The purpose of this study was to develop and optimize different processing, extraction, amplification, and sequencing methods for metagenomic next-generation sequencing (mNGS) of cerebrospinal fluid (CSF) specimens. We applied mNGS to 10 CSF samples with known standard-of-care testing (SoC) results (8 positive and 2 negative).
KEYWORDS: CSF, metagenomics, next-generation sequencing
ABSTRACT
The purpose of this study was to develop and optimize different processing, extraction, amplification, and sequencing methods for metagenomic next-generation sequencing (mNGS) of cerebrospinal fluid (CSF) specimens. We applied mNGS to 10 CSF samples with known standard-of-care testing (SoC) results (8 positive and 2 negative). Each sample was subjected to nine different methods by varying the sample processing protocols (supernatant, pellet, neat CSF), sample pretreatment (with or without bead beating), and the requirement of nucleic acid amplification steps using DNA sequencing (DNASeq) (with or without whole-genome amplification [WGA]) and RNA sequencing (RNASeq) methods. Negative extraction controls (NECs) were used for each method variation (4/CSF sample). Host depletion (HD) was performed on a subset of samples. We correctly determined the pathogen in 7 of 8 positive samples by mNGS compared to SoC. The two negative samples were correctly interpreted as negative. The processing protocol applied to neat CSF specimens was found to be the most successful technique for all pathogen types. While bead beating introduced bias, we found it increased the detection yield of certain organism groups. WGA prior to DNASeq was beneficial for defining pathogens at the positive threshold, and a combined DNA and RNA approach yielded results with a higher confidence when detected by both methods. HD was required for detection of a low-level-positive enterovirus sample. We demonstrate that NECs are required for interpretation of these complex results and that it is important to understand the common contaminants introduced during mNGS. Optimizing mNGS requires the use of a combination of techniques to achieve the most sensitive, agnostic approach that nonetheless may be less sensitive than SoC tools.
INTRODUCTION
Agnostic metagenomic next-generation sequencing (mNGS) for infectious diseases diagnostics is emerging as a promising universal pathogen detection method (1). This method has the capability to overcome many of the limitations of current microbiologic methods, such as culture, targeted nucleic amplification tests, and serologic assays, by providing a hypothesis-free, culture-independent, broad pathogen detection method directly from clinical specimens (2). Although the method demonstrates great potential, there remain many challenges to the development and application of these technologies in the clinical setting.
With mNGS, all nucleic acid from a clinical specimen is isolated and sequenced in parallel. This results in the isolation and amplification of host, pathogen, and other sources of extraneous nucleic acid (normal microbiota, reagent, and processing contaminants) from the specimen. As the specimen is of human origin and the human genome is much larger than microbial genomes, the results from mNGS typically generate >90% host reads, with a minute fraction assigned to microbial reads (3). Interpretation of these results can be rather difficult, especially in the absence of an obvious pathogen and appropriate controls.
Using mostly research applications of mNGS, many successful cases or retrospective case series are reported in the literature for the diagnosis of central nervous system (CNS) infections (3–10). Despite these successes, there are still many gaps in knowledge as it applies to mNGS methods, as most published studies to date utilize a variety of nonstandardized methodologies and for the most part lack suitable controls (1). For laboratories considering offering these tests, there is little published in terms of development and standardization of methods. Furthermore, development and optimization of mNGS in a diagnostic microbiology laboratory are resource intensive in terms of reagent and instrument costs and technologist time. Consequently, there are very few method comparison studies. These steps are required to identify the best approach prior to performing an extensive validation of the wet (laboratory methods) and dry (informatics) components of mNGS for implementation for clinical care (2). Based on this, a proof-of-concept study was performed to optimize and evaluate different processing, extraction, amplification, and sequencing methods to determine a workflow to further validate mNGS of cerebrospinal fluid (CSF) specimens for clinical care.
MATERIALS AND METHODS
Sample selection.
A total of 10 CSF samples with sufficient volume (2.6 ml) were selected for the method comparison study. Eight CSF samples that were previously determined to be positive using standard-of-care testing (SoC) in the Johns Hopkins Hospital Microbiology Laboratory for a pathogenic agent (yeasts, Gram-positive bacteria, Gram-negative bacteria, and viruses) were selected for testing. Residual CSF had been stored within 7 days of collection in nuclease-free 2.0-ml (Sarstedt) tubes at −80°C until the time of testing. Two negative samples were selected, one from a patient with hydrocephalus who was not suspected to have an infectious process and a second specimen that was negative by conventional microbiologic methods. Data Set S1 in the supplemental material summarizes the CSF cell counts and SoC microbiologic results. This study was reviewed and approved by the Johns Hopkins University School of Medicine Institutional Review Board.
Method comparison study.
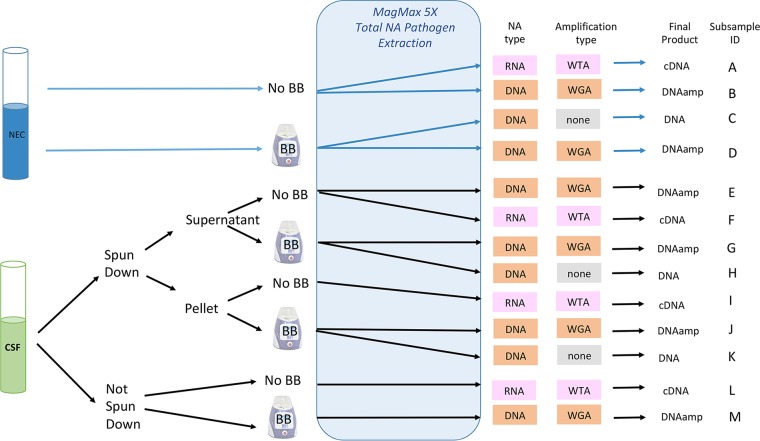
Each of the 10 CSF specimens (labeled samples 1 to 10) were subjected to nine different methods (subsamples E to M) by varying the sample processing protocols (supernatant, pellet, and neat CSF), sample pretreatment (with and without a bead-beating [BB] step), and the requirement of nucleic acid amplification steps using DNA sequencing (DNASeq) (with and without whole-genome amplification [WGA]) and RNA sequencing (RNASeq) with whole-transcriptome amplification [WTA] methods. In addition, for each sample, a negative extraction control (NEC; nuclease-free water) was run with each of the DNASeq and RNASeq methods for an additional four subsamples per specimen (subsamples A to D). All methods are described in further detail below. Figure 1 illustrates the various methods applied to the samples. At the completion of the method comparison study, a host depletion (HD) and limit of detection (LOD) study was performed.
FIG 1.
Overview of the method comparison study. Each CSF sample had four matched negative controls (A to D) and nine CSF subsamples (E to M). BB, bead beating; WGA, whole-genome amplification; WTA, whole-transcriptome amplification; DNA, DNASeq (no WGA); DNAamp, DNASeq with WGA; cDNA, RNASeq with WTA; NEC, negative extraction control; NA, nucleic acid; not spun down, neat CSF.
Sample processing, pretreatment, and NA extraction.
