Most cases of multidrug-resistant (MDR) tuberculosis (TB) are never diagnosed (328,300 of the ∼490,000 cases in 2016 were missed). The Xpert MTB/RIF assay detects resistance only to rifampin, despite ∼20% of rifampin-resistant cases being susceptible to isoniazid (a critical first-line drug).
KEYWORDS: FluoroType MTBDR, tuberculosis, drug resistance, diagnostics, molecular, Mycobacterium tuberculosis, isoniazid, rifampin
ABSTRACT
Most cases of multidrug-resistant (MDR) tuberculosis (TB) are never diagnosed (328,300 of the ∼490,000 cases in 2016 were missed). The Xpert MTB/RIF assay detects resistance only to rifampin, despite ∼20% of rifampin-resistant cases being susceptible to isoniazid (a critical first-line drug). Consequently, many countries require further testing with the GenoType MTBDRplus assay. However, MTBDRplus is not recommended for use on smear-negative specimens, and thus, many specimens require culture-based drug susceptibility testing. Furthermore, MTBDRplus requires specialized expertise, lengthy hands-on time, and significant laboratory infrastructure and interpretation is not automated. To address these gaps, we evaluated the accuracy of the FluoroType MTBDR (FluoroType) assay. Sputa from 244 smear-positive and 204 smear-negative patients with presumptive TB (Xpert MTB positive, n = 343) were tested. Culture and MTBDRplus on isolates served as reference standards (for active TB and MDR-TB, respectively). Sanger sequencing and MTBDRplus, both of which were performed on sputa, were used to resolve discrepancies. The sensitivity of FluoroType for the detection of M. tuberculosis complex was 98% (95% confidence interval [CI], 95 to 99%) and 92% (95% CI, 84 to 96%) for smear-positive and smear-negative specimens, respectively (232/237 versus 90/98 specimens; P < 0.009). The sensitivity and specificity for smear-negative specimens were 100% and 97%, respectively, for rifampin resistance; 100% and 98%, respectively, for isoniazid resistance; and 100% and 100%, respectively, for MDR-TB. FluoroType identified 98%, 97%, and 97% of the rpoB, katG, and inhA promoter mutations, respectively. FluoroType has excellent sensitivity with sputa equivalent to that of MTBDRplus with the isolates and can provide rapid drug susceptibility testing for rifampin and isoniazid. In addition, the capacity of FluoroType to simultaneously identify virtually all mutations in the rpoB, katG, and inhA promoter may be useful for individualized treatment regimens.
INTRODUCTION
The underdiagnosis of tuberculosis (TB) remains a major hurdle for the eradication of the disease. The World Health Organization (WHO) recommends that all individuals presenting with symptoms or signs of TB be tested with either the Xpert MTB/RIF (Xpert) assay or its successor (Ultra). Individuals diagnosed with rifampin-resistant TB are then initiated on an anti-multidrug-resistant (MDR) TB treatment regimen (1). It is therefore recommended that treatment subsequently be optimized following confirmatory testing for resistance to rifampin, isoniazid, and second-line anti-TB drugs (1). However, confirmatory testing is frequently not done in many settings and rifampin monoresistance thereby fails to be recognized (∼20%) (2, 3).
Only one widely used commercial assay available for MDR-TB exists: GenoType MTBDRplus (MTBDRplus; Hain Lifescience GmbH, Nehren, Germany). WHO has endorsed MTBDRplus, but it is recommended for use only on smear-positive specimens. Smear-negative patients, who are common in high-HIV infection-burden settings, hence first require culture-based drug susceptibility testing (DST), which causes significant delays in diagnosis and initiation of treatment. MTBDRplus is a line probe assay that produces amplicons in an open-tube format and therefore requires an experienced operator, a minimum of three separate rooms, and strict adherence to standard operating procedures to minimize the risk of amplicon cross contamination. These assays also require experienced readers for the interpretation of the hybridized bands, although this can be semiautomated at added expense.
The new assay was invented and constructed at Brandeis University and commercialized as the FluoroType MTBDR (FluoroType) assay by Hain Lifescience GmbH. It is designed as a qualitative in vitro test for the automated detection of the Mycobacterium tuberculosis complex and resistance to rifampin and isoniazid directly from sputum specimens. Detection of rifampin resistance is enabled by identification of mutations in the rifampin resistance-determining region of the rpoB gene. Isoniazid resistance is identified through mutations in katG and the inhA promoter region.
FluoroType uses a nonsymmetric PCR (previously described as a linear after the exponential [LATE] PCR), together with sets of lights-on/lights-off probes (4, 5). The design and construction of this assay have been discussed in detail (our unpublished data). Briefly the assay works as follows: primers amplify separate amplicons for the rpoB, inhA promoter, and katG gene targets plus an internal control. The single-stranded products are detected at the endpoint by melt curve analysis of the hybridized sets of lights-on/lights-off probes (5). The collective fluorescent signals from all probes comprise a temperature-dependent fluorescent signature in a single color. Each fluorescent signature is automatically interpreted by the Fluoro-Software IVD to reliably distinguish and classify sequences, even when they differ by a single nucleotide.
The aim of this study was to evaluate the diagnostic performance of FluoroType using DNA extracted from sputum specimens and to compare the results to those obtained with MTBDRplus.
MATERIALS AND METHODS
Clinical specimens.
A total of 448 sputum specimens were collected from Xpert MTB/RIF-positive and -negative patients at the National Health Laboratory Services, Green Point, Cape Town, South Africa. Sputum specimens were decontaminated with NaOH–N-acetyl-l-cysteine (final concentration, 1%) and resuspended in 2 ml phosphate-buffered saline. Thereafter, 50 μl was subjected to auramine O smear microscopy, and 0.5 ml used to inoculate an MGIT culture. DNA was extracted only from Xpert MTB/RIF-positive sputum sediments and the corresponding positive MGIT cultures using a GenoLyse kit (Hain Lifescience GmbH, Nehren, Germany), and drug susceptibility testing was done on both using the MTBDRplus assay prior to storage of the DNA extracts at −20°C. The residual sediments were stored at 4°C for a maximum of 48 h prior to transport and storage at Stellenbosch University at −20°C. A unique study number was assigned to each specimen, and patient identifiers were removed. A waiver of consent has been granted for this study by the Stellenbosch University Health Research Ethics Committee (reference no. N12/01/001).
