Carbapenem-resistant and hypervirulent Klebsiella pneumoniae strains have emerged recently. These strains are both hypervirulent and multidrug resistant and may also be highly transmissible and able to cause severe infections in both the hospital and the community.
KEYWORDS: carbapenem resistance, hypervirulence, plasmid, multiplex PCR, capsular polysaccharide
ABSTRACT
Carbapenem-resistant and hypervirulent Klebsiella pneumoniae strains have emerged recently. These strains are both hypervirulent and multidrug resistant and may also be highly transmissible and able to cause severe infections in both the hospital and the community. Clinical and public health needs require a rapid and comprehensive molecular detection assay to identify and track the spread of these strains and provide timely infection control information. Here, we develop a rapid multiplex PCR assay capable of distinguishing K. pneumoniae carbapenem-resistant isolates of sequence type 258 (ST258) and ST11, and hypervirulent ST23, ST65/ST375, and ST86 clones, as well as capsular types K1, K2, K locus type 47 (KL47), and KL64, and virulence genes rmpA, rmpA2, iutA, and iroN. The assay demonstrated 100% concordance with 118 previously genotyped K. pneumoniae isolates and revealed different populations of carbapenem-resistant and hypervirulent strains in two collections in China and the United States. The results showed that carbapenem-resistant and hypervirulent K. pneumoniae strains are still rare in the United States, whereas in China, ∼50% of carbapenem-resistant strains carry rmpA/rmpA2 and iutA virulence genes, which are largely associated with the epidemic ST11 strains. Similarly, a high prevalence of hypervirulent strains was found in carbapenem-susceptible isolates in two Chinese hospitals, but these primarily belong to ST23, ST65/ST375, and ST86, which are distinct from the carbapenem-resistant strains. Taken together, our results demonstrated that this PCR assay can be a useful tool for molecular surveillance of carbapenem-resistant and hypervirulent K. pneumoniae strains.
INTRODUCTION
During the past 3 decades, two largely nonoverlapping populations of Klebsiella pneumoniae strains have emerged. One is associated with multidrug resistance, including epidemic carbapenem-resistant sequence type 285 (ST258) and ST11 strains; and the other population consists of the hypervirulent capsular K1 or K2 strains, primarily belonging to ST23, ST86, and ST65 (or ST375, a single locus variant of ST65) (1, 2). Alarmingly, recent reports revealed the convergence of virulence and antimicrobial resistance, most commonly as a result of plasmid-mediated resistance traits and virulence gene transfer. A recent study conducted in China described an outbreak where all patients died of a carbapenem-resistant ST11 strain with increased virulence—the consequence of a pLVPK-like hypervirulent plasmid transferring into the epidemic carbapenem-resistant K. pneumoniae (CRKp) ST11 strain (3). The discovery of a virulence plasmid in epidemic CRKp strains raises significant concern that these “hybrid” organisms, which are both hypervirulent (HV) and multidrug resistant (MDR), may also be highly transmissible and able to cause severe fatal infections in both the hospital and the community (3, 4). As a result, clinical and public health needs require a rapid and comprehensive molecular detection assay to identify and track the spread of these strains and provide timely infection control information.
In response to this emerging problem, we developed three multiplex PCR amplification reactions (PCR-I, PCR-II, and PCR-III), which are capable of detecting and differentiating the epidemic carbapenem-resistant ST258 and ST11 strains. In addition, these reactions detect: (i) serotype K1- and K2-associated ST23, ST86, and ST65/ST375 strains, and (ii) two ST11-associated capsules, K locus type 47 (KL47) and KL64. Furthermore, this assay was also designed to detect four virulence genes in the pLVPK-like virulence plasmids (5).
