC genes located on the phi3 phage could therefore lead to missing the identification of other Sa3 prophages without IEC genes and has to be seen as a limitation of the study.
Due to our study design, which was based on spa typing and BURP clustering, only MRSA isolates clustering to spa-CC011 and closely related clades such as CC034, CC108, and CC1451 were included. A previous study by Strommenger et al. revealed that spa typing together with BURP clustering is a useful tool for typing of S. aureus isolates (51). However, limitations in using spa typing for the assignment of MRSA isolates to clonal complexes have been reported (18). This was observed for spa type t899, which was shown to be associated with ST398 or ST9 (52). Previous MLST of isolates in our study with spa type t899 showed that these isolates belonged to ST9 (8).
In conclusion, the acquisition of phage phi3 does not seem to be a major factor leading to the readaptation of LA-MRSA spa-CC011 back to the human host over a period of 16 years, as shown before for human-adapted methicillin-susceptible S. aureus (MSSA) CC398 (15, 16). Moreover, the absence of bacteriophage phi3 in the MRSA spa-CC011 genome does not impair the ability of MRSA spa-CC011 to colonize and infect the human host. Therefore, further surveillance of S. aureus CC398 dissemination is necessary, in particular, in regions with a high density of livestock production since the high prevalence of this lineage in animal husbandry correlates with its prevalence in persons with occupational livestock exposure and its occurrence in health care facilities (7, 53–55). These findings highlight the importance of monitoring programs for emerging pathogens, such as the S. aureus lineage CC398. Furthermore, investigations on the adaptation of livestock-associated MRSA spa-CC011 to the human host are of major importance because MRSA spa-CC011 presents a zoonotic threat for the human environment and for health care settings (56).
Supplementary Material
ACKNOWLEDGMENTS
We thank Melanie Bach, Martina Schulte, and Anja Hassing for excellent technical assistance. We also are grateful to the Technology Development group, in particular, Robert Sebra, from the Icahn Institute for Genomics and Multiscale Biology at the Icahn School of Medicine at Mount Sinai for PacBio sequencing and assembly.
We thank the GenDB support team for technical assistance and access to resources financially supported by the BMBF (FKZ 031A533) within the de.NBI network. This work was supported by grants of the German Federal Ministry of Education and Research (BMBF) within the framework of the MedVet-Staph (01KI1301A) and #1 Health-PREVENT (01KI1727A) to KB and RK.
We do not have any conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00140-18.
REFERENCES
- 1.van Cleef BA, Monnet DL, Voss A, Krziwanek K, Allerberger F, Struelens M, Zemlickova H, Skov RL, Vuopio-Varkila J, Cuny C, Friedrich AW, Spiliopoulou I, Pászti J, Hardardottir H, Rossney A, Pan A, Pantosti A, Borg M, Grundmann H, Mueller-Premru M, Olsson-Liljequist B, Widmer A, Harbarth S, Schweiger A, Unal S, Kluytmans JAJ. 2011. Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg Infect Dis 17:502–505. doi: 10.3201/eid1703.101036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graveland H, Duim B, van Duijkeren E, Heederik D, Wagenaar JA. 2011. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int J Med Microbiol 301:630–634. doi: 10.1016/j.ijmm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis 11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köck R, Harlizius J, Bressan N, Laerberg R, Wieler LH, Witte W, Deurenberg RH, Voss A, Becker K, Friedrich AW. 2009. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among pigs on German farms and import of livestock-related MRSA into hospitals. Eur J Clin Microbiol Infect Dis 28:1375–1382. doi: 10.1007/s10096-009-0795-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuny C, Friedrich A, Kozytska S, Layer F, Nübel U, Ohlsen K, Strommenger B, Walther B, Wieler L, Witte W. 2010. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int J Med Microbiol 300:109–117. doi: 10.1016/j.ijmm.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Armand-Lefevre L, Ruimy R, Andremont A. 2005. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg Infect Dis 11:711–714. doi: 10.3201/eid1105.040866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köck R, Schaumburg F, Mellmann A, Köksal M, Jurke A, Becker K, Friedrich AW. 2013. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS One 8:e55040. doi: 10.1371/journal.pone.0055040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Alen S, Ballhausen B, Peters G, Friedrich AW, Mellmann A, Köck R, Becker K. 2017. In the centre of an epidemic: fifteen years of LA-MRSA CC398 at the University Hospital Münster. Vet Microbiol 200:19–24. doi: 10.1016/j.vetmic.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Schaumburg F, Köck R, Mellmann A, Richter L, Hasenberg F, Kriegeskorte A, Friedrich AW, Gatermann S, Peters G, von Eiff C, Becker K, and the study group. 2012. Population dynamics among methicillin-resistant Staphylococcus aureus isolates in Germany during a 6-year period. J Clin Microbiol 50:3186–3192. doi: 10.1128/JCM.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argudin MA, Tenhagen BA, Fetsch A, Sachsenröder J, Käsbohrer A, Schroeter A, Hammerl JA, Hertwig S, Helmuth R, Bräunig J, Mendoza MC, Appel B, Rodicio MR, Guerra B. 2011. Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Appl Environ Microbiol 77:3052–3060. doi: 10.1128/AEM.02260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feßler A, Scott C, Kadlec K, Ehricht R, Monecke S, Schwarz S. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J Antimicrob Chemother 65:619–625. doi: 10.1093/jac/dkq021. [DOI] [PubMed] [Google Scholar]
- 12.Ballhausen B, Jung P, Kriegeskorte A, Makgotlho PE, Ruffing U, von Müller L, Köck R, Peters G, Herrmann M, Ziebuhr W, Becker K, Bischoff M. 2014. LA-MRSA CC398 differ from classical community acquired-MRSA and hospital acquired-MRSA lineages: functional analysis of infection and colonization processes. Int J Med Microbiol 304:777–786. doi: 10.1016/j.ijmm.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 13.van Alen S, Kaspar U, Idelevich EA, Köck R, Becker K. 2018. Increase of zinc resistance in German human derived livestock-associated MRSA between 2000 and 2014. Vet Microbiol 214:7–12. doi: 10.1016/j.vetmic.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Diene SM, Corvaglia AR, François P, van der Mee-Marquet N. 2017. Prophages and adaptation of Staphylococcus aureus ST398 to the human clinic. BMC Genomics 18:133. doi: 10.1186/s12864-017-3516-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlemann AC, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, Lipkin WI, Lindsay JA, DeLeo FR, Lowy FD. 2012. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio 3:e00027-12. doi: 10.1128/mBio.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Mee-Marquet N, Corvaglia AR, Valentin AS, Hernandez D, Bertrand X, Girard M, Kluytmans J, Donnio PY, Quentin R, François P. 2013. Analysis of prophages harbored by the human-adapted subpopulation of Staphylococcus aureus CC398. Infect Genet Evol 18:299–308. doi: 10.1016/j.meegid.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 17.McCarthy AJ, van Wamel W, Vandendriessche S, Larsen J, Denis O, Garcia-Graells C, Uhlemann AC, Lowy FD, Skov R, Lindsay JA. 2012. Staphylococcus aureus CC398 clade associated with human-to-human transmission. Appl Environ Microbiol 78:8845–8848. doi: 10.1128/AEM.02398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Concepción Porrero M, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsay JA. 2010. Genomic variation and evolution of Staphylococcus aureus. Int J Med Microbiol 300:98–103. doi: 10.1016/j.ijmm.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Schijffelen MJ, Boel CHE, van Strijp JAG, Fluit AC. 2010. Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. doi: 10.1186/1471-2164-11-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung JML, Lloyd DH, Lindsay JA. 2008. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 154:1949–1959. doi: 10.1099/mic.0.2007/015289-0. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy AJ, Lindsay JA. 2013. Staphylococcus aureus innate immune evasion is lineage-specific: a bioinfomatics study. Infect Genet Evol 19:7–14. doi: 10.1016/j.meegid.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Brüssow H, Desiere F. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol 39:213–223. doi: 10.1046/j.1365-2958.