DNA extraction.
DNA was extracted from the remaining Xpert MTB/RIF-positive and -negative sputum sediments using a FluoroLyse kit (Hain Lifescience GmbH) according to the manufacturer's instructions. Briefly, a 500-μl aliquot of the residual resuspended sputum sediment was centrifuged for 15 min at 10,000 × g to pellet the bacilli. The resulting pellet was resuspended in 100 μl lysis buffer (F-LYS) (mixed with 2 μl of the amplification control [β-IC]) by vortexing. The mixture was then incubated for 5 min at 95°C. Subsequently, 100 μl of the neutralization buffer (F-NB) was added, and the mixture was vortexed for 5 s and then centrifuged for 5 min at full speed (14,000 × g). The supernatant was then transferred to a new tube and stored at −20°C until further use. A tube containing no clinical material was included during the extraction protocol as a negative control.
FluoroType MTBDR assay.
FluoroType tests were done using the FluoroCycler96 instrument. PCR mixes were freshly prepared by combining 6 μl amplification mix A (AM-A) and 14 μl amplification mix B (AM-B). Thereafter, 20 μl of the FluoroLyse-extracted DNA was added to the PCR mix. Controls included 20 μl of the negative control prepared during the DNA extraction process or 20 μl of the positive control (β-C+). The PCR mixes were immediately loaded into the FluoroCycler96 instrument to avoid photobleaching.
The FluoroType analyzer software validates the positive and negative controls, whether M. tuberculosis complex DNA is identified through the presence of a positive signature for rpoB, and what the specific mutations for all three genes are. It also reports unknown signatures (but different from the wild type) as “MUT” (undifferentiated mutation) and indeterminate results (the software cannot differentiate between the wild type and a mutant) as “IND.” In cases in which the positive, negative, or amplification control failed, “invalid” is reported by the software. Heteroresistance and/or mixed infections were counted as correctly identified if the underlying mutation was detected.
Analysis.
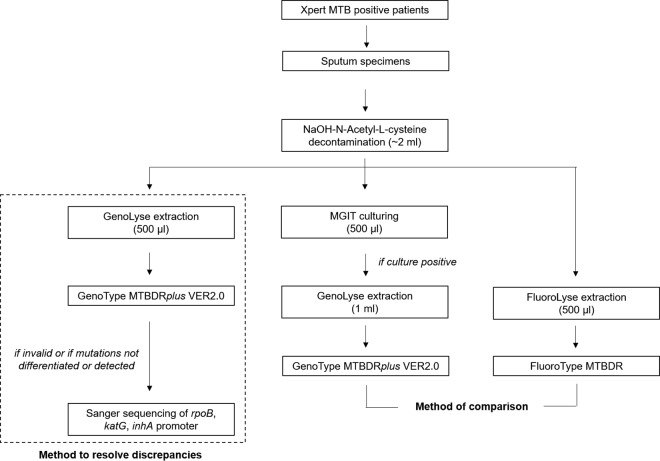
For the detection of the M. tuberculosis complex, the FluoroType result was compared to the presence or absence of the TUB band from the MTBDRplus result obtained for the corresponding cultured isolates. For the detection of rifampin and isoniazid resistance, the FluoroType results were compared to the MTBDRplus hybridization patterns of the probes corresponding to the rpoB and katG genes and the inhA promoter region obtained from the corresponding cultured isolates (as not all samples have a valid MTBDRplus result from a specimen) (Fig. 1). For discrepant results, MTBDRplus results for DNA extracted from the sputum specimens with the GenoLyse kit were used as the reference. In cases in which no valid MTBDRplus result was obtained from sputum or MTBDRplus failed to differentiate or detect specific mutations, Sanger sequencing was also used as a reference method. Targeted Sanger sequencing for the resistance-determining regions of rpoB, katG, and the inhA promoter was done as previously described (6). In case of invalid results, the FluoroType assay was repeated and the second result was used for the final analysis.
FIG 1.
Description of methods used for comparison and discrepancy analysis for the detection of resistance to rifampin and isoniazid using the FluoroType MTBDR assay.
Statistical analysis (two-sample test of proportions) was done using Stata statistical software, release 14.0 (StataCorp. 2015; StataCorp LP, College Station, TX).
RESULTS
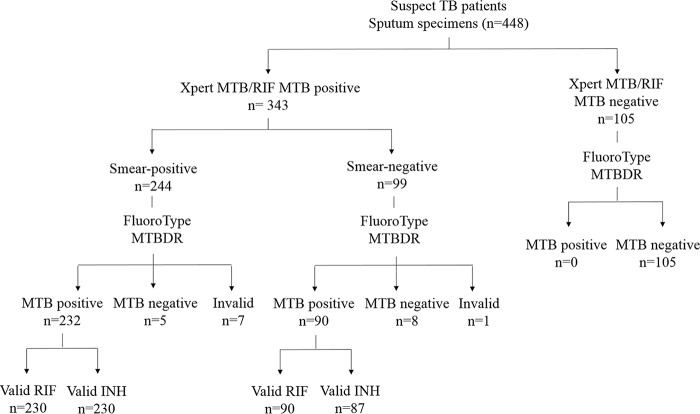
A total of 448 sputum specimens were included in the study; 343 of these were collected from patients diagnosed as Xpert positive, while 105 sputum specimens were collected from patients diagnosed as Xpert negative (smear and culture negative). The 343 sputum specimens from Xpert-positive patients comprised 244 sputum smear-positive and 99 sputum smear-negative specimens (Fig. 2). The Xpert-positive samples comprised 132 (38%) rifampin- and isoniazid-sensitive specimens, 35 (10%) rifampin-monoresistant specimens, 13 (4%) isoniazid-monoresistant specimens, and 163 (48%) MDR specimens, as characterized by MTBDRplus and sequencing.
FIG 2.