MATERIALS AND METHODS
For the development of the multiplex PCR design, we investigated 105 completely sequenced K. pneumoniae genomes from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/genomes/; searched May 2017). These genomes were assigned into 37 different STs, including sequence types 11, 14, 15, 16, 17, 23, 37, 45, 65, 66, 67, 86, 101, 147, 258, 278, 340, 512, etc., using the Kleborate pipeline (https://github.com/katholt/Kleborate). Clone-specific sequences were then extracted by the Panseq (6) script, using the default settings. The clone-specific sequences were then compared against ∼1,400 K. pneumoniae draft genome sequences obtained from the NCBI FTP server to examine the sequence specificity.
Using this analytical approach, we identified the PCR target sequences for ST11/258, ST23, ST65/ST375, and ST86. The pilon-like gene, pilV (locus_tag KPNJ1_01960), located in the ST258-specific integrative and conjugative element ICEKp258.2, was selected as a marker for ST258. This gene is also present in ST379, ST418, and ST512 strains (closely related to ST258), which belong to the previously described clonal group 258 (CG258)-tonB79 cluster (7). ST11 is a single locus variant to ST258 and shares ∼80% genome sequences with ST258 (8). Panseq analysis failed to identify unique targets in ST11 strains compared to other STs. As a result, we selected two additional genes, kphp (KPNJ1_00468) and pilL (KPNJ1_00782) that were positive for both ST258 and ST11 and were located on a truncated prophage and an integrative and conjugative element (ICEKp258.1), as the markers for both ST258 and ST11.
Taken together, CG258-tonB79 cluster strains (e.g., ST258, ST379, ST418, and ST512) are identified as being positive for kphp, pilL, and pilV, while other CG258 strains (e.g., ST11, ST340, and ST437) are only positive for kphp and pilL. In addition, we have also included a K. pneumoniae subsp. pneumoniae species-specific marker khe (KPNJ1_05155) as an internal PCR control to evaluate the species and the quality of the DNA extraction. Similarly, specific target sequences for ST23 (KP1_1127), ST86 (D364_23960), and ST65/ST375 (P243_2648) were selected for the development of the primers for each individual ST.
PCR-II was designed to target the capsular polymerase gene, wzy, in K1 (wzyK1) and K2 (wzyK2), as well as in K locus 47 (KL47) (wzyKL47) and KL64 (wzyKL64), two of the most prevalent capsular types found in ST11 strains in China (5). K locus is a novel nomenclature for K. pneumoniae capsule synthesis loci that is based on the whole-genome data (9). In general, KL1 to KL77 correspond to the loci associated with each of the 77 serologically defined K types, while KL101 and higher are defined from DNA sequence data on the basis of gene content (9) (https://github.com/katholt/Kaptive). The wzy gene is unique to each capsular type and can be used as a genetic marker to differentiate the different capsular types. In our previous study, we developed a multiplex PCR for the detection of the ST258-associated capsular types ST258-cps1 (corresponding to KL106) and ST258-cps2 (KL107) (10). Here, we selected four major wzy genes, and, in combination with our previously developed PCR assay (10), we are able to detect the common ST11- and ST258-associated capsular types, as well as the K1 and K2 serotypes.
PCR-III was designed to detect four virulence genes, namely, rmpA, rmpA2, iroN, and iutA, previously described on the pLVPK virulence plasmid. Among them, rmpA/rmpA2 are the mucoid phenotype regulator genes, whereas iroN and iutA are two genes linked to the siderophore-mediated iron-acquisition operons iroBCDN and iucABCD-iutA, respectively (11). These genes have essential roles in the virulence process in K. pneumoniae pathogenesis (1). In addition, the IncHIB replicon gene in pLVPK was included as another PCR target to evaluate the association between the virulence genes and the plasmid replicon.
