Escherichia coli is the most common cause of human and canine urinary tract infection (UTI). Clonal groups, often with high levels of antimicrobial resistance, are a major component of the E. coli population that causes human UTI.
KEYWORDS: Escherichia coli, canine, molecular epidemiology, urinary tract infection, veterinary microbiology
ABSTRACT
Escherichia coli is the most common cause of human and canine urinary tract infection (UTI). Clonal groups, often with high levels of antimicrobial resistance, are a major component of the E. coli population that causes human UTI. While little is known about the population structure of E. coli that causes UTI in dogs, there is evidence that dogs and humans can share fecal strains of E. coli and that human-associated strains can cause disease in dogs. In order to better characterize the E. coli strains that cause canine UTI, we analyzed 295 E. coli isolates obtained from canine urine samples from five veterinary diagnostic laboratories and analyzed their multilocus sequence types, phenotypic and genotypic antimicrobial resistance profiles, and virulence-associated gene repertoires. Sequence type 372 (ST372), an infrequent human pathogen, was the predominant sequence type in dogs at all locations. Extended-spectrum β-lactamase-producing isolates with blaCTX-M genes were uncommon in canine isolates but when present were often associated with sequence types that have been described in human infections. This provides support for occasional cross-host-species sharing of strains that cause extraintestinal disease and highlights the importance of understanding the role of companion animals in the overall transmission patterns of extraintestinal pathogenic E. coli.
INTRODUCTION
Escherichia coli is the bacterial species most commonly isolated from human urinary tract infections (UTIs). UTIs are the third most common infection experienced by people (after respiratory and gastrointestinal infections), and as a result, a UTI is a very frequent indication for the prescription of antimicrobial drugs (1, 2). The antimicrobial resistance of extraintestinal pathogenic E. coli (ExPEC) isolates associated with human UTI has increased dramatically since the early 2000s, in large part due to the emergence of the dominant clonal strain sequence type 131 (ST131), which alone accounts for up to one-third of all UTIs (3, 4). Even when ST131 is not the predominant strain of E. coli within a given patient population, other clonal E. coli strains are similarly dominant across a national, continental, or even global scale (5, 6).
E. coli is also the most frequently isolated organism from canine UTIs, and some of the clonal types associated with human UTIs have been isolated from dogs, including ST131 (7–13). Case reports also suggest the potential for zoonotic transmission of UTI-associated E. coli; e.g., a longitudinal study within a household of five humans and a dog showed that there were at least six between-host transfers of the ExPEC strain that caused an episode of cystitis in the dog during the study period (14). Additionally, exposure to dogs and/or dog feces has been identified as a risk factor for the development of drug-resistant E. coli UTI in women (15).
Relatively little is known about the epidemiology of ExPEC in companion animals. A recent study in Scotland comparing multilocus sequence types of antimicrobial drug-resistant and -susceptible canine UTI E. coli isolates failed to identify a predominant sequence type (ST) among the 33 isolates included in the study (16). A study of UTI caused by E. coli in cats identified ST73 as the predominant feline strain (17). A better understanding of the epidemiology of ExPEC in dogs is essential to understanding the role of disseminated clones in UTI, especially those with high levels of antimicrobial resistance. Additionally, an understanding of the clonal distribution in dogs may inform us about the epidemiology of ExPEC in general, including strains that cause significant human morbidity.
We tested the hypothesis that uropathogenic E. coli in dogs has a clonal population structure with a predominant canine-specific clade. To test this hypothesis, we designed a multisite survey of antimicrobial resistance, virulence factors, and sequence types of E. coli isolates associated with UTI in dogs in the United States. We prospectively obtained E. coli isolates obtained from canine urine samples from five veterinary diagnostic laboratories, assessed the population structure of these isolates by multilocus sequence typing (MLST), determined the prevalence of a group of virulence-associated genes linked with ExPEC, and explored the resistance genotypes of isolates resistant to third-generation cephalosporins.
MATERIALS AND METHODS
Sampling time frame.