Description of specimens collected for the study broken down according to smear status, detection of the M. tuberculosis complex using FluoroType MTBDR, and determination of rifampin and isoniazid susceptibility using FluoroType MTBDR. Definitions of abbreviations: RIF, rifampin; INH, isoniazid; MTB, M. tuberculosis complex.
Detection of M. tuberculosis complex using FluoroType MTBDR.
The sensitivity of the FluoroType assay for the detection of M. tuberculosis was 97.9% in smear-positive specimens and 91.8% in smear-negative specimens (Table 1) when using the presence or absence of the TUB band from the MTBDRplus culture-based result. In the smear-positive sample set, 5/244 of the specimens were not detected as M. tuberculosis complex positive. For 10 specimens, invalid results were obtained. After repeat testing, seven specimens remained with invalid results. In the smear-negative sample set (which included both culture-positive and culture-negative specimens), false-negative results were scored for 8 specimens. For 11 specimens, an invalid result was obtained. After repeat testing, only 1 specimen had an invalid result. The inhibition rate (number of tests reported as “invalid” by the analyzer software) for FluoroType was 1.8% (8/448) for sputum specimens. Table 1 also shows the performance of MTBDRplus done on the same set of sputum specimens. No significant difference was observed between FluoroType and MTBDRplus for the performance on smear-positive specimens (97.8% [95% confidence interval {CI}, 95.2 to 99.3%; 232/237] versus 99.2% [95% CI, 96.8 to 99.9%; 242/244], P = 0.238) and smear-negative specimens (91.8% [95% CI, 84.1 to 96.2%; 90/98] versus 92.3% [95% CI, 85.5 to 96.9%; 92/99], P = 0.775).
TABLE 1.
Performance of FluoroType MTBDR and Genotype MTBDRplus for detection of M. tuberculosis complex from smear-positive and smear-negative sputum specimens compared to culture and Genotype MTBDRplus results obtained from the corresponding cultured isolatesa
| Specimen | FluoroType MTBDRb |
MTBDRplusc |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity |
Specificity |
% PPV (95% CI) | % NPV (95% CI) | No. of invalid tests | Sensitivity |
% PPV (95% CI) | No. of invalid tests | ||||
| % (95% CI) | No. of specimens positive/total no. of samples tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | ||||||
| All (n = 448) | 96.11 (93.3–97.8) | 322/335 | 100 (95.6–100) | 105/105 | 100 (98.5–100) | 89 (81.6–93.8) | 8 | 97 (94.5–98.8) | 334/343 | 100 (98.6–100) | 0 |
| Sputum smear positive (n = 244) | 97.9 (94.9–99.2) | 232/237 | 100 (98.4–100) | 7 | 99.2 (96.8–99.9) | 242/244 (P = 0.24d) | 100 (98.0–100) | 0 | |||
| Sputum smear negative (n = 204) | 91.8 (84.1–96.2) | 90/98 (P = 0.009e) | 100 (96.6–100) | 105/105 | 100 (96–100) | 92.9 (86.5–96.9) | 1 | 92.3 (85.5–96.9) | 92/99 (P = 0.001,e P = 0.78d) | 100 (95–100) | 0 |
Definitions of abbreviations: CI, confidence interval; NPV, negative predictive value; PPV, positive predictive value.
Fluorotype MTBDR assay done on sputum specimens.
Genotype MTBDRplus done on sputum specimens.
P values comparing FluoroType MTBDR and Genotype MTBDRplus results within smear groups.
Assay-specific P values comparing results for smear-positive and smear-negative specimens.
Detection of rifampin resistance using FluoroType MTBDR.
For the detection of rifampin resistance from smear-positive specimens, an initial sensitivity and specificity of 96.9% and 97%, respectively, were achieved when the FluoroType results were compared to the MTBDRplus results for the corresponding cultured isolates (Table 2). This was not significantly different from the performance of MTBDRplus on the same set of sputum specimens (Table 2). Three of 3 false-resistant and 3 of 4 false-sensitive results of FluoroType were confirmed by the MTBDRplus result or Sanger sequencing using the GenoLyse extract from the sputum specimens (Table 3). The remaining false-sensitive result by FluoroType was caused by heteroresistance, identified by Sanger sequencing (Table 3). Therefore, the final sensitivity and specificity of FluoroType for the detection of rifampin resistance from smear-positive specimens were 99.2% and 100%, respectively (Table 4). Only 2/232 smear-positive specimens (0.86%) showed an indeterminate FluoroType result.
TABLE 2.