Prior to extraction, samples were divided into two 1-ml and one 600-μl aliquot (neat CSF). Each 1-ml sample was centrifuged in a 15-ml tube at 3,000 rpm for 5 min. The supernatant was removed, and 600 μl was set aside for testing; 300 μl, including pelleted material remaining in the 15-ml tubes, was resuspended and combined in a separate tube for a total sample volume of 600 μl. Each of the 600-μl aliquots (neat CSF, supernatant, pellet) were further divided, with 200 μl transferred to an extraction processing tube (subsamples E, F, I, and L) and 400 μl transferred to a FastPrep B lysis tube, which was processed on a FastPrep24 5G homogenizer (MP Biomedicals) for 30 s at 6 m/s. Homogenized samples were allowed to settle for 20 min, and then 200 μl was transferred to extraction tubes (subsamples G, H J, K, and M). Each extraction run was accompanied by two NECs (nuclease-free water with [subsamples D and C] and without [subsamples A and B] FastPrep bead beating).
Total nucleic acid extraction was performed manually using the MagMAX pathogen RNA/DNA kit (ThermoFisher) per the manufacturer's recommendations, with the following specified parameters: manual extraction method, sample input of 200 μl, and all manipulations performed at room temperature.
Amplification.
Nucleic acid amplification was performed using the REPLI-g WGA & WTA kit (Qiagen) per the manufacturer's package insert, with the omission of the cell lysis step, which had been performed previously using the MagMAX kit (see above). Subsamples A, F, I, and L underwent whole-transcriptome amplification prior to RNASeq (referred herein as the cDNA method). Subsamples B, D, E, G, J, and M underwent whole-genome amplification prior to DNASeq (referred herein as the DNAamp method). Subsamples C, H, and K did not undergo any amplification prior to library preparation for DNASeq (referred herein as the DNA method).
Library preparation.
Library preparation was performed with the Nextera XT kit (Illumina) according to the guidance generated from the Illumina Protocol Selector (https://support.illumina.com/custom-protocol-selector.html). Briefly, samples were quantified by a Qubit 3.0 fluorometer and diluted to a standard input of 0.2 ng/μl; 5 μl input DNA was added to 10 μl Tagment DNA buffer and 5 μl Amplicon Tagment mix in a 96-well PCR plate, which was then gently mixed and placed on a thermal cycler (TC) for 5 min at 55°C with a brief subsequent hold at 10°C. Neutralization buffer was then added in 5-μl aliquots to each sample well; the plate was centrifuged for 1 min and incubated at room temperature for 5 min.
The PCR plate was transferred to the Illumina-supplied index plate fixture, and samples were dual-indexed with 5 μl of unique i5 and i7 index adaptors; 15 μl Nextera PCR master mix was added to each sample well, and samples were mixed by gentle pipetting. The plate was transferred to a TC programmed with the following parameters: 72°C for 3 min, 95°C for 30 s, followed by 12 cycles of 95°C for 10 s, 55°C for 30 s, 72°C for 30 s, and 72°C for 5 min, and a 10°C hold.
Library cleanup was performed using Agencourt AMPure XP beads (Beckman Coulter, Indianapolis, IN), at a 3:5 ratio, followed by two washes with 80% ethanol and suspension in Illumina's resuspension buffer. The DNA concentration of each sample was then measured by Qubit, and the DNA fragment size was determined using a 4200 TapeStation (Agilent, Santa Clara, CA) using high-sensitivity D5000 ScreenTapes and reagents (Agilent).
If required, there were stopping points built into the workflow, allowing for storage at −80°C after extraction, at −20°C after REPLI-g nucleic acid amplification, or after library cleanup.
mNGS.
For each sample, two 150-cycle v3 flow cells were run: DNAamp subsamples (B, D, E, G, J, M) were run on flow cell 1, and cDNA (A, F, I, L) and unamplified DNA (C, H, K) subsamples were processed on flow cell 2. Samples were normalized to a concentration range between 2 and 4 nM, pooled, denatured by 0.2 N NaOH, and diluted by Illumina HT1 buffer to a 20 pM solution. A final loading volume of 600 μl was then loaded into an Illumina MiSeq cartridge and run overnight. To assess the quality control (QC) of the MiSeq run, we set a baseline acceptable Q30 (metric to assess base calling accuracy) score of 80% (i.e., 80% of each run's calls have a 1 in 1,000 chance of being incorrect).
Bioinformatics.
We used Illumina Experiment Manager (version 1.12.0) and Illumina MiSeq Reporter (version 2.6.2.1) software to trim adapters and then analyzed all sequences using Kraken (database v.2016-01-13) (11). Kraken classifies overlapping 31-kmer bp sequences by mapping them to the lowest common ancestor to provide the most accurate taxonomic classification possible (i.e., species, genus, family, order, etc.). Kraken results were visualized using Pavian (12). KrakenHLL (version 0.3.2) was applied for read classification. KrakenHLL enhances Kraken classifications with unique kmer counts, which can be used as proxy for genome coverage. Taxa that are detected with high unique kmer counts are more reliable, as more of the genomic sequence is covered (13). As a comparator, genomic coverage of positives was also determined by alignment.
Analysis of results.
Paired Kraken results for the NEC and CSF samples were compared to each other by normalizing the counts to reads per million (RPM). A positive cutoff of ≥10 times the RPM of the NEC was used as the reporting threshold for reads in the CSF samples, as previously described (7). In cases where the NEC contained zero raw reads of the organism, a value of “1” was inserted and adjusted to RPM to ensure a positive integer as the cutoff. Any sample without an organism meeting the positive threshold was further assessed by identifying the top five microorganisms with the highest unique kmer counts and was also assessed for genomic coverage by alignment. Any organism, excluding common nonpathogenic contaminants, in the top five with ≥2% genomic coverage was considered positive. To further assess the impact of the various methods on achieving a positive result, >10 times the positive cutoff value and >100 times the positive cutoff value were determined to compare the yields. Additionally, Lin's concordance correlation coefficient (rho_c) (14), based on no assumption of an underlying analysis of variance (ANOVA) model, was calculated for comparing each neat specimen versus its pelleted and supernatant versions. Results from all samples were compiled for calculating agreement by using rho_c, Bland and Altman limits, and unweighted Cohen's kappa coefficients in these settings. Analyses were performed using Stata v.14 (Stata Corp., TX, USA).
Host depletion.
For any sample (samples 3, 4, 5, and 10) for which sufficient volume was remaining (600 μl) at the completion of the method comparison study (as described above), a host depletion study was performed using the method applied to subsample L (method L; neat CSF, no BB, cDNA) and the method applied to subsample M (method M; neat CSF, BB, DNAamp). We evaluated the use of saponin (differential host cell lysis), followed by treatment with Turbo DNase (Invitrogen). Saponin (Sigma) was reconstituted in nuclease-free water to a 1% solution, which was then filtered through a 0.22-µm sterile syringe filter. Sixty microliters of saponin was added to 600 μl of CSF, vortexed for 10 s, and incubated at room temperature for 5 min. For DNase treatment, 66 μl DNase solution (60 μl 1× Turbo DNase buffer and 6 μl Turbo DNase) was added to the extracted total nucleic acid, mixed by inversion (20 to 30 times), and incubated at 37°C for 30 min. Internal controls were included in these studies: Nostoc DNA bacteriophage N-1 (ATCC 27893-B15) and MS2 RNA bacteriophage (ZeptoMetrix 0810066) were diluted 1:10 in nuclease-free water and spiked into both the NEC and the specimens to assess analytical sensitivity.
Preliminary LOD study.