E. coli isolates obtained from canine urine samples were prospectively collected from the Washington Animal Disease Diagnostic Laboratory, Washington State University, Pullman, WA (WADDL; 9 April to 15 May 2015 and 1 October 2015 to 17 May 2016), North Dakota State University Veterinary Diagnostic Laboratory, Fargo, ND (NDSU VDL; 13 to 23 April 2015 and 1 October 2015 to 17 May 2016), Indiana Animal Disease Diagnostic Laboratory, Purdue University, West Lafayette, IN (IADDL; 10 April to 19 May 2015 and 23 September 2015 to 17 May 2016), The Ohio State University Veterinary Medical Center Microbiology Laboratory, Columbus, OH (OSU VMC; 1 October 2015 to 17 May 2016), and the University of California Davis Veterinary Medical Teaching Hospital Laboratory, Davis, CA (UCD VMTH; 1 October 2015 to 17 May 2016). All viable E. coli isolates saved during the respective study periods were included in the analysis. If a given patient had more than one laboratory submission during the study periods, only the first isolate was included in the analysis. Urine samples were submitted to the diagnostic laboratories for clinical diagnostic purposes. Generally, a single isolate was saved by the laboratories and was considered to be representative of the bacterial population causing the infection. However, if multiple E. coli colony morphologies (for example, both hemolytic and nonhemolytic colonies) were observed, a representative colony from each morphology was collected at the source laboratory and underwent molecular analysis; the isolate was included in the final analysis only if it was of a genotype that was distinct from that of the other isolate in the same patient.
Isolate preservation and DNA preparation.
Isolates were stabbed into LB agar, grown overnight at 37°C, and then stored at 4°C until shipment to Washington State University (WSU). Upon arrival at WSU, isolates were recovered on Columbia blood agar and then stored for further analysis by suspending a colony in brain heart infusion (BHI) broth with 20% buffered glycerol at −80°C. Template DNA for molecular procedures was obtained by suspending overnight growth from 1 ml Miller Luria-Bertani broth (Fisher Scientific, Fair Lawn, NJ) in sterile water, followed by heating (100°C, 10 min).
Laboratory identification and susceptibility testing.
E. coli isolates were obtained and identified using standard procedures at each laboratory. Identification at four laboratories (WADDL, IADDL, OSU VMC, and UCD VMTH) was performed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Biotyper; Bruker Daltonics, Billerica, MA, USA). At one laboratory (NDSU VDL), biochemical identification was based on colony morphology and lactose fermentation on MacConkey agar (Hardy Diagnostics, Santa Maria, CA, USA), positive spot indole test (Indole spot reagent; Hardy Diagnostics), and lactose and sucrose fermentation with no hydrogen sulfide production (Difco triple sugar iron agar; Becton Dickinson, Sparks, MD, USA). Susceptibility testing was performed at the primary source laboratories for all isolates by determining the MICs of drugs by broth microdilution using commercial susceptibility plates (Sensititre; Thermo Fisher Scientific, Waltham, MA, USA) and following manufacturer-recommended protocols. The plate formats used for susceptibility testing varied by time and laboratory location and included Thermo Scientific Sensititre products COMPAN1F, COMPAN2F, CMV1BURF, COMPGN1F, and CMV3AGNF. The MICs for the following drugs were included in the analysis: ampicillin, amoxicillin-clavulanic acid, trimethoprim-sulfamethoxazole, and enrofloxacin, based on their importance for the treatment of canine and/or human UTIs; these drugs were consistently tested at comparable MIC ranges across all plate types used for this study (18, 19). Cefpodoxime or ceftiofur was used to assess phenotypic susceptibility to third-generation cephalosporins. Susceptibility interpretation was based on CLSI VET01S 3rd edition breakpoints for Enterobacteriaceae, with canine urine breakpoints used when available (20). These canine breakpoints are very similar to human Enterobacteriaceae breakpoints for ampicillin, amoxicillin-clavulanic acid, and cefpodoxime; human breakpoints are used for trimethoprim-sulfamethoxazole (SXT) susceptibility interpretation in dogs (20, 21). Bovine interpretations were used for ceftiofur since there are no canine-specific breakpoints (20). Isolates with missing susceptibility data were tested by broth microdilution after shipment to Washington State University. If the only information missing was susceptibility information to third-generation cephalosporins, disk diffusion testing with ceftiofur (30-μg disk) using standard methods was performed (22).
MLST.
