LETTER
Candida auris is an emerging, multidrug-resistant pathogen associated with a high mortality rate. Since this yeast's first identification and classification by our research group in 2009 (1), there have been several outbreaks linked to this pathogen in health care facilities around the world (2–10). It has been reported that most clinical isolates are resistant to azoles, and about half of the isolates also are resistant to more than one class of antifungal agent, limiting the therapeutic options (2–8, 10). Moreover, the pathogen can persist on environmental surfaces for weeks, resulting in the yeast's spread among patients in health care facilities (11). Therefore, accurate identification of C. auris is critical for controlling this pathogen's prevalence around the globe and preventing further outbreaks.
Traditional methods have proven to be unsuitable for accurate identification of C. auris. Automated identification systems popularly used in clinical laboratories, like the Vitek 2 YST card (bioMérieux, Marcy I'Etoile, France) or API20C AUX (bioMérieux), commonly misidentify C. auris as Candida haemulonii or Rhodotorula glutinis, respectively (2, 4–7, 12), and MicroScan misidentifies C. auris as any of several different Candida species (12). On the other hand, specialized methods can provide accurate identification. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is useful for identifying C. auris, if a proper reference database is available (13–15). Moreover, sequencing of the genes for the D1/D2 region of large subunit ribosomal DNA (rDNA) or of the internal transcribed spacer (ITS) region of rDNA is a reliable option. Real-time PCR assays also are useful for detection of C. auris (16, 17). However, these methods may not be suitable for local or small clinical settings due to financial and technical issues.
As shown in the present study, we have successfully devised and assessed the reliability of a loop-mediated isothermal amplification (LAMP)-based identification approach specific to C. auris, enabling distinction of the pathogen from closely related species and other fungi.
To design the LAMP primers, the genome sequences of four Candida species, C. auris (PRJNA342691), C. tropicalis (GCF_000006335.2), C. albicans (GCA_000182965.3), and C. lusitaniae (LYUB00000000.2), were aligned and compared using Mauve (version 20150226) (18). An 869-bp DNA fragment of the C. auris genome (accession no. XM_018317007) that encodes a pyruvate:ferredoxin oxidoreductase domain (19) was identified as sharing low similarity with other Candida species. This DNA fragment was amplified using EmeraldAmp PCR master mix (TaKaRa Bio, Inc., Shiga, Japan) in combination with C. auris JCM15448T as a template and a pair of primers, AurisF (5′-GCTATGCCGCTAGCAACG-3′) and AurisR (5′-CACTACAGCAGGATCAACGG-3′). The resulting amplicon was purified with a QIAquick PCR purification kit (Qiagen, Venlo, The Netherlands), cloned into the pTAC-2 vector using a DynaExpress TA PCR cloning kit (BioDynamics Laboratory, Inc., Tokyo, Japan) to create plasmid pTAC-2Auris, and subsequently sequenced using an ABI PRISM 3130xl genetic analyzer (Applied Biosystems, Foster City, CA, USA). A candidate LAMP primer set (LAMPAuris) (Table 1) was designed using the sequence of this DNA fragment and PrimerExplorer V5 software (https://primerexplorer.jp/lampv5e/index.html), specifically targeting a 192-bp fragment (corresponding to bp 774 to 965 of the XM_018317007 sequence).
TABLE 1.
LAMP oligonucleotide primer sequences specific to Candida auris
| Primer | Sequence (5′→3′) |
|---|---|
| AurisFIP | AGGCTACTGAGCTTGCTGGTGTAACCAAACCAACAGGAGAGG |
| AurisBIP | ACGGTTTCAGGGTTAGCATGGCTCAACAAAGTCGCTGGTACA |
| AurisLoop-F | CATCTCGAAGGCCTCGGT |
| AurisLoop-B | CACATACTCGAACGGAGTC |
| AurisF3 | GGGAAAGGAACCCTGACCT |
| AurisB3 | GGACACAGCATTCGAAGTGT |
LAMP amplification reactions were run at 56°C for 90 min using a Loopamp turbidimeter RT-160C (Eiken Chemical, Co., Ltd., Tochigi, Japan). Reactions were terminated by deactivating the DNA polymerase at 80°C for 5 min. Each 25-μl reaction mixture consisted of 12.5 μl of 2× reaction mix (Eiken Chemical Co., Ltd.), 1 μl of each primer (40 μM FIP, 40 μM BIP, 20 μM Loop-F, 20 μM Loop-B, 5 μM F3, and 5 μM B3), 1 μl of Bst DNA polymerase, 2 μl of sample DNA solution, and 3.5 μl distilled water.