The multiplex PCR primers in this study were designed using MPprimer (12), and the oligonucleotides sequences are listed in Table 1. DNA templates were extracted using the boiling lysis method described previously (10). The PCR was performed in 15-μl reaction volumes consisting of 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), 2 mM MgCl2, and 250 μM of each deoxynucleoside triphosphate (dNTP), in 1× PCR buffer (Applied Biosystems) on an iCycler thermal cycler (Bio-Rad). The primer concentrations of each multiplex PCR are listed in Table 1. The optimal cycling conditions are as follows: an initial denaturation step of 94°C for 10 min, followed by 35 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 60 s, and a final extension step of 72°C for 5 min. Amplicons were visualized after running at 100 V for 1.5 h in 1.5% agarose gels containing GelStar nucleic acid gel stain (Ionza Ltd., Allendale, NJ) (Fig. 1).
TABLE 1.
Oligonucleotide primers used in this study
| PCR reaction or primer | Sequence (5′–3′) | Length (bp) | Concentration (μM) | Target (locus_tag) |
|---|---|---|---|---|
| PCR-I | ||||
| Khe-F3 | CCGGAGCGTTTTTCAATCGGCG | 441 | 200 | KPNJ1_05155 |
| Khe-R3 | CGCTTCGCCCCTCACCTGAAAT | |||
| pilV-F3 | CGATGGCGCTGGCGACGATTAT | 627 | 200 | KPNJ1_01960 |
| pilV-R3 | CCCGATGGGCAAGAACATGCGT | |||
| kphp-F3 | TGGCGGGTAATGCCCGATCAGT | 236 | 200 | KPNJ1_00468 |
| kphp-R3 | AGGCCGCTTTCCATAAGCCGTT | |||
| pilL-F2 | CGGTATTTGCTCTGCGTGATAG | 350 | 200 | KPNJ1_00782 |
| pilL-R2 | TGGTTATACAGAACGGCATTGG | |||
| ST65-F1 | GCTATGCCAATGCCGGAACCGT | 146 | 200 | P243_2648 |
| ST65-R1 | ACCCACCCCCGTAAAGATGGCA | |||
| ST23-F1 | ACGCATGCCTACAGGACATCGAA | 518 | 200 | KP1_1127 |
| ST23-R1 | TCCAGTAGGAGCAGACAGCGGA | |||
| ST86-F2 | TATTCTTTCTAGGGGCCGAATG | 778 | 400 | D364_23960 |
| ST86-R2 | TCACCAGCTCACTTGAACATTA | |||
| PCR-II | ||||
| wzyK1-F1 | ACGGAGCAATATGGCCAGTCCG | 174 | 200 | KP1_3714 |
| wzyK1-R1 | GCCAACAATTCCCGTTTCTGCTGC | |||
| wzyK2-F1 | TATTCGAGCTGGCGCCGCCTTA | 451 | 200 | D364_12775 |
| wzyK2-R1 | CGTTCCCCTTGTTGCCACGGAT | |||
| wzyKL64-F1 | TCAGTTCCGACCCTGATGCAGGTA | 268 | 200 | BT120_10560 |
| wzyKL64-R1 | GCCAGAGCAACTATCATCCAAAGCCA | |||
| wzyKL47-F1 | GGACGCACAGTTTCCCAATTCGC | 392 | 200 | CJU70_21765 |
| wzyKL47-R1 | GCCCACATGAACCCACTTGGCA | |||
| PCR-III | ||||