Multilocus sequence types were assigned to all isolates as previously described by Achtman et al. (23). Briefly, PCR products of seven housekeeping genes were directly sequenced either bidirectionally (adk) or unidirectionally (fumC, gyrB, icd, mdh, purA, and recA) with the Sanger sequencing services of Functional Biosciences, Inc. (Madison, WI). Previously undescribed alleles were sequenced in both directions to confirm the nucleotide sequence. MLST types were assigned using BioNumerics 6.6 (Applied Maths, Austin, TX) and EnteroBase (http://enterobase.warwick.ac.uk). During the study period, EnteroBase stopped accepting new alleles and sequence types based on Sanger sequencing, so these new alleles and STs are listed in the supplemental material (Table S1). Those STs that could not be assigned were assigned temporary placeholder ST identifiers for the purpose of this analysis.
Virulence genotyping.
A multiplex PCR and capillary electrophoresis protocol was developed with the following eight targets: papA, papC, sfa-foc, afa-dra, iutA, kpsMTII, ireA, and fyuA. Primer sequences were as previously described, with the addition of a new kpsMTII primer (5′-AGGCCGATGAACAGGGTGACCA-3′) (Table S2), with fluorescent-labeled forward primers (Applied Biosystems, Foster City, CA) (24–26). The method was validated using a control set of previously characterized isolates provided by James Johnson (Veterans Affairs Medical Center, Minneapolis, MN) (24).
Each PCR (25 μl) contained 3.0 μl PCR H2O, 2.5 μl 10× PCR buffer, 2.0 μl 50 mM MgCl, 2.0 μl dinucleoside triphosphates (dNTPs), 0.25 μl Platinum Taq polymerase (Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA), 14.25 μl of the primer pool, and 1 μl of DNA lysate as the template. One microliter of a 1:10 diluted PCR product was added to 18.5 μl formamide and 0.5 μl GeneScan LIZ 1200 ladder (Applied Biosystems, Foster City, CA, USA) and was run on an ABI 3730 analyzer at the WSU Molecular Biology and Genomics Core (Pullman, WA). GeneMarker version 2.4.0 (SoftGenetics, State College, PA, USA) was used to analyze capillary electrophoresis data using expected peak sizes based on previous reports (24–26). Isolates were classified as ExPEC if they had at least two of the following virulence gene combinations: papA or papC, sfa-foc, afa-dra, iutA, and kpsMTII, as described by Johnson et al. (27). Virulence gene scores were calculated by assigning one point for the detection of each of the eight virulence-associated genes assessed.
Broad-spectrum β-lactamase genotyping.
Isolates showing decreased susceptibility to a third-generation cephalosporin (ceftiofur MIC ≥4 or zone of inhibition <24 mm, cefpodoxime MIC ≥8) were screened for the presence of blaCMY-2 and blaCTX-M genes (28). Primers for blaCMY-2 detection were designed in-house based on a published blaCMY-2 sequence, that with GenBank accession no. JN714983.1 (CMY2F, 5′-CCTCTTTCTCCACATTTGCTGC-3′, and CMY2R, 5′-AAGTGCAGCAGGCGGATACC-3′), and blaCTX-M was detected using previously described primers (29). Isolates that were positive on pan-blaCTX-M PCR were grouped using previously described blaCTX-M grouping primers (30). Isolates positive for blaCTX-M group 9 were sequenced in both directions using group 9 sequencing primers (31). Group 1 isolates were sequenced bidirectionally using upstream and downstream blaCTX-M group 1 primers (32). Sequencing was performed by Functional Biosciences, Inc. (Madison, WI), and sequences were analyzed with the Sequencher 5.0 software (Ann Arbor, MI, USA) and were compared to published sequences of blaCTX-M using the NCBI Basic Local Alignment Search Tool (reference GenBank accession numbers LC259308, GQ274933, LC259307, KF155155, KX023260, and KU510263). Phenotypic extended-spectrum β-lactamase (ESBL) production, using cefotaxime (30 μg), cefotaxime-clavulanic acid (30/10 μg), ceftazidime (30 μg), and ceftazidime-clavulanic acid (30/10 μg) disks (BD BBL Sensi-Disc; Thermo Fisher Scientific, Waltham, MA, USA), was confirmed in all blaCTX-M-positive isolates, and none of the screened isolates that were negative for blaCTX-M and blaCMY-2 showed an ESBL phenotype (21).
Typing of fimH.
ST131 isolates underwent typing of the fimH gene to assess similarity to human ST131 strains. A forward primer described by Weissman et al. (33) and a reverse primer from Johnson and Stell (24) were used to amplify the fimH region. The sequences of the 498-bp fimH typing region were used to assign fimH allele numbers using FimTyper 1.0 (available at https://cge.cbs.dtu.dk/services/FimTyper/) and sequence alignment using Sequencher 5.0 (Ann Arbor, MI, USA) (34). Sequencing was performed by Functional Biosciences, Inc. (Madison, WI, USA).