Performance of FluoroType MTBDR and Genotype MTBDRplus for detection of rifampin and isoniazid resistance from smear-positive and smear-negative sputum specimens compared to GenoType MTBDRplus results obtained from DNA extracts from the corresponding cultured isolatesa
| Specimen | FluoroType MTBDRb |
Genotype MTBDRplusc |
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rifampin |
Isoniazid |
Rifampin |
Isoniazid |
|||||||||||||||||||||||||
| No. of indeterminate results | Sensitivity |
Specificity |
Positive LR (95% CI) | Negative LR (95% CI) | No. of indeterminate results | Sensitivity |
Specificity |
Positive LR (95% CI) | Negative LR (95% CI) | No. of indeterminate tests | Sensitivity |
Specificity |
Positive LR (95% CI) | Negative LR (95% CI) | No. of indeterminate results | Sensitivity |
Specificity |
Positive LR (95% CI) | Negative LR (95% CI) | |||||||||
| % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | |||||||||||||
| All (n = 322) | 97.8 (94.1–99.3) | 178/182 | 95.6 (90.4–98.2) | 132/138 | 22.5 (10.3–49.2) | 0.02 (0.009–0.06) | 98.8 (95.1–99.8) | 158/160 | 97 (92.6–98.9) | 152/157 | 32 (13.5–75.8) | 0.01 (0.003–0.05) | 97.3 (93.5–99) | 181/186 | 91.5 (85.3–95.3) | 129/141 | 11.4 (6.7–19.6) | 0.03 (0.01–0.07) | 96.4 (92–98.5) | 162/168 | 96.8 (92.3–98.8) | 151/156 | 30.1 (12.7–71.3) | 0.04 (0.02–0.08) | ||||
| Sputum smear positive (n = 232) | 2 | 96.9 (92.4–99.2) | 127/131 | 97 (91.4–99.4) | 96/99 | 32.3 (10.5–97.5) | 0.03 (0.01–0.08) | 2 | 98.3 (94.1–99.8) | 117/119 | 97.3 (92.3.–99.4) | 108/111 | 36.4 (11.9–111.1) | 0.02 (0.004–0.07) | 0 | 97.1 (92.2–99.1) | 132/136 (P = 0.95d) | 95.3 (88.8–98.3) | 101/106 | 20.58 (8.74–48.4) | 0.03 (0.01–0.08) | 0 | 97.6 (92.7–99.4) | 123/126 (P = 0.69d) | 99.1 (92.7–99.4) | 115/116 | 113.3 (16.1–797.3) | 0.03 (0.008–0.07) |
| Sputum smear negative (n = 90) | 0 | 100 (93–100) | 51/51 (P = 0.21e) | 92.3 (79.1–98.4) | 36/39 | 13 (4.4–38.6) | 0 (0–NaN) | 3 | 100 (91.4–100) | 41/41 (P = 0.40e) | 95.7 (85.2–99.5) | 44/46 | 23.3 (5.93–89.2) | 0 (0–NaN) | 6 | 98 (88–99.9) | 49/50 (P = 0.73,e P = 0.31d) | 80 (62.5–90.9) | 28/35 | 4.9 (2.5–9.5) | 0.03 (0.003–0.18) | 9 | 92.8 (79.4–98.1) | 39/42 (P = 0.15,e P = 0.08d) | 90 (75.4–96.7) | 36/40 | 9.3 (3.7–23.6) | 0.08 (0.03–0.24) |
Definitions of abbreviations: CI, confidence intervals; LR, likelihood ratio; NaN, the calculation could not be performed because the values entered included one or more instances of zero.
FluoroType MTBDR assay done on sputum specimens.
Genotype MTBDRplus done on sputum specimens.
P values comparing FluoroType MTBDR and Genotype MTBDRplus results within smear groups.
Assay specific P values comparing results for smear-positive and smear-negative specimens.
TABLE 3.
Resolution of discrepant FluoroType MTBDR rifampin and isoniazid resistance results using Sanger sequencing and Genotype MTBDRplus results obtained from DNA extracted from sputum specimensa
| Antimicrobial | Smear result | Initial classification | MTBDRplus resultb | FluoroType MTBDR resultc | Method used to resolve discrepancyd | Final result | Reclassification |
|---|---|---|---|---|---|---|---|
| Rifampin | Positive | False resistant (n = 3) | WT | D516V | MTBDRplus | WT + D516V | True resistant |
| WT | MUT | Sequencing | Q513P | True resistant | |||
| WT | ins(514-515) | Sequencing | TTC ins(514-515) | True resistant | |||
| False sensitive (n = 4) | S531L | WT | MTBDRplus | WT | True resistant | ||
| WT + S531L | WT | MTBDRplus | WT | True resistant | |||
| D516V | WT | MTBDRplus | WT | True resistant | |||
| WT + D516V | WT | Sequencing | WT + D516V | False sensitive | |||
| Negative | False resistant (n = 3) | WT | S531L | MTBDRplus | S531L | True resistant | |
| WT | MUT | Sequencing | WT + D516Y | True resistant | |||
| WT | S531L | MTBDRplus | WT | False resistant | |||
| Isoniazid | Positive | False resistant (n = 3) | WT | S315T2 | MTBDRplus | WT + S315T1 | True resistant |
| WT | S315T1 | MTBDRplus | WT + S315T1 | True resistant | |||
| WT | MUT | MTBDRplus | WT | False resistant | |||
| False sensitive (n = 2) | S315T2 | WT | MTBDRplus | WT | True sensitive | ||
| S315T1 | WT | MTBDRplus | WT | True sensitive | |||
| Negative | False resistant (n = 2) | WT | S315T1 | MTBDRplus | S315T1 | True resistant | |
| WT | C-15T | No valid result obtained | False resistant |
Definitions of abbreviations: WT, wild type; MUT, undifferentiated mutation; ins(514-515), insertion between codons 514 and 515.
GenoType MTBDRplus result from DNA extracted from cultured isolate.
FluoroType MTDR result from DNA extracted from sputum specimen.
Discrepancies were resolved by using either the sequencing or GenoType MTBDRplus results from DNA extracted from sputum specimens.
TABLE 4.
Corrected diagnostic performance of the FluoroType MTBDR assay for detection of rifampin and isoniazid resistance using Sanger sequencing and GenoType MTBDRplus as reference methodsa
| Specimen | Rifampin |
Isoniazid |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of indeterminate results | Sensitivity |
Specificity |
Positive LR (95% CI) | Negative LR (95% CI) | No. of indeterminate results | Sensitivity |
Specificity |
Positive LR (95% CI) | Negative LR (95% CI) | |||||
| % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | |||||||
| All (n = 322) | 99.5 (96.6–100) | 183/184 | 99.3 (95.6–100) | 135/136 | 135.3 (19.2–953.3) | 0.01 (0.0008–0.04) | 100 (97.1–100) | 161/161 | 98.7 (95–99.8) | 154/156 | 78 (19.7–309.1) | 0 (0–NaN) | ||
| Sputum smear positive (n = 232) | 2 | 99.2 (95.8–100) | 130/131 | 100 (96.6–100) | 99/99 | Undefined (NaN–infinity) | 0.01 (0.001–0.05) | 2 | 100 (97–100) | 119/119 | 99.1 (95.1–100) | 110/111 | 111.1 (15.8–781.1) | 0 (0–NaN) |
| Sputum smear negative (n = 90) | 0 | 100 (93–100) | 53/53 | 97.3 (85.8–99.9) | 36/37 | 37.03 (5.4–255.8) | 0 (0–NaN) | 3 | 100 (91.6–100) | 42/42 | 97.8 (88.2–99.9) | 44/45 | 45.5 (6.48–312.5) | 0 (0–NaN) |
Definitions of abbreviations: CI, confidence intervals; LR, likelihood ratio; NaN, the calculation could not be performed because the values entered included one or more instances of zero.