| rmpA-F1 | ATGTGGCTTGACGTTTCGGGGG | 160 | 200 | LV255 |
| rmpA-R1 | GCCGTGGATAATGGTTTACAATTCGGC | |||
| rmpA2-F1 | GGATGTGGCTTGACATTTCGGGGG | 227 | 200 | LV136 |
| rmpA2-R1 | TTCATGGATGCCCTCCCTCCTG | |||
| HI1B-F1 | TCGCTACTGCGATTGGGGGTCT | 351 | 600 | LV040 |
| HI1B-R1 | GAAATGGGTGTGCTGGAGCCGT | |||
| iroN-F1 | CCGCAAAGAGACGAACCGCCTT | 546 | 100 | LV243 |
| iroN-R1 | CGGGCAATCCCCGCTTTGACTT | |||
| iutA-F1 | AATCACCTGGGGGCTGGATGCT | 683 | 270 | LV107 |
| iutA-R1 | CCGCACCTTCCACGCCGTAAAT |
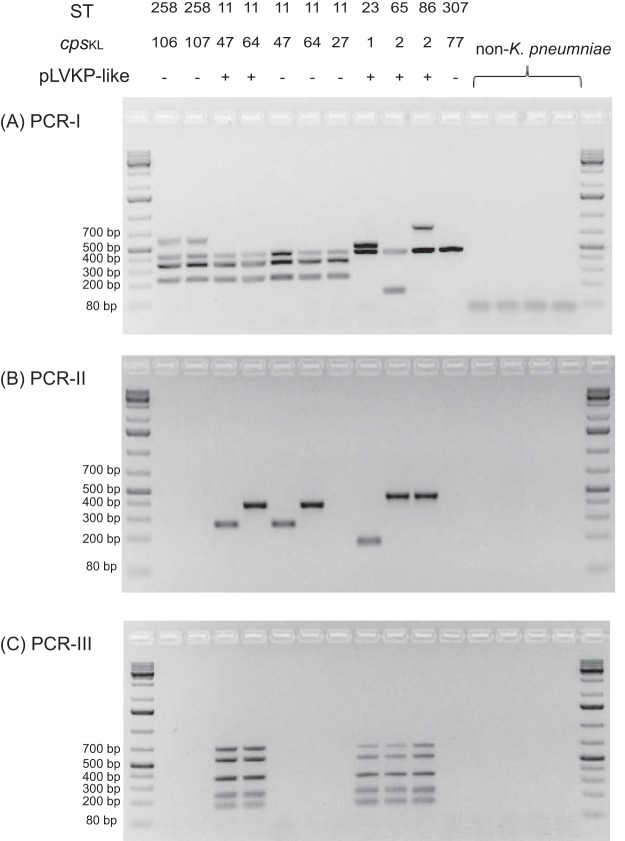
FIG 1.
Agarose gel electrophoresis of multiplex PCR products.
Hypervirulence in K. pneumoniae has been significantly associated with a hypermucoviscous phenotype (1, 2). In this study, the hypermucoviscosity phenotype was examined using the string test (13) by stretching a bacterial colony on a 5% sheep blood agar plate from overnight culture; the formation of a viscous string >5 mm in length was considered to be positive.
RESULTS AND DISCUSSION
The validation of the multiplex PCR assays initially used 118 K. pneumoniae strains selected from our archived strain collection at the Public Health Research Institute (PHRI) Tuberculosis (TB) Center, which had been previously characterized by multilocus sequence typing (MLST) or whole-genome sequencing, including isolates of ST11 (n = 10), ST23 (n = 8), ST86 (n = 6), ST65 (n = 6), ST258 (n = 25), ST340 (n = 2), ST375 (n = 2), ST379 (n = 5), ST412 (n = 1), ST437 (n = 5), ST512 (n = 8), and other STs (n = 40). The K. pneumoniae-specific khe target was amplified for all 118 K. pneumoniae strains, generating a 441-bp amplification product (10). In PCR-I, the ST258, ST379, ST412, and ST512 strains, members of CG258-tonB79 cluster, all gave four bands of 236 (pkph), 441 (khe), 627 (pilV), and 350 (pilL) bp, while the ST11, ST340, and ST437 (CG258 non-tonB79 cluster) isolates were positive for kphp (236 bp), khe (441 bp), and pilL (350 bp) but not for pilV (627 bp) (Fig. 1). Similarly, ST23, ST65/ST375 and ST86 strains were all positive for khe (441 bp), and their clone-specific targets (KP1_1127 for ST23, 518 bp; D364_23960 for ST86, 778 bp; and P243_2648 for ST65/ST375, 146 bp). For the 40 other STs, only the internal khe was amplified. The PCR-I results showed 100% concordance with the MLST or whole-genome sequencing (WGS) results.