A limit-of-detection (LOD) study was completed by spiking 104 to 101 CFU or copies/ml of Staphylococcus aureus, Haemophilus influenzae, Mycobacterium smegmatis, Aspergillus fumigatus (hyphae), Cryptococcus neoformans, cytomegalovirus (CMV), and coxsackievirus A9 into pooled negative CSF. The bacterial and fungal organisms were quantified by plate counts, whereas the viruses were prequantified by digital droplet PCR. The LOD was evaluated for the methods with and without host depletion. In addition, the impact of a single freeze-thaw cycle was also evaluated, as specimens are likely to be frozen prior to processing for batch testing rather than in real time.
Quality control.
A matched NEC was run with each processing method for the parallel CSF sample. The sample being run acted as a de facto positive control, as it was known to be a strong positive for host DNA and/or RNA. For QC reagents and consumables, we performed an aggregated QC run with each new lot in order to capture any variability within reagents. After a point-of-failure risk analysis, we decided that the following reagent groups required QC with each new lot: 5× MagMax pathogen RNA/DNA kit, REPLI-g cell WGA & WTA kit, Nextera XT library prep reagents, and MiSeq reagents. We used a whole-cell community standard (WCC) with matched DNA standard (Zymo Research, Irvine, CA), which provided documented concentrations of eight bacteria and two yeasts (Salmonella enterica, Pseudomonas aeruginosa, Lactobacillus fermentum, Bacillus subtilis, Listeria monocytogenes, Enterococcus faecalis, Staphylococcus aureus, Escherichia coli, Saccharomyces cerevisiae, and Cryptococcus neoformans). The WCC was diluted 1:4 in molecular biology-grade nuclease-free water and processed in the same volumes/conditions as if it were neat CSF. The QC run was comprised of DNA standard straight (S1) and amplified (S2), the WCC straight DNA (positive extraction control [PEC]), amplified DNA (positive template control [PTC]-DNA), and amplified RNA (PTC-RNA); included in this run were two negative template controls (cDNA-NTC and DNAamp-NTC) made up of nuclease-free water which underwent WTA/WGA. Positive results for the PEC were judged versus S1, while the two PTCs were compared to their respective NTCs, as described above. The WCC subsamples were compared to S1, S2, and NTCs in order to determine the robustness of the extraction (S1) and amplification (S2) portions of the assay. QC runs were compared to each other to judge the reproducibility of the NGS library prep and sequencing run.
RESULTS
mNGS results compared to expected results.
Table 1 summarizes the results that met the positive cutoff for mNGS compared to the expected standard-of-care results for each of the methods. DNASeq results without WGA (DNA method; subsamples H and K) were not included in Table 1, as very few samples met the ≥10 times the RPM of the NEC positive cutoff (2/8; see Table 1 footnotes); many were subsequently found to be positive by unique kmer analysis (see Data Set S8 in the supplemental material). The raw data for the DNA, DNAamp, and cDNA methods consisting of the top 50 to 100 organisms per run are summarized in Data Sets S2, S3, and S4, respectively. Each sample is discussed in further detail below.
TABLE 1.
Comparison of mNGS results to expected standard-of-care results
| Sample | Expected standard-of-care result | mNGS resultsa |
|||||
|---|---|---|---|---|---|---|---|
| Pellet |
Neat CSF |
Supernatant |
|||||
| Subsample I (no BB, cDNA) | Subsample J (BB, DNAamp)b | Subsample L (no BB, cDNA) | Subsample M (BB, DNAamp) | Subsample F (no BB, cDNA) | Subsample G (BB, DNAamp)c | ||
| 1 | Varicella-zoster virus | Human herpesvirus 3 (GC, 0.3%; UKM, 602), Raoultella ornithinolytica, Corynebacterium singulare | Human herpesvirus 3 (GC, 32.7%; UKM, 33,001) | Human herpesvirus 3 (GC, 1.9%; UKM, 2,319) | Human herpesvirus 3 (GC, 43.7%; UKM, 45,575) | Human herpesvirus 3 (GC, 4.0%; UKM, 4,503), Polaromonas naphthalenivorans | Human herpesvirus 3 (GC, 33.9%; UKM, 37,640), Cronobacter dublinensis |
| 2 | Cryptococcus neoformans | Cryptococcus neoformans (GC, 10.4%; UKM, 209,888), TTV | Cryptococcus neoformans (GC, 70.1%; UKM, 1,229,464), Cryptococcus gattii | Cryptococcus neoformans (GC, 3.2%; UKM, 66,648), TTV | Cryptococcus neoformans (GC, 62.3%; UKM, 1,125,775), Cryptococcus gattii | TTV | Cryptococcus neoformans (GC, 73.5%; UKM, 136,619), Cryptococcus gattii |
| 3 | Pseudomonas aeruginosa | Positived | Negative | Positived | Negative | Positived | Negative |
| 4 | Enterovirus | Negative | Negative | Positivee | Negative | Negative | Negative |
| 5 | Staphylococcus aureus | Staphylococcus aureus (GC, 98.4%; UKM, 2,182,976) | Staphylococcus aureus (GC, 98.3%; UKM, 2,284,188) | Staphylococcus aureus (GC, 42.4%; UKM, 883,874) | Staphylococcus aureus (GC, 96.4%; UKM, 2,214,990) | Staphylococcus aureus (GC, 17.6%; UKM, 426,194) | Staphylococcus aureus (GC, 96.4%; UKM, 2,241,949) |
| 6 | Pseudomonas aeruginosa | Pseudomonas aeruginosa (GC, 22.4%; UKM, 23,386) | Pseudomonas aeruginosa (GC, 7.4%; UKM, 516,222) | Pseudomonas aeruginosa (GC, 8.0%; UKM, 35,293) | Positivef | Pseudomonas aeruginosa (GC, 37.6%; UKM, 20,810), Staphylococcus epidermidis, Micrococcus luteus | Pseudomonas aeruginosa (GC, 13.7%; UKM, 926,777) |
| 7 | Cryptococcus neoformans | Negative | Negative | Negative | Negative | Dermacoccus nishinomiyaensis, Deinococcus geothermalis | Negative |
| 8 | Negative | Negative | Negative | Negative | Negative | Negative | Negative |
| 9 | Negative | Negative | Negative | Negative | Negative | Negative | Negative |
| 10 | JC polyomavirus | JC polyomavirus (GC, 87.2%; UKM, 4,059) | JC polyomavirus (GC, 96.8%; UKM, 12,006) | JC polyomavirus (GC, 96.7%; UKM, 5,063) | JC polyomavirus (GC, 96.8%; UKM, 11,766) | JC polyomavirus (GC, 92.7%; UKM, 4,471) | JC polyomavirus (GC, 96.8%; UKM, 12,041) |
BB, samples were processed with a bead-beating step on the FastPrep instrument; DNAamp, a whole-genome amplification step was performed prior to DNASeq; TTV, torque teno virus; UKM, unique kmer; GC, genomic coverage; human herpesvirus 3, varicella-zoster virus. Entries in bold are the expected result that met the initial positive cutoff of ≥10 times the RPM of the negative extraction control (NEC).
Subsamples K (neat, BB, DNA with no amplification) for samples 2 (Cryptococcus neoformans/C. gattii) and 5 (S. aureus) were the only two samples that met the ≥10 times the RPM of the NEC positivity threshold. However, both subsamples K and H for samples 1, 2, 3, 5, 6, and 10 were positive by secondary analysis using the unique kmer approach.
Subsample H (supernatant, BB, DNA with no amplification), the only positive, was sample 5 for S. aureus by the ≥10 times the RPM of the NEC positivity threshold.