In the smear-negative sample set, an initial sensitivity and specificity of 100% and 92.3%, respectively, were achieved when the FluoroType rifampin susceptibility results were compared to the MTBDRplus results obtained from cultured isolates (Table 2). This was not significantly different from the MTBDRplus result from the same set of sputum specimens (Table 2). Three false-resistant results of FluoroType were confirmed to be true resistant by the MTBDRplus assay done on the DNA extracted directly from the sputum specimens by use of the GenoLyse kit (Table 3). The final sensitivity and specificity of the FluoroType assay for the detection of rifampin resistance from smear-negative specimens were 100% and 97.3%, respectively, with no indeterminate results (Table 4). MTBDRplus done directly on the DNA extracted from the sputum specimens showed a higher indeterminate rate than FluoroType (6.5% [95% CI, 1.5 to 11.7%; 6/91] versus 0% [95% CI, 0 to 0%; 0/90], P = 0.01).
Detection of isoniazid resistance using FluoroType MTBDR.
For the detection of isoniazid resistance from smear-positive specimens, an initial sensitivity and specificity of 98.3% and 97.3%, respectively, were achieved when the FluoroType result was compared to the MTBDRplus result obtained from the corresponding cultured isolates (Table 2). This was not significantly different from the results of MTBDRplus done on the same set of sputum specimens (Table 2). Using the MTBDRplus result from the sputum specimens, 2/3 false-resistant and 2/2 false-sensitive results of FluoroType were confirmed to be true resistant and true sensitive, respectively (Table 3). Therefore, the overall sensitivity and specificity of FluoroType for the detection of isoniazid resistance from smear-positive specimens were 100% and 99.1%, respectively (Table 4), with 2 (0.86%) specimens showing an indeterminate result.
In the smear-negative sample set, an initial sensitivity and specificity for the detection of isoniazid resistance of 100% and 95.7%, respectively, were achieved when the FluoroType results were compared to the results from the MTBDRplus assay done on the corresponding cultured isolates (Table 2). This was not significantly different from the MTBDRplus result done on sputum (Table 2). Two false-resistant results were detected by FluoroType, while 3/90 specimens (3.33%) showed indeterminate results (Table 2 and 3). One of the false-resistant results was confirmed to be true resistant by MTBDRplus assay analysis of DNA extracted from the sputum specimen by use of the GenoLyse kit, while for the second sample, no valid MTBDRplus or sequencing result could be obtained from the sputum specimen. Thus, the overall sensitivity and specificity of the FluoroType assay for the detection of isoniazid resistance in smear-negative specimens were 100% and 97.8%, respectively (Table 4).
Detection of MDR using the FluoroType MTBDR.
When using Sanger sequencing and MTBDRplus (from specimens and the corresponding isolates) as the reference methods, FluoroType showed a sensitivity and a specificity of 99.1 and 100%, respectively, with smear-positive sputum specimens for the detection of MDR-TB (Table 5). With smear-negative specimens, FluoroType showed a sensitivity and a specificity of 100% for the detection of MDR-TB (Table 5).
TABLE 5.
Performance of FluoroType MTBDR for detection of MDR-TB from smear-positive and smear-negative sputum specimens when using Sanger sequencing and GenoType MTBDRplus as reference methodsa
| Specimen | Sensitivity |
Specificity |
Positive LR (95% CI) | Negative LR (95% CI) | No. of indeterminate results | ||
|---|---|---|---|---|---|---|---|
| % (95% CI) | No. of specimens positive/total no. tested | % (95% CI) | No. of specimens positive/total no. tested | ||||
| All (n = 315) | 99.3 (95.8–100) | 148/149 | 100 (97.2–100) | 166/166 | Infinity (NaN–infinity) | Infinity (NaN–infinity) | 7 |
| Sputum smear positive (n = 228) | 99.1 (94.3–100) | 108/109 | 100 (96.1–100) | 119/119 | Infinity (NaN–infinity) | 0.09 (0.001–0.06) | 4 |
| Sputum smear negative (n = 87) | 100 (89.1–100) | 40/40 | 100 (90.6–100) | 47/47 | Infinity (NaN–infinity) | Infinity (NaN–infinity) | 3 |
Definitions of abbreviations: ind, indeterminate; CI, confidence interval; LR, likelihood ratio; NaN, the calculation could not be performed because the values entered included one or more instances of zero.
Detection of specific resistance-associated mutations by the FluoroType MTBDR assay.
The specific resistance-associated mutations in rpoB, katG, and the inhA promoter detected by the FluoroType assay were compared to the resistance mutations identified by MTBDRplus and Sanger sequencing (Tables 6 to 8). Since the FluoroType assay can output only one result per gene, heteroresistance and/or mixed infections were regarded as correctly interpreted if the software was able to identify the specific resistance-associated mutation. Overall, the FluoroType assay correctly identified 97.5% (314/322), 96.6% (311/322), and 96.6% (311/322) of the rpoB, katG, and inhA promoter mutations, respectively (Tables 6 to 8).
TABLE 6.
Accuracy of FluoroType MTDR to detect specific mutations in rpoB identified by Genotype MTBDRplus and sequencinga
| rpoB genotype | No. of isolates for which the genotype was: |
% of isolatesb | ||||||
|---|---|---|---|---|---|---|---|---|
| Identified by MTBDRplus or sequencing | Correctly identified by FluoroType MTBDR | Incorrectly identified by FluoroType MTBDR |
||||||
| Indeterminate | False resistance | Incorrect mutation | Undifferentiated mutation | Only WT was recorded | ||||
| WT | 138 | 135 | 2 | 1 | 97.8 | |||
| S531L | 135 | 133 | 2 | 98.5 | ||||
| Q513P | 1 | 0 | 1 | 0 | ||||
| Het/mixed | 11 | 9 | 1 | 1 | 81.1 | |||
| Totalc | 322 | 314 | 97.5 | |||||
Definitions of abbreviations: WT, wild type; Het/mixed, heteroresistance and/or mixed infection.