In PCR-II, K. pneumoniae ST23 isolates were all positive for wzyK1, defined by the 174-bp amplicon, while ST86, ST65, and ST375 isolates were positive for wzyK2, with a 451-bp amplicon. In addition, two isolates, belonging to ST14 and ST25, were also positive for wzyK2, consistent with the literature, which previously reported that the K2 capsule can be found on different STs (14). Among the 10 ST11 K. pneumoniae isolates, 2 were positive for wzyKL47, defined by a 392-bp amplicon, and 3 were positive for wzyKL64, with a 268-bp amplicon, while the remaining 5 ST11 strains were negative for wzyKL47 and wzyKL64. Further sequencing of the wzi alleles (15) from these ST11 strains revealed that all wzyKL47-positive strains carry wzi209, while wzyKL64-positive strains harbor wzi64. In contrast, the remaining five wzyKL47- and wzyKL64-negative ST11 isolates carry wzi24, wzi27, wzi39, wzi50, and wzi273, which belong to other capsular serotypes.
PCR-III was designed to detect virulence genes found in the pLVPK-like plasmids. Our results showed (Fig. 1) that the ST23, ST65/ST375, and ST86 strains all have positive amplification for five bands, corresponding to the four virulence genes (rmpA, rmpA2, iutA, and iroN) and the IncHIB plasmid replicon. Other ST strains were not found to carry rmpA, rmpA2, or iroN, but 3 ST258 strains and one ST147 strain harbored iutA, while 10 strains (2 ST258, 2 ST11, 3 ST147, 2 ST14, and 1 ST340) carried the IncHIB replicon. The PCR-III results were further confirmed by simplex PCRs, using previously published primers (3, 16), followed by Sanger sequencing, demonstrating 100% concordance with the multiplex results.
Following the validation, we retrospectively screened 208 clinical CRKp strains from two collections in the United States and China, using the multiplex PCR assay. The U.S. collections consisted of 97 CRKp isolates collected in 9 hospitals in the New Jersey and New York City in 2013 (17). The Chinese collection contained 66 CRKp and 45 carbapenem-susceptible K. pneumoniae (CSKp) collected in two hospitals in eastern China between 2014 and 2016 (Table 2). All isolates were subjected to the string test.
TABLE 2.
PCR screening results for K. pneumoniae isolates
| PCR screening target | No. (%) of isolatesb |
||
|---|---|---|---|
| U.S. collection, CRKp (n = 97) | Chinese collection |
||
| CRKp (n = 66) | CSKp (n = 45) | ||
| Multilocus STs | |||
| ST258 | 78 (80.4) | 0 (0) | 0 (0) |
| ST11 | 0 (0) | 52 (78.8) | 1 (2.2) |
| ST23 | 0 (0) | 0 (0) | 15 (33.3) |
| ST65/375 | 0 (0) | 0 (0) | 4 (8.9) |
| ST86 | 0 (0) | 0 (0) | 2 (4.4) |
| Other STs | 19 (19.6) | 14 (21.2) | 23 (51.1) |
| cps type | |||
| wzyKL106a | 20 (20.6) | 0 (0) | 0 (0) |
| wzyKL107a | 43 (44.3) | 0 (0) | 0 (0) |
| wzyKL47 | 0 (0) | 29 (43.9) | 0 (0) |
| wzyKL64 | 0 (0) | 22 (33.3) | 1 (2.2) |
| wzyK1 | 0 (0) | 0 (0) | 15 (33.3) |
| wzyK2 | 0 (0) | 0 (0) | 8 (17.8) |
| p-LVPK genes | |||
| IncHIB | 8 (8.2) | 29 (43.9) | 36 (80.0) |
| rmpA/rmpA2 | 0 (0) | 32 (48.5) | 39 (86.7) |
| iutA | 0 (0) | 32 (48.5) | 31 (68.9) |
| iroN | 0 (0) | 10 (15.2) | 29 (64.4) |
| Carbapenemases | |||
| KPC-2 | 44 (45.3) | 55 (83.3) | 0 (0) |
| KPC-3 | 48 (49.5) | 0 (0) | 0 (0) |
| OXA-232 | 1 (1.0) | 0 (0) | 0 (0) |
| NDM-1 | 0 (0) | 6 (9.1) | 0 (0) |
| NDM-5 | 0 (0) | 3 (4.5) | 0 (0) |
| IMP-26 | 0 (0) | 1 (1.5) | 0 (0) |
| Hypermucoviscosity | 5 (5.2) | 28 (42.4) | 32 (71.1) |
Conducted by previously published multiplex PCR (10).