The sample did not initially meet the positive cutoff. However, following the unique kmer analysis approach, the clinical sample was interpreted as positive for P. aeruginosa. Unique kmer and genome coverage ranged by subsamples from 1,039 to 27,333 and 0.01 to 7.4%, respectively. See Fig. 2 for further details.
Following the method comparison study, host depletion studies were performed. The sample processed with saponin/DNase yielded a positive result, with 43,905 enterovirus reads covering 14.9% of the genome.
The sample did not meet the initial positive threshold but was found to be positive using the UKM (top hit) and genomic coverage (8%) approach.
(i) Sample 1: VZV (human herpesvirus 3).
Sample 1 was positive by a varicella-zoster virus (VZV) real-time PCR assay with a cycle threshold (CT) value of 34. All mNGS methods were positive for VZV. A minimum of 8 reads per million (RPM) was achieved with the pelleted CSF without bead beating by the cDNA method and up to 406 RPM using neat CSF with bead beating by the DNAamp method. The methods without the bead-beating step resulted in the lowest RPM yields compared to those with a bead-beating step.
(ii) Sample 2: C. neoformans (high-level positive).
Sample 2 was positive for heavy yeast and light polymorphonuclear leukocytes (PMNs) on Gram stain. Both the bacterial and fungal cultures demonstrated moderate growth of C. neoformans. All mNGS methods detected C. neoformans reads, but the cDNA method from the supernatant processed without bead beating did not reach the positive cutoff. Although C. neoformans/C. gattii were among the top five organisms with the highest unique kmer count, the genomic coverage was only 0.5 to 0.8% and therefore did not meet the secondary positivity assessment for subsample F. Of those that were considered positive, the specimens that had the bead-beating step performed reached RPM yields that were 4 to 35 times greater than those that were not processed by bead beating. The method achieving the highest RPM of 152,366 was the pelleted CSF with bead beating by DNAamp. In addition to C. neoformans, many of the samples had reads assigned above the positive cutoff aligning to C. gattii, resulting in an identification of C. neoformans/C. gattii. Torque teno virus (TTV) was also identified among all samples that did not have a bead-beating step introduced.
(iii) Sample 3: P. aeruginosa (low-level positive).
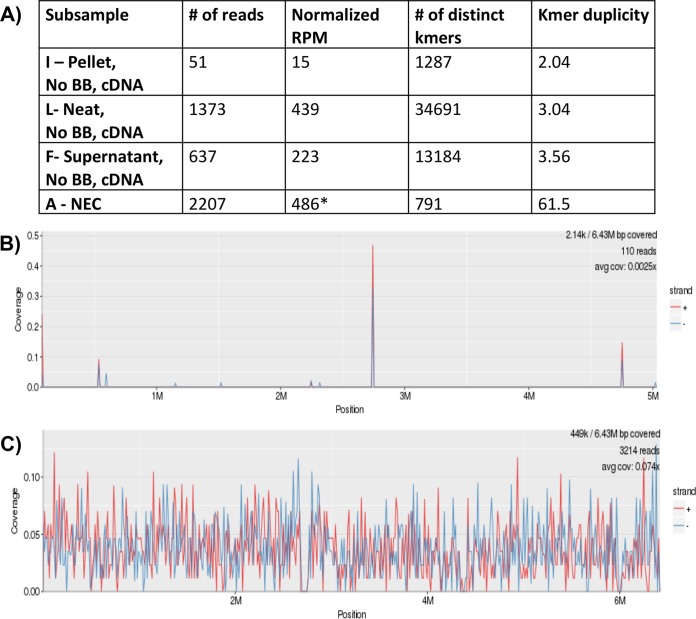
Sample 3 had very light PMNs on Gram stain with no organisms seen. Bacterial culture revealed light growth of P. aeruginosa. P. aeruginosa reads were detected by all mNGS methods, including samples that were processed with host depletion. However, due to a high level of P. aeruginosa found in the NECs, none of the samples met the positive cutoff. The pelleted CSF sample processed with bead beating by the DNA method achieved the highest RPM of 997. To further analyze the sample, KrakenHLL was run on the cDNA subsamples (F, I, and L) to see how many of the kmers were unique among both the NEC and the subsamples (Fig. 2A). The kmer duplicity was highest among the NEC at 61.5, compared to a duplicity rate of 2 to 3.5 among the CSF subsamples. Furthermore, when the reads from the NEC were aligned with the P. aeruginosa genome, only a few areas of the genome were covered, in comparison to 7.4% of the genome being covered by the reads in the neat CSF subsample L (Fig. 2B and C). Based on these results, these subsamples were considered positive with the inclusion of the KrakenHLL and genomic coverage analysis.
FIG 2.
Unique kmer and alignment analysis methods for CSF sample 3 positive for Pseudomonas aeruginosa. (A) Comparison of the numbers of unique kmers and kmer duplicity assigned to the P. aeruginosa reads among sample 3 cDNA subsamples (F, I, and L) with those of the negative extraction control (subsample A-NEC) by KrakenHLL. (B and C) Demonstration of the reads aligned to the P. aeruginosa genome found in the NEC (B) compared to those in the neat CSF sample without bead beating (BB) using cDNA methods (subsample L with 7.4% genome coverage) (C). An asterisk indicates a positive cutoff for these samples of 4,855 (10 times the RPM of the NEC).
(iv) Sample 4: enterovirus.
Sample 4 was a low-level positive by a reverse transcriptase real-time PCR (RT-PCR) assay with a CT value of 35.6. All subsamples were found to be negative, and there were no reads assigned to enterovirus. To confirm sample positivity, the RT-PCR was repeated on the same samples used to perform mNGS, and sample 4 repeated as a low-level positive. Sequencing depth was a concern in cases where each subsample produced from 2,458,734 up to 6,040,108 reads due to the multiplexing of samples on the same flow cell (e.g., predominately less than the 5 million reads suggested in the literature) (2). An additional aliquot of sample was obtained and processed in triplicate by the cDNA method. The replicates were pooled (to reach sufficient amounts of cDNA for library preparation) and run on a flow cell with a matching NEC; the total number of reads were 19,516,247, but again, no enterovirus reads were identified. However, sufficient volume was remaining to perform HD studies with saponin/DNase on the neat CSF specimen. This resulted in the detection of enterovirus (43,905 reads) by the cDNA method (method L) with 14.9% genomic coverage of echovirus E7.
(v) Sample 5: S. aureus.
Sample 5 had moderate Gram-positive cocci on Gram stain with moderate growth of S. aureus by culture. All methods detected S. aureus reads above the positive cutoff for the samples. The sample achieving the highest RPM was the pelleted CSF processed without bead beating by cDNA. Among the DNAamp samples, the pelleted sample with bead beating achieved the highest yields.
Sufficient volume was remaining to perform HD studies with saponin/DNase on the neat CSF specimen. HD had no effect on detection of S. aureus by the DNAamp method with BB (6.72% reads with HD versus 7.01% reads without HD), but HD resulted in no detection by the cDNA method (no BB), as the percent reads decreased from 11.77% without HD (positive result) to 0.009% with HD (does not meet the positive threshold).
(vi) Sample 6: P. aeruginosa (high-level positive).