Patient information.
Patient information that accompanied the urine submission to the source laboratory or that was available to the source laboratory via electronic medical record was collected. These parameters included age, sex, breed, spay/neuter status, method of urine collection, history of urinary tract infection, previous urine submissions to the source laboratory, if the animal was being treated with antimicrobials when the sample was collected, if the animal received antimicrobial therapy in the month prior to sample collection, comorbidities (neurologic, neoplastic, and/or urogenital), treatment with potentially immunosuppressive drugs (corticosteroids, chemotherapy, other immunomodulating drugs), if the sample was submitted by a specialist or general practice veterinarian, urinalysis findings, and if the patient had clinical signs associated with urinary tract infection. Because of the large number of different breeds represented in the patient population, breeds were divided into the following three groups based on average adult weight: small breed (≤9.1 kg), medium breed (9.2 to 27.3 kg), and large breed (≥27.4 kg). Urinalysis parameters included information on the presence of pyuria (>5 leukocytes/high-powered field on urine sediment exam), hematuria (observed grossly or >5 erythrocytes/high-powered field on urine sediment exam), isosthenuria (urine specific gravity, 1.008 to 1.012), and proteinuria (more than a trace protein on urine dipstick or >25 mg/dl protein by photometer). Laboratory parameters, such as the quantity of E. coli isolated, number of bacterial species isolated, and number of colony morphologies of E. coli isolated, were also recorded.
For the purpose of this study, urinary tract infection was defined as the isolation of E. coli from a urine sample. Most (86%) of the dogs that had clinical data available had at least one of the following: clinical signs consistent with UTI (dysuria, pollakiuria, and increased urgency), pyuria, or hematuria. However, we cannot rule out subclinical bacteriuria in a portion of these patients, nor can we completely rule out the possibility of clinically insignificant contamination in free-catch or catheter-collected urine samples, especially in some of the patients that had missing clinical data. However, based on the isolates that had clinical data, it appears that most of the isolates truly represent urinary tract infections and not bacteriuria.
Statistical analysis.
Statistical analyses were performed with R version 3.4.3 (35) and R Studio version 1.1.423 (Boston, MA, USA). Diversity was calculated using the R packages simboot version 0.2-6 and vegan version 2.4-4 (36, 37). Phylogenetic inference using a global optimal eBURST (goeBURST) was performed using PHYLOViZ 2.0 (38). Nonparametric tests were used to compare virulence gene scores, as the scores were not normally distributed (Shapiro-Wilk normality test, P < 0.001). Mann-Whitney U tests were used for pairwise comparison of virulence gene distributions, and a Kruskal-Wallis test was used to compare multiple groups. All hypothesis tests were considered significant when P values were ≤0.05.
Logistic regression.
Multivariate models were created to assess the association between blaCTX-M and blaCMY-2 status with the presence of each of the eight individual virulence-associated genes papA, papC, sfa-foc, afa-dra, iutA, kpsMTII, ireA, and fyuA. Predictor variables were selected in a stepwise manner, using backwards selection.
Poisson regression with robust standard errors.
Univariate log binomial analyses were used to identify potential predictor variables for use in multivariate models. Those predictors with a Wald P value of ≤0.3, along with biologically plausible predictors and risk factors described in the literature, were used to build multivariate models (39, 40). Predictor variables were selected in stepwise manner, using backwards selection with a 10% change-in-estimate criterion for inclusion in the final model. Multivariate models were also compared using Akaike's information criterion (AIC), with a preference for models with a lower AIC. If predictors were highly correlated, the predictor variable with the lower P value in the univariate analysis was selected for inclusion in the multivariable model, and/or each predictor was assessed individually in the multivariate model and AIC values were compared. Multivariate models were created using modified Poisson regression with robust standard errors, and missing data were estimated using multiple imputations (n = 200) for the dogs that had at least baseline clinical data (age, sex, and breed) available (41). The missing data estimated by multiple imputation included spay/neuter status, quantification of E. coli, if two colony types were present in the urine, history of urinary tract infection, history of antimicrobials within the month before submission, if the animal was on antimicrobials at the time of urine collection, if there were previous urine submissions to the laboratory, if the animal received immunosuppressive therapies, if the patient had other comorbidities (cancer, including transitional cell carcinoma, or neurologic disease), if the animal had clinical signs associated with UTI, and urinalysis findings. Summaries of the raw data are in the supplemental material (Table S3). Data were assumed to be missing at random for this analysis. Multiple imputations were performed using R package Amelia version 1.7.4 (42, 43). Robust standard errors were calculated with R packages lmtest version 0.9.35 and sandwich version 2.4.0 (44, 45). A set of imputations and a multivariable model were created for each of two outcomes: (i) isolation of ExPEC status E. coli and (ii) isolation of blaCTX-M-positive E. coli.