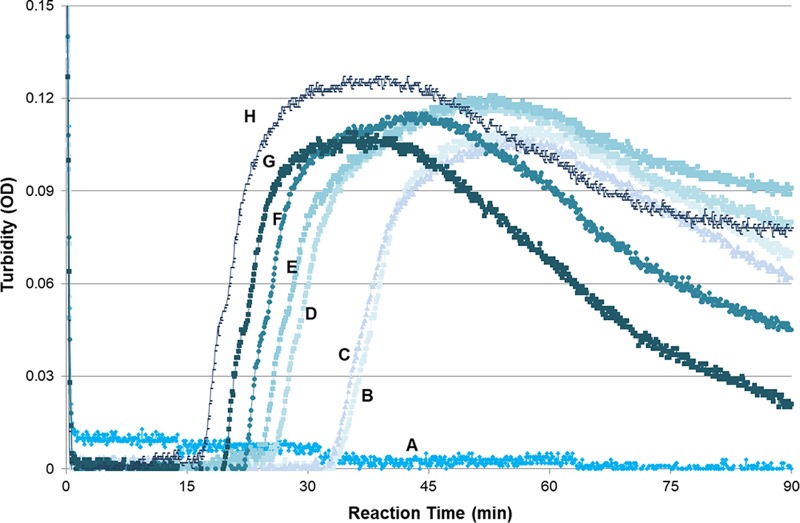
To determine the detection limit of the LAMPAuris primer set, pTAC-2Auris was serially diluted (1 × 100 to 1 × 1010 copies/μl) and used as the template in triplicate reactions. LAMPAuris was able to detect pTAC-2Auris when the plasmid was present at concentrations as low as 2 × 101 copies per reaction (Fig. 1), demonstrating the high sensitivity of the reaction.
FIG 1.
Sensitivity of the LAMPAuris approach. The reaction was run at 56°C for 90 min; the sensitivity of 2 × 101 copies per reaction was confirmed in triplicate reactions. A, negative control (no reaction); B, 2 × 100 copies/reaction (34 min); C, 2 × 101 copies/reaction (33 min); D, 2 × 102 copies/reaction (26 min); E, 2 × 103 copies/reaction (24 min); F, 2 × 104 copies/reaction (24 min); G, 2 × 106 copies/reaction (22 min); H, 2 × 108 copies/reaction (17 min).
To evaluate the specificity of the LAMPAuris primer set toward C. auris, a panel of 63 strains consisting of 39 species, including 21 filamentous fungi and 18 yeasts, were tested (Tables 2 and 3). For each of the filamentous fungi, total DNA was extracted and purified as described previously (20). For each of the yeast strains, small portions of colonies grown on Sabouraud dextrose agar (SDA) plates were suspended in 25 μl distilled water, heated at 100°C for 15 min, and briefly centrifuged. For each LAMPAuris reaction, an aliquot of 2 μl of the purified DNA (filamentous fungi) or heated supernatants (yeasts) was used as a template. All of the 20 C. auris strains yielded amplification signals using the LAMPAuris primer set. In contrast, none of the filamentous fungi or the other yeast species, including those for which C. auris is commonly misidentified, yielded any amplification signal. To validate the quality of the DNA templates used in the LAMPAuris reactions, LAMP reactions using a panfungal LAMP primer set (20) were run separately; amplifications were detected with templates from each of the tested species (data not shown).
TABLE 2.
Candida auris strains and their origins tested in this study
| Country of isolation | Cladea | Strain |
|---|---|---|
| Japan | JCM15448T, LSEM52-3449, LSEM53-3540, LSEM53-3541 | |
| South Korea | CBS12372, CBS12373 | |
| India | CBS12766, CBS12767, CBS12768, CBS12769, CBS12770, CBS12771, CBS12772, CBS12773, CBS12774, CBS12775 | |
| United Kingdom | Japan/Korea | NCPF8984 |
| India/Kuwait/Malaysia | NCPF8971, NCPF8985 | |
| South Africa | NCPF8977 |
Clade as specified by Borman et al. (9).
TABLE 3.