Genotypes identified with 100% accuracy by FluoroType MTBDR: H526D (9/9), H526Y (7/7), D516V (4/4), D516Y (3/3), H526N (3/3), L533P (2/2), insertion between codons 514 and 515 (1/1), L511P (2/2), H526L (2/2), H526R (1/1), S531F (1/1), Q513K (1/1), and H526P (1/1).
The total includes the results for genotypes identified with 100% accuracy.
TABLE 7.
Accuracy of FluoroType MTDR to detect specific mutations in katG identified by Genotype MTBDRplus and sequencing
| katG genotypea | No. of isolates for which the genotype was: |
% of isolates | ||||||
|---|---|---|---|---|---|---|---|---|
| Identified by Genotype MTBDRplus and sequencing | Correctly identified by FluoroType MTBDR | Incorrectly identified by FluoroType MTBDR |
||||||
| Indeterminate | False resistance | Incorrect mutation | Undifferentiated mutation | Only WT was recorded | ||||
| WT | 257 | 254 | 2 | 1 | 98.8 | |||
| S315T1 | 42 | 42 | 100.0 | |||||
| S315T2 | 14 | 11 | 1 | 2 | 78.6 | |||
| S315I | 1 | 0 | 1 | 0.0 | ||||
| Het/mixed | 8 | 4 | 3 | 1 | 50.0 | |||
| Total | 322 | 311 | 96.6 | |||||
Definitions of abbreviations: WT, wild type, Het/mixed, heteroresistance and/or mixed infection.
TABLE 8.
Accuracy of FluoroType MTDR to detect specific mutations in inhA promoter identified by Genotype MTBDRplus and sequencinga
| inhA promoter genotype | No. of isolates in which the genotype was: |
% of isolates | ||||||
|---|---|---|---|---|---|---|---|---|
| Identified by Genotype MTBDRplus and sequencing | Correctly identified by FluoroType MTBDR | Incorrectly identified by FluoroType MTBDR |
||||||
| Indeterminate | False resistance | Incorrect mutation | Undifferentiated mutation | Only WT was recorded | ||||
| WT | 211 | 207 | 3 | 1 | 98.1 | |||
| T−15C | 100 | 99 | 1 | 99.0 | ||||
| G−17T | 4 | 0 | 1 | 3 | 0.0 | |||
| T−8A | 1 | 1 | 100.0 | |||||
| T−8C | 1 | 1 | 100.0 | |||||
| Het/mixed | 5 | 3 | 1 | 1 | 60.0 | |||
| Total | 322 | 311 | 96.6 | |||||
Definitions of abbreviations: WT, wild type; Het/mixed, heteroresistance and/or mixed infection.
DISCUSSION
This was the first study to evaluate the diagnostic performance of the FluoroType assay with smear-positive and -negative sputum specimens. Our key findings are as follows: (i) FluoroType showed increased sensitivity for the detection of the M. tuberculosis complex in smear-negative sputum specimens, (ii) FluoroType was highly comparable to MTBDRplus for the detection of resistance to rifampin and isoniazid irrespective of smear gradation, (iii) FluoroType showed a high accuracy for the identification of mutations in rpoB, katG, and the inhA promoter, and (iv) FluoroType is a simple, rapid closed-tube molecular test that can provide operator-independent results. These findings suggest that FluoroType can be used as a replacement test for MTBDRplus. (v) Interpretation of FluoroType data is fully automated, thereby removing operator interpretation subjectivity.
FluoroType showed increased sensitivity (91.8%) for the detection of the M. tuberculosis complex in smear-negative sputum specimens compared to that previously shown for MTBDRplus (40 to 100%) (7–12). In this study, no significant difference in the performance of MTBDRplus (sensitivity, 92.3% [95% CI, 85.5 to 96.9; 92/99], P = 0.76) from that of FluoroType with smear-negative sputum specimens was observed. This is not unexpected, as all specimens tested were diagnosed as Xpert positive, and similar agreement between MTBDRplus and Xpert has been reported (13). We did not test the performance of FluoroType with Xpert TB-negative TB culture-positive specimens, as the specimen inclusion criteria were based on the current diagnostic algorithm used in South Africa. The rate of detection of M. tuberculosis complex in smear-positive sputum specimens was 97.9%, with seven invalid results. This might be due to the DNA degradation explained by the long-term storage of the DNA of the isolates for several months at −20°C prior to performance of the FluoroType assay. In a routine laboratory, DNA extracts should be analyzed immediately.
The FluoroType MTBDR assay showed a high sensitivity for the detection of mutations conferring resistance to rifampin and isoniazid irrespective of smear gradation. This is an important finding, as the only commercially available WHO-endorsed test for MDR-TB is currently recommended for use on smear-positive specimens only. While no difference in the performance between MTBDRplus and FluoroType for the detection of rifampin and isoniazid resistance was observed, a higher indeterminate rate was observed for MTBDRplus for the detection of rifampin resistance in smear-negative specimens. Within the group of 322 sputum specimens that tested positive for the M. tuberculosis complex with the FluoroType MTBDR assay, 17 FluoroType results were found to be discrepant from the MTBDRplus results obtained for the corresponding cultured isolate. When using Sanger sequencing and the MTBDRplus result for the sputum specimen, only 4 results discrepant by FluoroType were proven to be truly discordant. For the remaining 13 results discrepant by FluoroType, 6 resistant results matched the MTBDRplus result from sputum, implying the loss of the resistant population of the cultured isolate; 2 mutations (rpoB Q513P and a TTC insertion in rpoB between codons 514 and 515) were undetected by MTBDRplus but found by Sanger sequencing; and 5 sensitive results matched the MTBDRplus result from sputum, implying a transcription error for the resistant cultured isolates.
Our results showed a sensitivity and a specificity for the detection of rifampin resistance in specimens similar to those obtained by culture of the isolates in combination with phenotypic DST as the reference method (14). However, Hillemann et al. showed a lower sensitivity for the detection of isoniazid resistance (91.7%), which may be explained by different frequencies of noncanonical resistance markers in the respective study populations (14).