Sample 6 had very light PMNs and Gram-negative bacilli on Gram stain with moderate growth of P. aeruginosa in culture. All methods had detectable reads assigned to P. aeruginosa. However, a higher number of reads in the NEC for the DNAamp methods made it more difficult to detect true positives, although all samples were considered positive just above the threshold, except the neat CSF processed with bead beating. The neat CSF processed with BB had P. aeruginosa listed as the top hit among unique kmer counts, with 8% genomic coverage further qualifying the sample as positive by secondary analysis. All cDNA subsamples were positive well above the threshold since there were no reads aligned to P. aeruginosa in the NEC for the cDNA method. The neat CSF with no bead beating by cDNA had the highest yield.
(vii) Sample 7: C. neoformans (low-level positive).
Sample 7 demonstrated very light yeast and PMNs on Gram stain with no growth in bacterial or fungal cultures. The sample was positive by the cryptococcal antigen with a titer of 1:160. No reads from any of the methods aligned with C. neoformans. To see if additional sequencing depth would allow detection, the subsample M (neat CSF, bead beating, DNAamp) was run on a dedicated flow cell, achieving 24,669,500 reads, again with no C. neoformans reads identified.
(viii) Samples 8 and 9: negative for infectious etiologies.
Samples 8 and 9 were CSF specimens found to be negative by standard-of-care testing. Both samples and all methods were negative for a pathogen based on the positive cutoff and unique kmer analysis.
(ix) Sample 10: JCV.
Sample 10 was positive by real-time PCR for JC polyomavirus (JCV) with a CT of 25.1. All methods were positive for JCV for the method comparison study. The DNAamp methods achieved higher yields than the cDNA methods. The sample with the highest RPM was the supernatant subsample that did not include a bead-beating step.
Sufficient volume was remaining to perform HD studies with saponin/DNase on the neat CSF specimen. Detection of JCV with HD resulted in 62 times (0.99% reads versus 62.3% reads) greater detection by DNAamp with BB but resulted in no detection by cDNA (no BB), as the percent reads decreased from 0.3% without HD to 0.02% with HD.
Common contaminants.
We assessed the 25 most common contaminants identified among our NECs and CSF specimens processed with and without host depletion (Data Set S5). We found that certain contaminants were present in both DNAamp and cDNA samples, such as Thermus scotoductus, Burkholderia fungorum, Thermus thermophilus, Cutibacterium (Propionibacterium) acnes, and Pseudomonas fluorescens, among others. We also identified contaminants that were unique to either the DNAamp or cDNA method and are likely due to unique reagents/steps in each of the methods. For the DNAamp samples, we found Pseudomonas spp., Acidithiobacillus caldus, Acidovorax spp., and Burkholderia spp., which are likely introduced during the bead-beating or subtraction steps. For the cDNA samples, the most common contaminants include Escherichia coli, Bordetella hinzii, Leptothrix cholodnii, Comamonas testosteroni, Verminephrobacter eiseniae, and Pseudomonas spp., which are likely introduced using the REPLI-g protocol to convert RNA to cDNA. The cDNA samples exhibited more contaminating reads reaching the positive cutoff (Table 1). Often, these reads were not found by the DNAamp methods, suggesting amplification bias in the WTA method.
Sample processing.
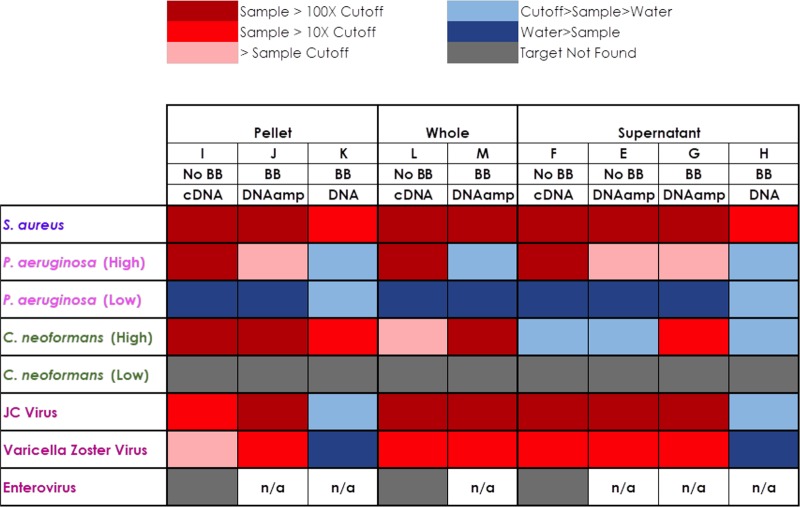
We compared sample processing methods of the CSF specimens by comparing neat, pelleted, and the supernatant processed samples (Fig. 3; Data Sets S2, S3, and S4). Not surprisingly, we found that the results differed by pathogen type and pathogen burden, as demonstrated by differences in punctual estimates of the agreement (rho_c) between the sample processing types (Data Set S7). For the positive bacterial samples, S. aureus was easily detected in all samples (>100 times the positive cutoff), whereas the low-level P. aeruginosa sample was best detected in neat samples, followed by supernatant and then pelleted samples. The C. neoformans sample was best detected in the pelleted sample, followed by neat and supernatant samples, whereas the viruses (JCV and VZV) were best detected in neat and supernatant samples (Fig. 3).
FIG 3.
Heat map summarizing the results comparing processing, bead-beading, and sequencing methods for the expected positive samples. BB, the samples were processed with a bead-beating step on the FastPrep instrument; DNAamp, a whole-genome amplification step was performed prior to DNASeq; n/a, not applicable.
Sample pretreatment: bead beating.
We compared processing of positive-control material (Data Set S6) and positive CSF specimens to determine the effect of bead beating on the different pathogen groups. We found that bead beating produced a higher number of reads for Gram-positive bacteria, enveloped viruses, and yeasts, whereas it decreased the number of reads of nonenveloped viruses and Gram-negative organisms (Fig. 3). In general, bead beating did not pair well with RNASeq analysis (data not presented) and was not pursued in this study.
Whole-genome amplification for DNASeq samples.
In the paired samples for which WGA was applied prior to DNASeq, enrichment of the pathogen reads ranged from 1 to 9 times enrichment. For sample 3, in contrast, the DNASeq sample without amplification had up to 10 times more pathogen reads than the samples that underwent WGA. Furthermore, overall, the DNASeq samples without amplification did have the representative pathogen reads present but were not able to meet the positive threshold of ≥10 times the RPM of the NEC (n = 8), except for samples 2 (C. neoformans) and 5 (S. aureus), which were high-burden samples.
Statistical analysis.
Overall, we found that the punctual estimates of the agreement (rho_c) with the RPM in the neat processed samples were higher for the pelleted version than for the supernatant for the DNAamp with bead beating and cDNA methods with some exceptions. For DNAamp with bead beating, punctual rho_C values were lower in pelleted samples for samples 5 (S. aureus), 9 (negative), and 10 (JCV) and, for cDNA without bead beating, for samples 1 (VZV), 3 (P. aeruginosa), 6 (P. aruginosa), and 10 (JCV). In general, total rho_c values were lower in the cDNA method but consistent with those found in DNAamp with bead beating (favoring more agreement between neat and pelleted samples) (Data Set S7). These differences are likely due to differences in the processing methods favoring one organism type over the other.