RESULTS
A total of 295 isolates were analyzed, with 59 isolates from the Washington Animal Disease Diagnostic Laboratory, 44 isolates from the North Dakota State University Veterinary Diagnostic Laboratory, 64 isolates from the Indiana Animal Disease Diagnostic Laboratory, Purdue University, 31 isolates from The Ohio State University Veterinary Medical Center Microbiology Laboratory, and 97 isolates from the University of California Davis Veterinary Medical Teaching Hospital Laboratory. Specialist veterinarians submitted 170 of the samples, and general practice veterinarians submitted 120 (41.3%) of the cases; the practice type of the veterinarian was unknown in 5 cases.
Population structure.
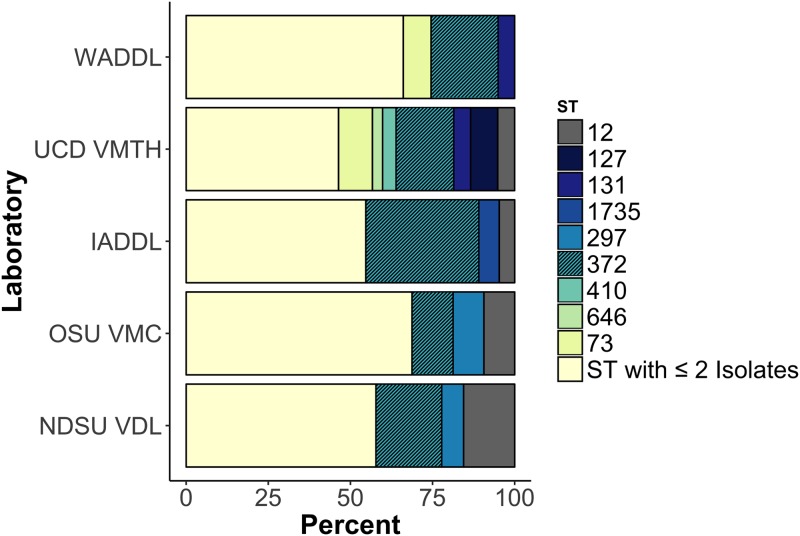
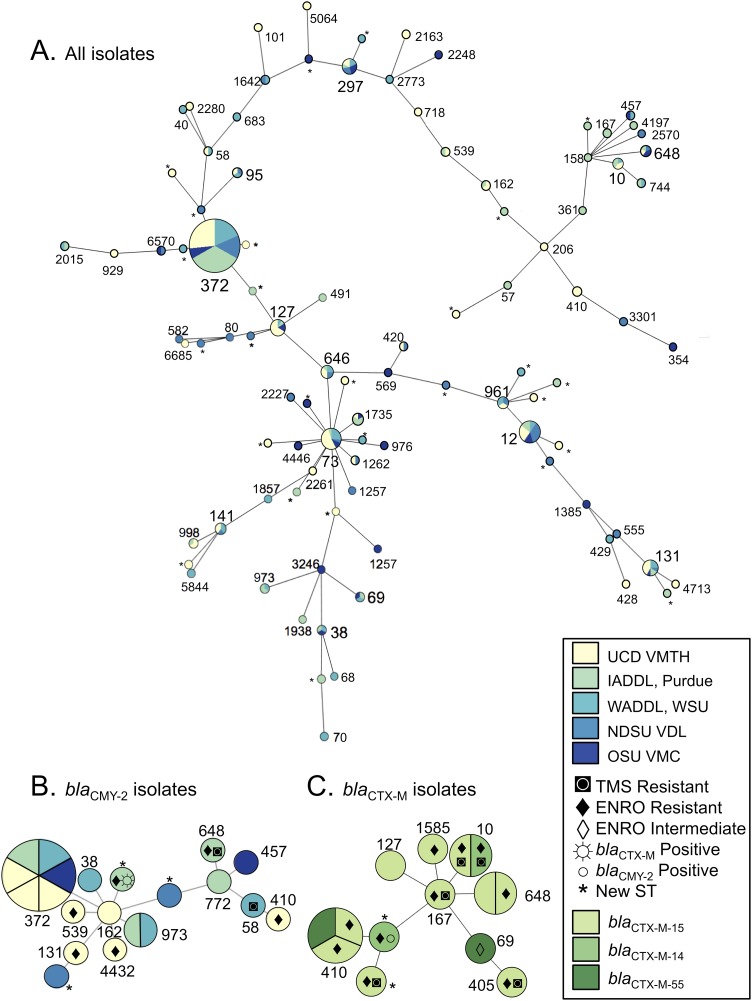
ST372 was the predominant ST at all five locations, comprising 21.7% of the 295 isolates (Fig. 1). Almost half of the isolates fell within the six most frequent STs, ST372, ST12, ST73, ST127, ST131, and ST297 (Table S3). Half of the 12 ST131 isolates were of the H30 subclone (Table 1). Thirty isolates (10.1%) were classified as new sequence types, and each new ST was detected only once. Thirty-three STs included at least two isolates, and 72 (24.3%) isolates were singletons that represented the only isolate of their respective ST; the Simpson's diversity index (1 − D) of the STs of all isolates was 0.95. Thirty-three STs included at least two isolates, and 72 (24.3%) isolates were singletons that represented the only member of their respective ST. The phylogenetic relatedness of the isolates is pictured in Fig. 2A. While ST372 was predominant at all locations, there was variability in ST distribution across locations (Fig. 1).
FIG 1.