Besides the discrimination of rifampin and/or isoniazid resistance, the software also allows the reliable identification of most significant associated mutations in rpoB, katG, and the inhA promoter region. The software was able to detect the correct mutation genotype for rpoB, katG, and the inhA promoter for 97.5%, 96.6%, and 96.6% of the samples, respectively, when using sequencing and MTBDRplus as reference methods. Isolates with a few uncommon mutations (e.g., rpoB Q513P, rpoB TTC insertion between codons 514 to 515, and inhA promoter G−17T) were correctly evaluated as mutants but without differentiation of the specific mutation. As specified by the manufacturer, the software-based differentiation of these rare mutations will be possible with the updated versions of the analyzer software. Knowing the exact mutation is crucial, as resistance is not a binary phenomenon and different mutations that are identified by this assay confer a range of resistance phenotypes with large variations in MICs (15–18). Five disputed rpoB mutations (L511P, D516V, H526L, H526N, and L533P) that have previously been associated with controversial rifampin phenotypes were correctly identified by FluoroType (17–23). Infections caused by strains harboring the mutations D516V, L533P, and H526L may still be treatable with rifabutin or high-dose rifampin (17, 22, 24, 25). Similarly, infections caused by strains harboring mutations in the promoter region of inhA may still be treated with high-dose isoniazid (26, 27). These mutations also provide additional information regarding MDR treatment, as they also confer resistance to ethionamide (28, 29).
The FluoroType assay is a rapid molecular test that can provide operator-independent results within 3 h, which is a time to the result significantly faster than that of the MTBDRplus assay, as it does not require any hands-on time after PCR amplification (2 to 3 h for setup and running of the GT-Blotter for MTBDRplus hybridization). In addition, the MTBDRplus assay requires manual or semiautomated scoring of hybridization patterns, whereas the results of the FluoroType assay are analyzed and reported independently by the analyzer software. The FluoroType assay is also performed in a single closed tube, preventing the release of amplicons and thereby eliminating the risk of laboratory cross contamination and reducing the laboratory infrastructure required to perform molecular-based DST. A limitation of the study was the use of MTBDRplus as a reference method. Phenotypic DST would have been the optimal reference method; however, the aim of this study was to evaluate the performance of FluoroType against a WHO-endorsed molecular assay that is routinely used in our setting. Evaluation of future diagnostics studies could use whole-genome sequencing or targeted deep next-generation sequencing to resolve discrepancies, given their superior sensitivity to detect resistant subpopulations.
These findings suggest that the FluoroType MTBDR assay could be implemented as a replacement for the MTBDRplus assay, thereby greatly simplifying the rapid diagnosis of rifampin and isoniazid resistance.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) under award number RO1 A1099532. This work is supported by the National Research Foundation, the South African Medical Research Council (SA MRC), and the Stellenbosch University Faculty of Medicine Health Sciences. G.T. acknowledges funding from the European and Developing Countries Clinical Trials Partnership (EDCTP).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the SA MRC. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Data analysis and interpretation were done independently from Hain Lifescience GmbH.
FluoroType MTBDR kits were a gift from Hain Lifescience GmbH.
R.M.W. and M.D.V. have received conference support from Hain Lifescience GmbH. J.E.R. and L.J.W. have received research grants from Hain Lifescience GmbH to develop the FluoroType MTBDR. J.E.R. and L.J.W. have royalties paid through Brandeis University from Hain Lifescience GmbH for the licensing of two U.S. patents.
We thank the National Health Laboratory Services, Green Point, Cape Town, South Africa, and Hain Lifescience GmbH.
REFERENCES
- 1.WHO. 2016. WHO treatment guidelines for drug-resistant tuberculosis, 2016 update. WHO, Geneva, Switzerland. [Google Scholar]
- 2.WHO. 2017. Global tuberculosis report 2017. WHO, Geneva, Switzerland. [Google Scholar]
- 3.Cox H, Dickson-Hall L, Ndjeka N, Van't Hoog A, Grant A, Cobelens F, Stevens W, Nicol M. 2017. Delays and loss to follow-up before treatment of drug-resistant tuberculosis following implementation of Xpert MTB/RIF in South Africa: a retrospective cohort study. PLoS Med 14:e1002238. doi: 10.1371/journal.pmed.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez JA, Pierce KE, Rice JE, Wangh LJ. 2004. Linear-after-the-exponential (LATE)-PCR: an advanced method of asymmetric PCR and its uses in quantitative real-time analysis. Proc Natl Acad Sci U S A 101:1933–1938. doi: 10.1073/pnas.0305476101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice JE, Reis AH Jr, Rice LM, Carver-Brown RK, Wangh LJ. 2012. Fluorescent signatures for variable DNA sequences. Nucleic Acids Res 40:e164. doi: 10.1093/nar/gks731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black PA, de Vos M, Louw GE, van der Merwe RG, Dippenaar A, Streicher EM, Abdallah AM, Sampson SL, Victor TC, Dolby T, Simpson JA, van Helden PD, Warren RM, Pain A. 2015. Whole genome sequencing reveals genomic heterogeneity and antibiotic purification in Mycobacterium tuberculosis isolates. BMC Genomics 16:857. doi: 10.1186/s12864-015-2067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomasicchio M, Theron G, Pietersen E, Streicher E, Stanley-Josephs D, van Helden P, Warren R, Dheda K. 2016. The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates. Sci Rep 6:17850. doi: 10.1038/srep17850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott LE, McCarthy K, Gous N, Nduna M, Van Rie A, Sanne I, Venter WF, Duse A, Stevens W. 2011. Comparison of Xpert MTB/RIF with other nucleic acid technologies for diagnosing pulmonary tuberculosis in a high HIV prevalence setting: a prospective study. PLoS Med 8:e1001061. doi: 10.1371/journal.pmed.