When considering positive cutoffs for the DNAamp method with bead beating, the neat sample exhibited an agreement of 0.922 (0.770 to 1.000) with the pelleted sample and 0.856 (95% confidence interval [CI95%], 0.658 to 1.000) with the supernatant. For the case of cDNA with no bead beating, kappa coefficients were moderate for both pelleted (0.488; CI95, 0.257 to 0.718) and supernatant (0.431; CI95%, 0.146 to 0.715). These differences are due to more contaminating reads meeting the positive cutoff for the cDNA methods than for the DNAamp methods, as highlighted in Table 1.
Reproducibility.
To assess the reproducibility of results, we used three positive-control runs that were performed during this study period (Data Set S6). We found that the PECs (whole-cell community standard spiked into nuclease-free water, extracted in parallel for DNASeq and RNASeq) for both DNAamp and cDNA results yielded reads with a standard deviation of 0 to 5% of reads per run for each organism.
Preliminary LOD studies.
A preliminary limit-of-detection (LOD) study was completed to assess the analytical sensitivity of methods applied to subsamples L and M with and without host depletion and to assess the impact of a freeze-thaw cycle (Table 2). All organisms were detected at 1 × 101, except for the viruses, by the DNAamp (M) method without host depletion. More variability was observed for the cDNA method (L) without host depletion, where A. fumigatus, H. influenzae, and M. smegmatis were detected at 1 × 101, S. aureus was detected at 1 × 102, and C. neoformans and CMV were detected at 1 × 103. Coxsackievirus A9 was not detected in any of the dilutions by either the DNAamp or the cDNA method (LOD > 104). Not surprisingly, host depletion significantly decreased host reads for both the DNA and cDNA methods. However, host depletion did detrimentally affect the recovery of S. aureus and C. neoformans but increased the recovery of H. influenzae. Overall, there was no obvious impact of a freeze-thaw cycle on recovery of the microorganisms.
TABLE 2.
Impact of host depletion and freeze-thaw on limit-of-detection studiesa
| Method and organism | % reads at indicated LODb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 104 |
103 |
102 |
101 |
|||||||||
| No HD, fresh | HD, fresh | No HD, fresh | HD, fresh | No HD, frozen | HD, frozen | No HD, fresh | HD, fresh | No HD, frozen | HD, frozen | No HD, fresh | HD, fresh | |
| DNAamp | ||||||||||||
| Host | 40.79 | 0.9034 | 82.53 | 4.94 | 76.88 | 8.57 | 96.21 | 58.05 | 93.61 | 20.00 | 97.48 | 58.77 |
| S. aureus | 0.2 | 0.01 | 0.09 | 0.005 | 0.05 | 0.01 | 0.0027 | ND | 0.007 | 0.008 | 0.0001 | ND |
| H. influenzae | 35.4 | 93.8 | 11.5 | 79.3 | 14.3 | 67.8 | 0.10 | 1.78 | 1.76 | 28.47 | 0.0073 | 0.28 |
| M. smegmatis | 0.02 | 0.006 | 0.003 | 0.002 | 0.006 | 0.0008 | 0.0019 | 0.0018 | 0.0007 | 0.0015 | 0.0001 | 0.0003 |
| A. fumigatus | 20.7 | 1.9 | 4.2 | 0.8 | 6.2 | 7.3 | 1.299 | 0.2694 | 1.1 | 0.07 | 0.0965 | 0.45 |
| C. neoformans | 0.08 | 0.0001 | 0.02 | 0.0002 | 0.002 | ND | 0.0048 | 0.0003 | 0.002 | ND | 0.0048 | ND |
| CMV | 0.007 | 0.002 | 0.0009 | 0.0003 | 0.002 | 0.003 | ND | ND | ND | ND | ND | ND |
| Coxsackievirus A9 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| cDNA | ||||||||||||
| Host | 41.2 | 2.1 | 72.4 | 10.3 | 46.9 | 1.1 | 1.43 | 0.0236 | 31.0 | 0.5 | 32.7 | 0.12 |
| S. aureus | 0.007 | 0.003 | 0.001 | 0.1 | 0.0001 | ND | ND | ND | 0.0002 | ND | ND | ND |
| H. influenzae | 32.1 | 60.8 | 11.0 | 49.6 | 4.5 | 13.5 | 0.67 | 0.35 | 0.6 | 1.3 | 0.0589 | 0.0294 |
| M. smegmatis | 1.5 | 3.3 | 0.003 | 0.02 | 0.008 | 0.005 | 0.0144 | 0.0153 | 0.007 | ND | 0.0001 | 0.0001 |
| A. fumigatus | 11.6 | 12.9 | 3.7 | 0.5 | 1.6 | 0.04 | 0.8522 | 0.0386 | 0.06 | 0.01 | 0.0392 | 0.0027 |
| C. neoformans | ND | 0.0002 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CMV | 0.0688 | 0.0050 | ND | ND | ND | ND | ND | ND | 0.0002 | ND | ND | ND |
| Coxsackievirus A9 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
ND, not detected; HD, host depletion; CMV, cytomegalovirus; fresh, processed without a freeze-thaw cycle.
LOD are given in CFU or copies/ml.
DISCUSSION
We performed a proof-of-concept study to optimize and evaluate different processing, extraction, amplification, and sequencing methods to determine a workflow to further validate mNGS of CSF specimens for clinical care. We found that no single method was perfect for all pathogen types but rather that a combination of techniques is required to achieve an agnostic approach. Development of mNGS requires balancing multiple factors to achieve maximal yields at each step of optimization without detrimentally affecting the recovery of certain microbial groups.
When comparing specimen processing techniques (neat versus pellet versus supernatant) we demonstrated that the results differed by pathogen type and pathogen burden. Overall, the neat CSF was appropriate for the detection of all pathogen types. However, similar to organism-specific processing that occurs in the clinical microbiology laboratory for culture or molecular biology-based methods, the pelleted specimen was more appropriate for bacteria and C. neoformans whereas the supernatant was appropriate for viruses. As anticipated, the higher the burden, the more likely the pathogen was detected in all specimen processing types. However, scenarios where the organism is of high burden and visualized on primary stains (i.e., Gram stain) are cases where mNGS is likely not to be cost-effective in comparison to SoC, unless the patient has been on treatment previously and the organism is not viable for culture or if it is an uncultivatable pathogen.
The method that had the largest impact on detection of the various pathogens was the use of a bead-beating step. We found that bead beating increased the yield of reads for detecting Gram-positive bacteria, enveloped viruses, and yeasts but detrimentally affected the recovery of nonenveloped viruses and Gram-negative organisms. The best examples of this were the C. neoformans-positive sample where we achieved up to 35 times greater yields with a bead-beating step, whereas only the P. aeruginosa subsamples (sample 6) without bead beating were able to meet the positive cutoff compared to those that underwent bead beating. Our positive-control runs further confirmed these trends.
We demonstrated that WGA prior to DNASeq runs resulted in no to minimal enrichment (1 to 9 times) of pathogen reads compared to samples without amplification in the absence of host depletion. This is not surprising, since an “unbiased” amplification of genomic DNA occurs through multiple displacement amplification technology, which theoretically should amplify all nucleic acid in the sample, host, or pathogen. WGA coupled with upfront host depletion would be ideal to achieve pathogen enrichment by first depleting host nucleic acid. A study found that WGA was required with synovial fluids to obtain sufficient amounts of DNA for library preparation when paired with host depletion steps (15). Similarly, a study applying mNGS to bone and joint infections found that without host depletion or WGA, only 23% of samples had sufficient DNA concentrations to perform DNASeq mNGS (16). Furthermore, we found that our NEC sequenced by DNASeq without upfront WGA resulted in random high-level sequencing of contaminants and sample carryover from multiplexing, making it difficult for pathogen reads to meet the established positive cutoff. Thus, DNASeq without WGA was no longer pursued in our method development. The one advantage to DNASeq without upfront WGA was that sample results were “cleaner,” with fewer contaminants in the sample reads due to fewer steps and reagents introduced in the methodology. Furthermore, a secondary analysis using unique kmer counts for the DNASeq approach did result in increased detections (6 of 8 positive results) when the NEC was not considered part of the assessment for positivity.