Strains tested in this study and LAMPAuris results
| Speciesa | Strain | LAMPAuris result |
|---|---|---|
| Acremonium curvulum | NBRC32242 | − |
| Aspergillus fumigatus | TIMM0108 | − |
| Aspergillus niger | TIMM0115 | − |
| Candida albicans | LSEM11-828 | − |
| Candida auris | 20 strains as described in Table 2 | + |
| Candida duobushaemulonii* | CBS7799 | − |
| Candida famata* | NBRC0083, NBRC0623 | − |
| Candida glabrata | CBS138, NBRC0005 | − |
| Candida guilliermondii* | TIMM0257 | − |
| Candida haemulonii* | JCM3762 | − |
| Candida krusei | TIMM3378 | − |
| Candida lusitaniae* | NBRC1019, NBRC10059 | − |
| Candida parapsilosis* | ATCC 22019 | − |
| Candida pseudohaemulonii | JCM12453 | − |
| Candida sake* | NBRC0435 | − |
| Candida tropicalis | ATCC 750, TIMM0313 | − |
| Chaetomium globosum | TSY-0369 | − |
| Cladosporium carrionii | TIMM3048 | − |
| Cunninghamella bertholletiae | TIMM3392 | − |
| Exophiala jeanselmei | TSY-0396 | − |
| Fusarium oxysporum | TSY-0351 | − |
| Fusarium solani | TSY-0403 | − |
| Malassezia furfur | CBS1878, LSEM51-3422 | − |
| Malassezia restricta | CBS7877 | − |
| Microsporum gypseum | NBRC5948 | − |
| Mucor circinelloides | TIMM3177 | − |
| Paecilomyces variotii | NBRC4855 | − |
| Penicillium citrinum | LSEM34-2305 | − |
| Pseudallescheria boydii | TIMM0886 | − |
| Rhodotorula glutinis* | LSEM 20-1447 | − |
| Rhodotorula minuta | TIMM6222 | − |
| Saccharomyces cervisiae | LSEM 14-1013 | − |
| Scopulariopsis brevicaulis | NBRC4843 | − |
| Scopulariopsis brumptii | NBRC6441 | − |
| Scytalidium lignicola | NBRC104988 | − |
| Trichophyton benhamiae | SM103 | − |
| Trichophyton mentagrophytes | TIMM2789 | − |
| Trichophyton rubrum | TIMM2659 | − |
| Trichophyton tonsurans | NBRC5928 | − |
Asterisks indicate species that C. auris has been commonly misidentified as.
We also tested the LAMPAuris method on a clinical sample. An ear swab specimen obtained from otitis caused by C. auris LC318417 (21) was tested. The swab was placed in a 2-ml microtube containing 1 ml of saline supplemented with 0.05% Tween 80 and then shaken for 10 min. The resulting suspension was centrifuged at 20,000 × g for 10 min. The generated pellet was washed with 100 μl saline and then subjected to total DNA extraction using the Kaneka Easy DNA extraction kit version 2 (Kaneka Co., Hyogo, Japan) according to the manufacturer's instructions. An aliquot of 2 μl of the extracted DNA was used as a template for the LAMPAuris reaction, yielding a LAMP-positive signal. The entire process of identifying this clinical sample required approximately 1 h, including the direct extraction of total DNA. Prior to use for DNA isolation, the swab was rubbed across the surface of an SDA plate, and this plate then was incubated at 37°C; the resulting small creamy colonies also were identified as C. auris by MALDI-TOF MS (Bruker Daltonics K.K., Kanagawa, Japan) and sequencing of rDNA.
The application of the LAMPAuris method to environmental surveillance was also assessed using mock environmental samples. The samples were prepared by mixing suspensions of Penicillium citrinum (LSEM34-2305), Malassezia furfur (LSEM51-3422), Staphylococcus aureus (ATCC 25923), and Bacillus subtilis (NBRC14132) as background species with C. auris (JCM15448T). First, the concentration of each species was adjusted to 106 cells/ml for fungi and 108 cells/ml for bacteria using a McFarland no. 1 turbidity standard, with additional cell counting for P. citrinum and C. auris. Each solution was then diluted and mixed to create two sets of environmental conditions with different concentrations of microbial cells; set A contained approximately 1 × 102 cells of each background fungal species (P. citrinum and M. furfur) and approximately 1 × 103 of each bacterial species (S. aureus and B. subtilis), and set B contained approximately 1 × 103 cells of each background fungal species and approximately 1 × 104 cells of each bacterial species. A dilution series (1 × 101 to 1 × 105 cells) of C. auris was added to the background solutions, and the mixtures were subjected to total DNA extraction using a Kaneka Easy DNA extraction kit as described above. The LAMPAuris method was tested in triplicate for each condition. As shown in Table 4, there was no apparent interference due to the presence of common skin flora or organisms common to the hospital environment.
TABLE 4.