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luetkemeyer AF, Kendall MA, Wu X, Lourenco MC, Jentsch U, Swindells S, Qasba SS, Sanchez J, Havlir DV, Grinsztejn B, Sanne IM, Firnhaber C, Adult AIDS Clinical Trials Group A5255 Study Team. 2014. Evaluation of two line probe assays for rapid detection of Mycobacterium tuberculosis, tuberculosis (TB) drug resistance, and non-TB mycobacteria in HIV-infected individuals with suspected TB. J Clin Microbiol 52:1052–1059. doi: 10.1128/JCM.02639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedrich SO, Venter A, Kayigire XA, Dawson R, Donald PR, Diacon AH. 2011. Suitability of Xpert MTB/RIF and GenoType MTBDRplus for patient selection for a tuberculosis clinical trial. J Clin Microbiol 49:2827–2831. doi: 10.1128/JCM.00138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorman SE, Chihota VN, Lewis JJ, van der Meulen M, Mathema B, Beylis N, Fielding KL, Grant AD, Churchyard GJ. 2012. GenoType MTBDRplus for direct detection of Mycobacterium tuberculosis and drug resistance in strains from gold miners in South Africa. J Clin Microbiol 50:1189–1194. doi: 10.1128/JCM.05723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crudu V, Stratan E, Romancenco E, Allerheiligen V, Hillemann A, Moraru N. 2012. First evaluation of an improved assay for molecular genetic detection of tuberculosis as well as rifampin and isoniazid resistances. J Clin Microbiol 50:1264–1269. doi: 10.1128/JCM.05903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnard M, Gey van Pittius NC, van Helden PD, Bosman M, Coetzee G, Warren RM. 2012. The diagnostic performance of the GenoType MTBDRplus version 2 line probe assay is equivalent to that of the Xpert MTB/RIF assay. J Clin Microbiol 50:3712–3716. doi: 10.1128/JCM.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillemann D, Haasis C, Andres S, Behn T, Kranzer K. 2018. Validation of the FluoroType MTBDR assay for detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J Clin Microbiol 56:e00072-18. doi: 10.1128/JCM.00072-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo H, Seet Q, Denkin S, Parsons L, Zhang Y. 2006. Molecular characterization of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis from the USA. J Med Microbiol 55:1527–1531. doi: 10.1099/jmm.0.46718-0. [DOI] [PubMed] [Google Scholar]
- 16.Morlock GP, Metchock B, Sikes D, Crawford JT, Cooksey RC. 2003. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 47:3799–3805. doi: 10.1128/AAC.47.12.3799-3805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamieson FB, Guthrie JL, Neemuchwala A, Lastovetska O, Melano RG, Mehaffy C. 2014. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J Clin Microbiol 52:2157–2162. doi: 10.1128/JCM.00691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang Y, Lu J, Wang Y, Song Y, Wang S, Zhao Y. 2013. Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893–900. doi: 10.1128/AAC.01024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Deun A, Aung KJ, Hossain A, de Rijk P, Gumusboga M, Rigouts L, de Jong BC. 2015. Disputed rpoB mutations can frequently cause important rifampicin resistance among new tuberculosis patients. Int J Tuberc Lung Dis 19:185–190. doi: 10.5588/ijtld.14.0651. [DOI] [PubMed] [Google Scholar]
- 20.Van Deun A, Barrera L, Bastian I, Fattorini L, Hoffmann H, Kam KM, Rigouts L, Rusch-Gerdes S, Wright A. 2009. Mycobacterium tuberculosis strains with highly discordant rifampin susceptibility test results. J Clin Microbiol 47:3501–3506. doi: 10.1128/JCM.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Deun A, Aung KJ, Bola V, Lebeke R, Hossain MA, de Rijk WB, Rigouts L, Gumusboga A, Torrea G, de Jong BC. 2013. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol 51:2633–2640. doi: 10.1128/JCM.00553-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrada ZL, Lin SY, Rodwell TC, Nguyen D, Schecter GF, Pham L, Janda JM, Elmaraachli W, Catanzaro A, Desmond E. 2016. Rifabutin and rifampin resistance levels and associated rpoB mutations in clinical isolates of Mycobacterium tuberculosis complex. Diagn Microbiol Infect Dis 85:177–181. doi: 10.1016/j.diagmicrobio.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andres S, Hillemann D, Rusch-Gerdes S, Richter E. 2014. Occurrence of rpoB mutations in isoniazid-resistant but rifampin-susceptible Mycobacterium tuberculosis isolates from Germany. Antimicrob Agents Chemother 58:590–592. doi: 10.1128/AAC.01752-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirgel FA, Warren RM, Bottger EC, Klopper M, Victor TC, van Helden PD. 2013. The rationale for using rifabutin in the treatment of MDR and XDR tuberculosis outbreaks. PLoS One 8:e59414. doi: 10.1371/journal.pone.0059414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rukasha I, Said HM, Omar SV, Koornhof H, Dreyer AW, Musekiwa A, Moultrie H, Hoosen AA, Kaplan G, Fallows D, Ismail N. 2016. Correlation of rpoB mutations with minimal inhibitory concentration of rifampin and rifabutin in Mycobacterium tuberculosis in an HIV/AIDS endemic setting, South Africa. Front Microbiol 7:1947. doi: 10.3389/fmicb.2016.01947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollela VR, Namburete EI, Feliciano CS, Macheque D, Harrison LH, Caminero JA. 2016. Detection of katG and inhA mutations to guide isoniazid and ethionamide use for drug-resistant tuberculosis. Int J Tuberc Lung Dis 20:1099–1104. doi: 10.5588/ijtld.15.0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller B, Streicher EM, Hoek KG, Tait M, Trollip A, Bosman ME, Coetzee GJ, Chabula-Nxiweni EM, Hoosain E, Gey van Pittius NC, Victor TC, van Helden PD, Warren RM. 2011. inhA promoter mutations: a gateway to extensively drug-resistant tuberculosis in South Africa? Int J Tuberc Lung Dis 15:344–351. [PubMed] [Google Scholar]
- 28.Larsen MH, Vilcheze C, Kremer L, Besra GS, Parsons L, Salfinger M, Heifets L, Hazbon MH, Alland D, Sacchettini JC, Jacobs WR Jr. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol Microbiol 46:453–466. doi: 10.1046/j.1365-2958.2002.03162.x. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]