The use of both the DNASeq and RNASeq methods was required to detect all pathogen types, as RNA viruses will be picked up only by the RNASeq method, which is theoretically capable of detecting pathogens that are transcriptionally active (17). That said, the majority (∼90%) of RNA isolated from these specimens is rRNA, which is helpful for the identification of microorganisms, whereas very little mRNA (∼1 to 2%) that could contribute to transcriptomics studies is present (18). Furthermore, the RNASeq method with WTA was more prone to high-level contaminants reaching the positive threshold in our study. This suggests that there is more bias introduced in the amplification step with the WTA method than with the WGA method. Nonetheless, the detection of the same pathogen(s) using both DNA and RNA methods provides increased confidence in the result. We have further added this to criteria that we consider when establishing the significance of our mNGS results. Similarly, Langelier et al. used this technique when assessing positivity with bronchoalveolar lavage (BAL) samples from immunocompromised hosts (17).
Overall, we were able to correctly evaluate 9 of 10 samples by mNGS compared to standard-of-care results. This required the inclusion of an NEC with each method type to determine the common background/contaminating reads introduced during processing to allow accurate sample assessment that met the positive threshold in the clinical specimen. It has been well established that reagents utilized for NGS methodologies contain contaminating nucleic acid that can interfere with interpretation of results, especially among low-microbial-biomass specimens, such as CSF (19–21). It is imperative that when performing mNGS for clinical use an NEC be included for each method or the results will be extremely difficult to interpret in the absence of an obvious pathogen and can be very misleading, as even negative samples can have high numbers of microbial organism reads (e.g., samples 8 and 9). Thus, a threshold to call the samples positive is required in relation to the NEC, such as the ≥10 times the RPM of the NEC utilized in this study and others (2, 7, 17, 22). We also demonstrated that the use of a unique kmer approach and an alignment approach can be helpful to decipher when organisms that can be contaminants are also queried as pathogens, such as in the P. aeruginosa case (sample 3). We proved that the reads in the water were duplicate reads localized to a few areas of the genome, whereas the clinical sample had a low duplicity rate and covered the genome. These results suggest that the NEC reads were a result of amplification of a few contaminant reads in the reagents, in comparison to the subsamples that represent a true positive where the genome of the organism is detected. Moreover, it is also important to understand the common contaminants identified by mNGS in each laboratory. This is especially helpful when these common contaminants appear to be significant based on a positive threshold and suggest that further studies are required (kmer or alignment approach).
In sample 2, an incidental finding of TTV among the C. neoformans-positive subsamples occurred when bead beating was not performed. TTV is a nonenveloped virus, so this result is consistent with the bead-beating step diminishing the detection of nonenveloped viruses. TTV is thought to encompass part of the normal human virome (23), where increased expression has been reported in immunocompromised patients (i.e., transplant recipients), and has been suggested as a biomarker to detect immunosuppression (24). The patient from which sample 2 was collected was HIV positive, which is consistent with the finding of both C. neoformans and TTV in the sample by mNGS studies. Sample 2 also highlights that certain very closely related pathogens (near neighbors) are hard to differentiate taxonomically based on a kmer approach where the DNAamp samples tended to have both C. neoformans and C. gattii listed as positive pathogens meeting the positive threshold and an identification of C. neoformans/C. gattii was provided.
In two cases, where standard-of-care results were positive, we were initially unable to confirm the result by mNGS. For the enterovirus case, we believe that the initial lack of positive results is attributable to the limit of detection (LOD) of a highly sensitive targeted RT-PCR in comparison to mNGS. A preliminary LOD for detection of enterovirus by our mNGS method by spike-in studies in negative CSF was determined to be >104 copies/ml. That LOD is consistent with that of another study in which more than 10,000 copies/ml of HIV and hepatitis C virus (HCV), as calculated by quantitative PCR (qPCR), were uniformly detected by mNGS, whereas sensitivity was much less with lower RNA abundance (25). Additionally, the specimen was handled suboptimally for detection of RNA viruses and ideally needed to be frozen as soon as possible for mNGS analysis, as detection rapidly decreases if the sample sits at refrigerator temperatures (data not shown). Finally, the cell count for the specimen was 217 cells/μl, which may have contributed to decreased analytical sensitivity, with 99% of reads of human origin. Ultimately, a host depletion step was required to increase the analytical sensitivity and enabled detection of enterovirus in this sample. Although host depletion allowed for recovery of the enterovirus, we did find that host depletion methods introduced another source of bias in our results.
The one false-negative mNGS case was a sample in which C. neoformans antigen was detected and an organism was evident on Gram stain but not cultured. This was a follow-up CSF sample for which previous cultures also grew C. neoformans. In the follow-up CSF, C. neoformans did not grow, likely consistent with the patient being treated. A similar observation was made during the multicenter evaluation of the BioFire FilmArray meningitis/encephalitis panel, where multiple samples were negative by this panel but were positive by antigen testing, which was attributed to antigen persistence after receipt of therapy rather than the presence of a live organism (26). We were unable to detect the pathogen by mNGS even with increased sequencing depth (>24,000,000 reads). This case highlights the importance of standard-of-care testing and that mNGS should be used as an adjunct rather than a replacement.
Based on the results from this study, we have decided to move forward to further assess a DNAamp method with upfront bead beating to select for more-difficult-to-lyse organisms and a cDNA method with host depletion (saponin/DNase) and WTA without a bead-beating step. This approach allows us to balance the advantages of bead beating and host depletion with broad-range detection of pathogens. We have also further adapted our assessment of mNGS results. We now include the following in our systematic analyses: a definition of a sample being positive by mNGS when the RPM are ≥10 times the NEC; the appearance of the pathogen reads in both DNA and RNA methods (except RNA viruses); and when no organisms meet the initial positivity criteria, further use of a unique kmer (top five organisms) and an alignment approach (≥2% genomic coverage) to assess positivity of samples. We found that higher unique kmer counts were congruent with higher genomic coverage. Also, genomic coverage can vary significantly based on the strain used for the alignment.
Although our study contributes to the literature, we have identified several limitations. First, we had a small sample size, making estimates of sensitivity imprecise. Luckily, we have performed some subsequent studies, and the observations listed herein have held true (data not shown). To maximize the resources assigned to this study, we often did not meet a read depth of a minimum of 5,000,000 reads passing filter per subsample, which had been previously been established by other studies (2). Despite this, we were still able to detect the pathogen in most instances, and in those where a pathogen was not detected, expanding the sequencing depth to >19,000,000 reads for certain subsamples still did not yield the pathogen. During the initial method comparison study, we still had not optimized our host depletion and internal control methods, which were included in a subsequent analysis of a subset of samples in this study. As the field is rapidly evolving, we are still refining our methods, including the host depletion of our DNAamp method. Lastly, as samples had been previously frozen prior to processing, lysis might have occurred during the freeze-thaw cycle and impacted some of our findings.