Dominant sequence types by laboratory source of isolates. Every ST that included three or more isolates at that given location is listed. The hatched boxes represent the dominant sequence type at all five locations, ST372.
TABLE 1.
Typing of fimH in the 12 ST131 canine UTI isolates, along with their laboratory source and antimicrobial susceptibility information
| fimH type | Source | Third-generation cephalosporin resistance gene | Susceptibility by antimicrobiala |
|||
|---|---|---|---|---|---|---|
| AMP | AMC | ENRO | SXT | |||
| H30 | IADDL | S | S | S | S | |
| WADDL | S | S | S | S | ||
| WADDL | R | R | R | R | ||
| UCD VMTH | blaCMY-2 | R | R | R | S | |
| UCD VMTH | R | S | R | S | ||
| UCD VMTH | R | S | S | S | ||
| H22 | WADDL | R | S | S | S | |
| IADDL | S | S | R | S | ||
| UCD VMTH | S | S | S | S | ||
| H41 | OSU VMC | R | S | S | S | |
| H89 | UCD VMTH | R | R | S | S | |
| H298 | NDSU VDL | R | S | R | S | |
AMP, ampicillin; AMC, amoxicillin-clavulanic acid; ENRO, enrofloxacin; SXT, trimethoprim-sulfamethoxazole; S, susceptible; R, resistance.
FIG 2.
Phylogenetic relatedness of the isolates in the study inferred by goeBURST. The size of each node is scaled to the number of isolates of that ST, and the distance between the nodes is scaled to the number of allele differences between STs. Nodes are labeled with ST number. (A) Phylogenetic relatedness of all isolates in the study. The color represents the laboratory source of the isolate. (B) blaCMY-2 isolates in the study and the coresistances are shown for each blaCMY-2 isolate. The color represents the laboratory source of the isolate. (C) blaCTX-M isolates in the study and the coresistances are shown for each isolate. The color represents the blaCTX-M gene in each isolate.
Antimicrobial resistance.