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- 24.Coleman D, Knights J, Russell R, Shanley D, Birkbeck TH, Dougan G, Charles I. 1991. Insertional inactivation of the Staphylococcus aureus beta-toxin by bacteriophage phi 13 occurs by site- and orientation-specific integration of the phi 13 genome. Mol Microbiol 5:933–939. doi: 10.1111/j.1365-2958.1991.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 25.van Wamel WJB, Rooijakkers SHM, Ruyken M, van Kessel KPM, van Strijp JAG. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J Bacteriol 188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Haas CJ, Veldkamp KE, Peschel A, Weerkamp F, van Wamel WJ, Heezius EC, Poppelier MJ, van Kessel KP, van Strijp JA. 2004. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial anti-inflammatory agent. J Exp Med 199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooijakkers SHM, van Wamel WJB, Ruyken M, van Kessel KPM, van Strijp JAG. 2005. Anti-opsonic properties of staphylokinase. Microbes Infect 7:476–484. doi: 10.1016/j.micinf.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 28.Gladysheva IP, Turner RB, Sazonova IY, Liu L, Reed GL. 2003. Coevolutionary patterns in plasminogen activation. Proc Natl Acad Sci U S A 100:9168–9172. doi: 10.1073/pnas.1631716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Bröker BM, Doskar J, Wolz C. 2009. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol 191:3462–3468. doi: 10.1128/JB.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stegger M, Lindsay JA, Sørum M, Gould KA, Skov R. 2010. Genetic diversity in CC398 methicillin-resistant Staphylococcus aureus isolates of different geographical origin. Clin Microbiol Infect 16:1017–1019. doi: 10.1111/j.1469-0691.2009.03003.x. [DOI] [PubMed] [Google Scholar]
- 31.Cuny C, Abdelbary M, Layer F, Werner G, Witte W. 2015. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet Microbiol 177:219–223. doi: 10.1016/j.vetmic.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 32.Deiters C, Günnewig V, Friedrich AW, Mellmann A, Köck R. 2015. Are cases of methicillin-resistant Staphylococcus aureus clonal complex (CC) 398 among humans still livestock-associated? Int J Med Microbiol 305:110–113. doi: 10.1016/j.ijmm.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Idelevich EA, Schüle I, Grünastel B, Wüllenweber J, Peters G, Becker K. 2014. Rapid identification of microorganisms from positive blood cultures by MALDI-TOF mass spectrometry subsequent to very short-term incubation on solid medium. Clin Microbiol Infect 20:1001–1006. doi: 10.1111/1469-0691.12640. [DOI] [PubMed] [Google Scholar]
- 34.Kaspar U, Kriegeskorte A, Schubert T, Peters G, Rudack C, Pieper DH, Wos-Oxley M, Becker K. 2016. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol 18:2130–2142. doi: 10.1111/1462-2920.12891. [DOI] [PubMed] [Google Scholar]
- 35.Becker K, Pagnier I, Schuhen B, Friedrich AW, Kipp F, Peters G, von Eiff C. 2006. Does nasal cocolonization by staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J Clin Microbiol 44:229–231. doi: 10.1128/JCM.44.1.229-231.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kriegeskorte A, Ballhausen B, Idelevich EA, Köck R, Friedrich AW, Karch H, Peters G, Becker K. 2012. Human MRSA isolates with novel genetic homolog, Germany. Emerg Infect Dis 18:1016–1018. doi: 10.3201/eid1806.110910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellmann A, Friedrich AW, Rosenkötter N, Rothgänger J, Karch H, Reintjes R, Harmsen D. 2006. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med 3:e33. doi: 10.1371/journal.pmed.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mellmann A, Weniger T, Berssenbrügge C, Keckevoet U, Friedrich AW, Harmsen D, Grundmann H. 2008. Characterization of clonal relatedness among the natural population of Staphylococcus aureus strains by using spa sequence typing and the BURP (based upon repeat patterns) algorithm. J Clin Microbiol 46:2805–2808. doi: 10.1128/JCM.00071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Pühler A. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahánková J, Pantůček R, Goerke C, Růžičková V, Holochová P, Doškař J. 2010. Multilocus PCR typing strategy for differentiation of Staphylococcus aureus siphoviruses reflecting their modular genome structure. Environ Microbiol 12:2527–2538. doi: 10.1111/j.1462-2920.2010.02226.x. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lekkerkerk WSN, van Wamel WJB, Snijders SV, Willems RJ, van Duijkeren E, Broens EM, Wagenaar JA, Lindsay JA, Vos MC. 2015. What is the origin of livestock-associated methicillin-resistant Staphylococcus aureus clonal complex 398 isolates from humans without livestock contact? An epidemiological and genetic analysis. J Clin Microbiol 53:1836–1841. doi: 10.1128/JCM.02702-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasigade J, Laurent F, Hubert P, Vandenesch F, Etienne J. 2010. Lethal necrotizing pneumonia caused by an ST398 Staphylococcus aureus strain. Emerg Infect Dis 16:1330. doi: 10.3201/eid1608.100317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valentin-Domelier AS, Girard M, Bertrand X, Violette J, François P, Donnio PY, Talon D, Quentin R, Schrenzel J, van der Mee-Marquet N, the Bloodstream Infection Study Group of the Réseau des Hygiénistes du Centre (RHC). 2011. Methicillin-susceptible ST398 Staphylococcus aureus responsible for bloodstream infections: an emerging human-adapted subclone? PLoS One 6:e28369. doi: 10.1371/journal.pone.0028369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez-Moreno MO, Centelles-Serrano MJ, Nogales-López J, Domenech-Spanedda MF, Lozano C, Torres C. 2017. Unusual presence of the immune evasion gene cluster in livestock-associated MRSA of lineage CC398 causing peridural and psoas abscesses in a poultry farmer. Enferm Infecc Microbiol Clin 35:651–654. doi: 10.1016/j.eimc.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Köck R, Siam K, Al-Malat S, Christmann J, Schaumburg F, Becker K, Friedrich AW. 2011. Characteristics of hospital patients colonized with livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) CC398 versus other MRSA clones. J Hosp Infect 79:292–296. doi: 10.1016/j.jhin.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 48.Kraushaar B, Hammerl JA, Kienöl M, Heinig ML, Sperling N, Thanh MD, Reetz J, Jäckel C, Fetsch A, Hertwig S. 2017. Acquisition of virulence factors in livestock-associated MRSA: lysogenic conversion of CC398 strains by virulence gene-containing phages. Sci Rep 7:2004. doi: 10.1038/s41598-017-02175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y, Nielsen LN, Hvitved A, Haaber JK, Wirtz C, Andersen PS, Larsen J, Wolz C, Ingmer H. 2017. Commercial biocides induce transfer of prophage Φ13 from human strains of Staphylococcus aureus to livestock CC398. Front Microbiol 8:2418. doi: 10.3389/fmicb.2017.02418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goerke C, Wirtz C, Fluckiger U, Wolz C. 2006. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol Microbiol 61:1673–1685. [DOI] [PubMed] [Google Scholar]
- 51.Strommenger B, Kettlitz C, Weniger T, Harmsen D, Friedrich AW, Witte W. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol 44:2533–2540. doi: 10.1128/JCM.00420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graveland H, Wagenaar JA, Broekhuizen-Stins MJ, Oosting-Schothorst I, Schoormans AH, van Duijkeren E, Huijsdens X, Mevius D, Heederik D. 2008. Methicillin-resistant Staphylococcus aureus (MRSA) in veal calf farmers and veal calves in the Netherlands, poster B84, p 62 Abstr ASM Conf Antimicrob Resist Zoonotic Bacteria Foodborne Pathog, Copenhagen, Denmark, 15 to 18 June 2018. [Google Scholar]
- 53.Wulf MW, Verduin CM, van Nes A, Huijsdens X, Voss A. 2012. Infection and colonization with methicillin-resistant Staphylococcus aureus ST398 versus other MRSA in an area with a high density of pig farms. Eur J Clin Microbiol Infect Dis 31:61–65. doi: 10.1007/s10096-011-1269-z. [DOI] [PubMed] [Google Scholar]
- 54.van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis 13:1834–1839. doi: 10.3201/eid1312.070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goerge T, Lorenz MB, van Alen S, Hübner NO, Becker K, Köck R. 2017. MRSA colonization and infection among persons with occupational livestock exposure in Europe: prevalence, preventive options and evidence. Vet Microbiol 200:6–12. doi: 10.1016/j.vetmic.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 56.Becker K, Ballhausen B, Kahl BC, Köck R. 2017. The clinical impact of livestock-associated methicillin-resistant Staphylococcus aureus of the clonal complex 398 for humans. Vet Microbiol 200:33–38. doi: 10.1016/j.vetmic.2015.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.