In conclusion, we found that optimizing mNGS requires integrating a combination of techniques and balancing several priorities to achieve an agnostic approach that did not always reproduce results obtained by routine microbiological methods. The neat CSF specimen was found to be the most appropriate specimen processing technique for all pathogen types. While bead beating and host depletion introduced bias, we found they were required for the detection of certain organism groups and decided to include bead beading as part of the DNASeq method and host depletion as part of the RNASeq method. Whole-genome amplification prior to DNASeq was beneficial for defining pathogens at the positive threshold, and a combined DNA and RNA approach yielded results with a higher confidence when the pathogen was detected by both methods. Currently, mNGS should be considered an adjunct to standard-of-care testing.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded in part by the 2017 Johns Hopkins Discovery Award.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00472-18.
REFERENCES
- 1.Simner PJ, Miller S, Carroll KC. 2018. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis 66:778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G, Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology, Microbiology Resource Committee of the College of American Pathologists. 2017. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med 141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 3.Salzberg SL, Breitwieser FP, Kumar A, Hao H, Burger P, Rodriguez FJ, Lim M, Quinones-Hinojosa A, Gallia GL, Tornheim JA, Melia MT, Sears CL, Pardo CA. 2016. Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system. Neurol Neuroimmunol Neuroinflamm 3:e251. doi: 10.1212/NXI.0000000000000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greninger AL, Messacar K, Dunnebacke T, Naccache SN, Federman S, Bouquet J, Mirsky D, Nomura Y, Yagi S, Glaser C, Vollmer M, Press CA, Kleinschmidt-DeMasters BK, Dominguez SR, Chiu CY. 2015. Clinical metagenomic identification of Balamuthia mandrillaris encephalitis and assembly of the draft genome: the continuing case for reference genome sequencing. Genome Med 7:113. doi: 10.1186/s13073-015-0235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann B, Tappe D, Hoper D, Herden C, Boldt A, Mawrin C, Niederstrasser O, Muller T, Jenckel M, van der Grinten E, Lutter C, Abendroth B, Teifke JP, Cadar D, Schmidt-Chanasit J, Ulrich RG, Beer M. 2015. A variegated squirrel bornavirus associated with fatal human encephalitis. N Engl J Med 373:154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- 6.Mai NTH, Phu NH, Nhu LNT, Hong NTT, Hanh NHH, Nguyet LA, Phuong TM, McBride A, Ha DQ, Nghia HDT, Chau NVV, Thwaites G, Tan LV. 2017. Central nervous system infection diagnosis by next-generation sequencing: a glimpse into the future? Open Forum Infect Dis 4:ofx046. doi: 10.1093/ofid/ofx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mongkolrattanothai K, Naccache SN, Bender JM, Samayoa E, Pham E, Yu G, Dien Bard J, Miller S, Aldrovandi G, Chiu CY. 2017. Neurobrucellosis: unexpected answer from metagenomic next-generation sequencing. J Pediatric Infect Dis Soc 6:393–398. doi: 10.1093/jpids/piw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naccache SN, Peggs KS, Mattes FM, Phadke R, Garson JA, Grant P, Samayoa E, Federman S, Miller S, Lunn MP, Gant V, Chiu CY. 2015. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, Sokolic R, Garabedian E, Candotti F, Buckley RH, Reed KD, Meyer TL, Seroogy CM, Galloway R, Henderson SL, Gern JE, DeRisi JL, Chiu CY. 2014. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson MR, Suan D, Duggins A, Schubert RD, Khan LM, Sample HA, Zorn KC, Rodrigues Hoffman A, Blick A, Shingde M, DeRisi JL. 2017. A novel cause of chronic viral meningoencephalitis: Cache Valley virus. Ann Neurol 82:105–114. doi: 10.1002/ana.24982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitwieser FP, Salzberg SL. 2016. Pavian: interactive analysis of metagenomics data for microbiomics and pathogen identification. bioRxiv doi: 10.1101/084715. [DOI] [PMC free article] [PubMed]
- 13.Breitwieser FP, Salzberg SL. 2017. KrakenHLL: confident and fast metagenomics classification using unique k-mer counts. bioRxiv doi: 10.1101/262956. [DOI] [PMC free article] [PubMed]
- 14.Lin LI. 1989. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45:255–268. doi: 10.2307/2532051. [DOI] [PubMed] [Google Scholar]
- 15.Thoendel M, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, Hanssen AD, Abdel MP, Patel R. 2016. Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. J Microbiol Methods 127:141–145. doi: 10.1016/j.mimet.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruppe E, Lazarevic V, Girard M, Mouton W, Ferry T, Laurent F, Schrenzel J. 2017. Clinical metagenomics of bone and joint infections: a proof of concept study. Sci Rep 7:7718. doi: 10.1038/s41598-017-07546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langelier C, Zinter MS, Kalatar K, Yanik GA, Christenson S, Odonovan B, White C, Wilson M, Sapru A, Dvorak CC, Miller S, Chiu CY, DeRisi JL. 2017. Metagenomic next-generation sequencing detects pulmonary pathogens in hematopoietic cellular transplant patients with acute respiratory illnesses. bioRxiv doi: 10.1101/102798. [DOI] [PMC free article] [PubMed]
- 18.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczesniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol 17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurence M, Hatzis C, Brash DE. 2014. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS One 9:e97876. doi: 10.1371/journal.pone.0097876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naccache SN, Greninger AL, Lee D, Coffey LL, Phan T, Rein-Weston A, Aronsohn A, Hackett J, Delwart EL, Chiu CY. 2013. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol 87:11966–11977. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murkey JA, Chew KW, Carlson M, Shannon CL, Sirohi D, Sample HA, Wilson MR, Vespa P, Humphries RM, Miller S, Klausner JD, Chiu CY. 2017. Hepatitis E virus-associated meningoencephalitis in a lung transplant recipient diagnosed by clinical metagenomic sequencing. Open Forum Infect Dis 4:ofx121. doi: 10.1093/ofid/ofx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernardin F, Operskalski E, Busch M, Delwart E. 2010. Transfusion transmission of highly prevalent commensal human viruses. Transfusion 50:2474–2483. doi: 10.1111/j.1537-2995.2010.02699.x. [DOI] [PubMed] [Google Scholar]
- 24.Beland K, Dore-Nguyen M, Gagne MJ, Patey N, Brassard J, Alvarez F, Halac U. 2014. Torque Teno virus load as a biomarker of immunosuppression? New hopes and insights J Infect Dis 210:668–670. doi: 10.1093/infdis/jiu210. [DOI] [PubMed] [Google Scholar]
- 25.Kandathil AJ, Breitwieser FP, Sachithanandham J, Robinson M, Mehta SH, Timp W, Salzberg SL, Thomas DL, Balagopal A. 2017. Presence of human hepegivirus-1 in a cohort of people who inject drugs. Ann Intern Med 167:1–7. doi: 10.7326/M17-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leber AL, Everhart K, Balada-Llasat JM, Cullison J, Daly J, Holt S, Lephart P, Salimnia H, Schreckenberger PC, DesJarlais S, Reed SL, Chapin KC, LeBlanc L, Johnson JK, Soliven NL, Carroll KC, Miller JA, Dien Bard J, Mestas J, Bankowski M, Enomoto T, Hemmert AC, Bourzac KM. 2016. Multicenter evaluation of BioFire FilmArray meningitis/encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol 54:2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.