CRKp, carbapenem-resistant K. pneumoniae; CSKp, carbapenem-susceptible K. pneumoniae.
In the U.S. carbapenem-resistant collection, the four virulence genes were not found, although 5 isolates (all ST258) displayed a hypermucoviscosity phenotype by the string test. A total of 78 (80.4%) of isolates belonged to ST258, and among them, 25.6% (n = 20) carried wzyKL106, and 55.1% (n = 43) harbored wzyKL107, while the remaining 15 ST258 isolates carried capsular types other than KL106 or KL107. The capsular types K1 and K2 were not detected in this collection. KPC-2 and KPC-3 were found in 45.3% and 49.5% of the isolates, respectively (18). In the carbapenem-resistant collection from China, 78.8% (n = 52) of isolates belonged to ST11 and harbored blaKPC-2. Virulence genes rmpA2, rmpA, iutA, and iroN were found in 32 (48.5%), 15 (22.7%), 32 (48.5%) and 10 (15.2%) isolates, respectively, and they were all from ST11 strains. Among the 32 rmpA2- and iutA-positive ST11 isolates, 21 harbored wzyKL47, the same capsular type reported previously (3), while 11 of them were found to carry wzyKL64, another major capsular type found in ST11 strains in China, indicating that the plasmid-borne virulence genes have spread to another major clade in ST11. Notably, 28 out of the 32 isolates harboring virulence determinants were also hypermucoviscous. Significantly, ST23, ST65/ST375, and ST86, and K1 or K2 capsular types, were not identified in this collection.
In contrast, in the carbapenem-susceptible collection of isolates from China, ST23, ST65/ST375, and ST86 accounted for 33.3% (n = 15), 8.9% (n = 4), and 4.4% (n = 2) of the total isolates, respectively. ST23 strains all carry the K1 capsular type, while ST65/ST375 and ST86 strains harbor the K2 capsular type, and they were all positive for the four virulence genes and the IncHIB replicon and showed the hypermucoviscous phenotype. Notably, a number of non-ST23, ST65/ST375, and ST86 strains, which carried non-K1 or K2 capsular types, were also hypermucoviscous and harbored the virulence genes, suggesting the presence of additional virulence clones among the CSKp isolates (Table 2).
Despite the compelling nature of these findings, our assay has some restrictions. First, our assay mainly detects the most common hypervirulent K1/K2 capsular types and their associated ST23, ST65/ST375, and ST86 clones, while other minor capsular serotypes with reported hypervirulence, including K5, K16, K20, K28, K54, K57, K63, and their STs (1), are not covered. Second, the assay only detected two common capsular types (KL47 and KL64) in ST11, but more than 10 capsular types have been found in ST11 strains (19), although virulence genes were not reported in ST11 strains with other capsular types. Nevertheless, our assay did demonstrate its applicability in identifying the epidemic carbapenem-resistant ST258 and ST11, and in differentiating them from the hypervirulent ST23, ST65/ST375, and ST86 strains. The molecular surveillance study suggested that carbapenem-resistant and hypervirulent strains are still rare in the United States, while in China ∼50% of CRKp strains, largely associated with the epidemic ST11 strains, may be hypervirulent. Similarly, a high prevalence of hypervirulent strains were found in carbapenem-susceptible isolates in two Chinese hospitals, but they primarily belonged to ST23, ST65/ST375, and ST86, which are distinct from the carbapenem-resistant strains.