Reaction time of LAMPAuris under mock environmental conditions
| No. of C. auris cells in mock sample | No. of C. auris cells/reaction (2 μl) | Mean reaction time in min (SD) in: |
||
|---|---|---|---|---|
| Saline | Set Aa | Set Bb | ||
| 1 × 101 | 1 × 10−1 | NRc | NR | NR |
| 1 × 102 | 1 × 100 | NR | NR | NR |
| 1 × 103 | 1 × 101 | 29 (2.8) | 38 (17) | 30 (2.9) |
| 1 × 104 | 1 × 102 | 22 (1.6) | 21 (0.8) | 22 (0) |
| 1 × 105 | 1 × 103 | 19 (1.4) | 20 (0.6) | 20 (0) |
In mock sample set A, Penicillium citrinum and Malassezia furfur cells were on the order of 102 cells, and Staphylococcus aureus and Bacillus subtilis cells were on the order of 103 cells.
In mock sample set B, P. citrinum and M. furfur cells were on the order of 103 cells, and S. aureus and B. subtilis cells were on the order of 104 cells.
NR, no reaction.
Our LAMP approach was proven to reliably identify all of the tested C. auris strains, distinguishing these isolates from other strains (even very closely related species) with a specificity of 100%. Our assay was able to detect a template provided at concentrations as low as 2 × 101 copies of target DNA per reaction. Moreover, the results were obtained within a short time, without any technical complications regarding the use of the amplification instrument. Direct LAMP from a clinical specimen was demonstrated; thus, this technique is expected to save clinicians the time required for cultivation and DNA extraction, allowing an early diagnosis. Recently, portable LAMP amplification equipment has been made commercially available. This availability is expected to facilitate the use of the LAMP assay, enabling large-scale and field surveillance detection. However, care should be taken when handling LAMPAuris product as opening the reaction tube could result in considerable contamination, as we mentioned previously (22). Overall, this assay should be particularly valuable for C. auris, a pathogen that is an important target of environmental control in health care facilities.
ACKNOWLEDGMENT
This research was supported in part by the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development (AMED 18fk0108008h0003).
REFERENCES
- 1.Satoh K, Makimura K, Hasui Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarma S, Kumar N, Sharma S, Govil D, Ali T, Mehta Y, Rattan A. 2013. Candidemia caused by amphotericin B and fluconazole resistant Candida auris. Indian J Med Microbiol 31:90–91. doi: 10.4103/0255-0857.108746. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 6.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo B, Melo AS, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, Meis JF, Colombo AL. 2016. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect 73:369–374. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Borman AM, Szekely A, Johnson EM. 2016. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. mSphere 1:e00189-. doi: 10.1128/mSphere.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borman AM, Szekely A, Johnson EM. 2017. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol 55:563–567. doi: 10.1093/mmy/myw147. [DOI] [PubMed] [Google Scholar]
- 10.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, Litvintseva AP. 2017. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. 2017. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55:638–640. doi: 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ, Chakrabarti A. 2015. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect 21:372–378. doi: 10.1016/j.cmi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 16.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. 2017. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol 55:2445–2452. doi: 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leach L, Zhu Y, Chaturvedi S. 2018. Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J Clin Microbiol 56:e01223-17. doi: 10.1128/JCM.01223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama T, Yamazaki T, Yo A, Tone K, Mahdi Alshahni M, Fujisaki R, Makimura K. 2017. Detection of fungi from an indoor environment using loop-mediated isothermal amplification (LAMP) method. Biocontrol Sci 22:97–104. doi: 10.4265/bio.22.97. [DOI] [PubMed] [Google Scholar]
- 21.Iguchi S, Mizushima R, Kamada K, Itakura Y, Yoshida A, Uzawa Y, Arai Y, Takaoka M, Sato S, Goto A, Karasawa T, Tsuruoka N, Totsuka D, Ono E, Nonaka M, Makimura K, Kikuchi K. 2018. The second Candida auris isolate from aural discharge in Japan. Jpn J Infect Dis 71:174–175. doi: 10.7883/yoken.JJID.2017.466. [DOI] [PubMed] [Google Scholar]
- 22.Uemura N, Makimura K, Onozaki M, Otsuka Y, Shibuya Y, Yazaki H, Kikuchi Y, Abe S, Kudoh S. 2008. Development of a loop-mediated isothermal amplification method for diagnosing Pneumocyctis pneumonia. J Med Microbiol 57:50–57. doi: 10.1099/jmm.0.47216-0. [DOI] [PubMed] [Google Scholar]