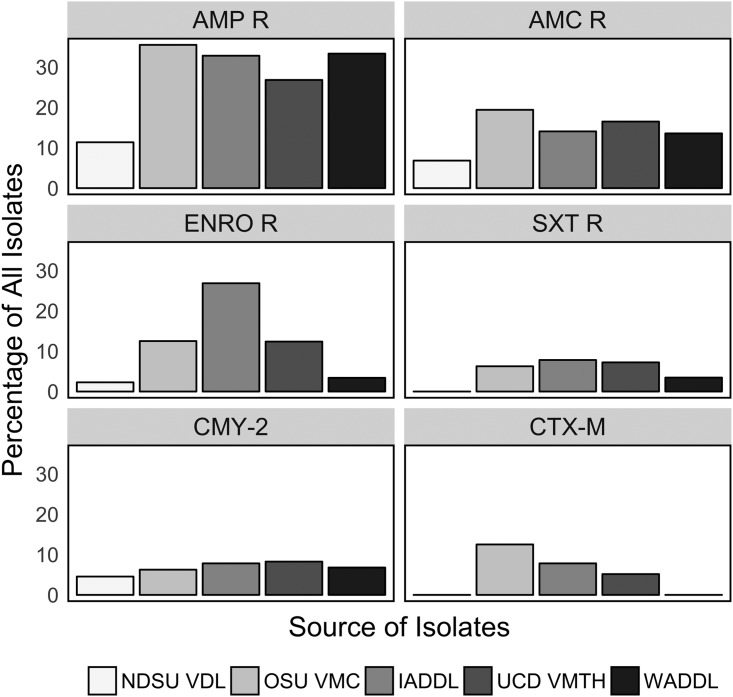
Antimicrobial susceptibility profiles varied, sometimes quite dramatically, among E. coli isolates obtained from the five laboratories (Fig. 3). The overall prevalences of resistance to the antimicrobials used most frequently to treat canine urinary tract infections were ampicillin, 26.8%; amoxicillin-clavulanic acid, 14.2%; enrofloxacin, 10.2%; and trimethoprim-sulfamethoxazole, 5.4%. Isolates carrying blaCTX-M often had coresistance to fluoroquinolones and folate pathway inhibitors (Fig. 2C). All isolates were susceptible to carbapenems. Most of the isolates of the predominant ST372 were susceptible to the major drug classes used to treat canine UTI included in this analysis (54/64 [84.3%]).
FIG 3.
Percentage of E. coli isolates obtained from canine urine from each source laboratory with phenotypic resistance (R) to given antimicrobials and the proportion of third-generation cephalosporin-resistant isolates positive for blaCMY-2 and blaCTX-M by the laboratory source of the isolate.
While phenotypic resistance to third-generation cephalosporins was observed in isolates obtained from all five laboratories, blaCTX-M-positive isolates were only identified at three of the laboratories, and the prevalences at those laboratories ranged from 5.2 to 12.9%. Most of the blaCTX-M isolates carried blaCTX-M-15 (n = 10), but blaCTX-M-14 (n = 2) and blaCTX-M-55 (n = 2) were also identified. The most frequent STs for the blaCTX-M-positive isolates were ST410 (n = 3), ST648 (n = 2), and ST10 (n = 2); the other blaCTX-M-positive isolates were each from a unique ST (Fig. 2C). None of the ST131 isolates were blaCTX-M positive.
All five laboratories obtained E. coli isolates positive for blaCMY-2, and prevalences were similar across locations (4.5 to 8.2%). A single isolate carried both blaCTX-M and blaCMY-2; the isolate was a new ST and positive for both blaCMY-2 and blaCTX-M-14. No blaCTX-M-positive ST372 isolates were detected. Six of 15 (40%) blaCMY-2 isolates were ST372; all of the additional blaCMY-2 isolates represented unique STs, except for two ST973 isolates.
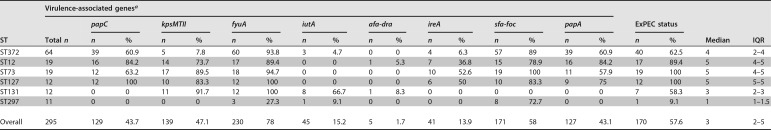
Virulence-associated genes.
One or more ExPEC virulence-associated genes were detected in 257 (86.8%) isolates, and 170 (57.4%) of isolates met the criterion (≥2 of papA or papC, sfa-foc, afa-dra, iutA, and kpsMTII) to be classified as ExPEC. The most frequently identified virulence-associated gene was fyuA (78%), and afa-dra was the least common (1.7%). The distribution of virulence-associated genes was varied among and within STs (Table 2 and Fig. S1). Virulence-associated gene scores varied from 0 to 7, with a median of 3 (interquartile range, 2 to 5). The virulence-associated gene scores were similar among isolates obtained from the five laboratories (Kruskal-Wallis rank sum test, P = 0.25).
TABLE 2.