In summary, we have developed and validated three novel multiplex PCR amplification reactions. In comparison to MLST and whole-genome sequencing, the PCRs can rapidly identify the epidemiological carbapenem-resistant K. pneumoniae ST258/ST11 strains and the hypervirulent ST23, ST65/ST375, and ST86 strains. Our results suggested that this PCR assay can be a useful tool for the molecular surveillance of carbapenem-resistant and hypervirulent K. pneumoniae strains.
ACKNOWLEDGMENTS
This work was in part supported by the following grants from the National Institutes of Health (R01AI090155, R21AI135250, R21AI117338, and CA008748), and by the Key Research and Development project of Jiangsu Provincial Science and Technology Department (BE2017654), Six Talent Peaks Project in Jiangsu Province (2016-WSN-112), Gusu Key Health Talent of Suzhou, Jiangsu Youth Medical Talents Program (QN-867) and the Science and Technology Program of Suzhou (SZS201715). In addition, research reported in this publication was also supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R21AI114508 (R.A.B.), R01AI100560 (R.A.B.), R01AI063517 (R.A.B.), and R01AI072219 (R.A.B.). This study was also supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, award 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research and Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 (R.A.B.).
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, Jeong BC, Lee SH. 2017. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol 7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Kreiswirth BN. 2018. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis 18:2–3. doi: 10.1016/S1473-3099(17)30517-0. [DOI] [PubMed] [Google Scholar]
- 5.Dong N, Zhang R, Liu L, Li R, Lin D, Chan EW, Chen S. 2018. Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China. Microb Genom. doi: 10.1099/mgen.0.000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, Thomas JE, Gannon VP. 2010. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11:461. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. 2014. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. mBio 5:e01355-14. doi: 10.1128/mBio.01355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2:e000102 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Chavda KD, Findlay J, Peirano G, Hopkins K, Pitout JD, Bonomo RA, Woodford N, DeLeo FR, Kreiswirth BN. 2014. Multiplex PCR for identification of two capsular types in epidemic KPC-producing Klebsiella pneumoniae sequence type 258 strains. Antimicrob Agents Chemother 58:4196–4199. doi: 10.1128/AAC.02673-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Shen Z, Qu W, Wang W, Lu Y, Wu Y, Li Z, Hang X, Wang X, Zhao D, Zhang C. 2010. MPprimer: a program for reliable multiplex PCR primer design. BMC Bioinformatics 11:143. doi: 10.1186/1471-2105-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decre D. 2013. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang HL, Chiang MK, Liou WJ, Chen YT, Peng HL, Chiou CS, Liu KS, Lu MC, Tung KC, Lai YC. 2010. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis 29:689–698. doi: 10.1007/s10096-010-0915-1. [DOI] [PubMed] [Google Scholar]
- 17.Satlin MJ, Chen L, Patel G, Gomez-Simmonds A, Weston G, Kim AC, Seo SK, Rosenthal ME, Sperber SJ, Jenkins SG, Hamula CL, Uhlemann AC, Levi MH, Fries BC, Tang YW, Juretschko S, Rojtman AD, Hong T, Mathema B, Jacobs MR, Walsh TJ, Bonomo RA, Kreiswirth BN. 2017. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother 61:e02349-16. doi: 10.1128/AAC.02349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J Clin Microbiol 49:579–585. doi: 10.1128/JCM.01588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wick RR, Heinz E, Holt KE, Wyres KL. 2018. Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J Clin Microbiol 56:e00197-18. doi: 10.1128/JCM.00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]