Distribution of eight virulence-associated genes among the six most frequently isolated STs across all five laboratory locations. Gene prevalence varied between and within STs
aThe distribution of the virulence-associated genes (VG) within these STs is illustrated in Fig. S1. ExPEC status is defined as the presence of ≥2 of the following: papA or papC, sfa-foc, afa-dra, iutA, and kpsMTII. Median virulence-associated gene scores and the interquartile range (IQR) of the virulence gene scores are also presented.
Virulence gene scores were lower in blaCTX-M-positive isolates (median, 1) than in blaCTX-M-negative isolates (median, 3; Mann-Whitney U test, P = 0.003). Fluoroquinolone resistance was also associated with lower virulence scores (median, 2) than those in fluoroquinolone-susceptible isolates (median, 4; Mann-Whitney U test, P = 0.001). Using a multivariate logistic regression model to examine the association between the eight virulence-associated genes assessed in this study and resistance to third-generation cephalosporins, we observed that papC (adjusted odds ratio [OR], 76; 95% confidence interval [95% CI], 1.4 to 2,089; P = 0.03) and iutA (adjusted OR, 21.9; 95% CI, 4.27 to 112; P ≤ 0.001) were both associated with blaCTX-M-positive isolates. The ferric aerobactin receptor gene iutA was also associated with blaCMY-2-positive isolates (adjusted OR, 3.44; 95% CI, 1.09 to 10.9; P = 0.04).
Risk factors.
Associations between patient characteristics, clinical signs, and comorbidities and the isolation of an isolate with ExPEC-defining virulence-associated genes or the isolation of a blaCTX-M-positive isolate were determined (Table 3). Male dogs were more likely to have UTIs due to ExPEC. Receipt of antimicrobials at the time of urine culture and presence of transitional cell carcinoma of the urinary bladder were positively associated with isolation of a blaCTX-M isolate. All other patient, laboratory, and clinicopathologic factors for which data were available were not associated with ExPEC or blaCTX-M status.
TABLE 3.
Patient characteristics associated with isolation of a blaCTX-M-positive E. coli isolate from the patient's urine sample or whether the E. coli isolate obtained from the patient's urine sample was classified as ExPEC
| Predictor | Crude relative risk (95% CI) | P value | Postimputation crude relative risk (95% CI) | P value | Postimputation adjusted relative risk (95% CI) (n = 237)a | P value |
|---|---|---|---|---|---|---|
| blaCTX-M outcome | ||||||
| On antibiotics at the time of urine submission | 12.6 (2.73–57.7) | 0.001 | 14.2 (4.18–48.2) | <0.001 | 11.1 (2.42–51.3) | <0.001 |
| Transitional cell carcinoma of the urinary bladder | 8.05 (2.06–31.5) | 0.003 | 9.22 (2.68–31.8) | <0.001 | 5.58 (1.06–29.8) | 0.02 |
| ExPEC outcome | ||||||
| Male | 1.36 (1.11–1.67) | 0.003 | No imputed data | 1.31 (1.04–1.65) | 0.01 |
Predictors for blaCTX-M outcome adjusted for age, sex, spay/neuter status, and laboratory of origin. Predictor for ExPEC outcome adjusted for age, spay/neuter status, breed size category, and laboratory of origin.
DISCUSSION
We investigated the population structure and antimicrobial susceptibility of E. coli isolates obtained from canine urine samples at five locations and demonstrated that dogs have a predominant ST and that dogs are susceptible to UTI caused by some of the common human ExPEC STs. We also describe E. coli virulence gene repertoires and associations between clinical parameters and problematic antimicrobial resistance genotypes.
Predominant STs among canine uropathogenic E. coli isolates.
All of the most frequently occurring UTI STs (ST372, ST12, ST73, ST127, ST131, and ST297) from the dogs in this study have previously been described in association with human extraintestinal disease, and some (ST12, ST73, ST127, and ST131) are major clonal groups in human ExPEC (46–52). The ST372 prevalence that we observed in dogs is similar to that of ST131 in human-uropathogenic E. coli studies and ST73 in feline E. coli UTIs (6, 17). This suggests that each host species may have a particular ST that comprises most of the E. coli uropathogens isolated despite the evidence of broad overlap of many STs across species. While ST131 was one of the major canine ST groups in this study, all canine ST131 isolates were blaCTX-M negative, contrasting with the major human ST131-H30-Rx blaCTX-M-15-carrying subclone that causes a high burden of human extraintestinal disease (5, 53, 54). However, fluoroquinolone-resistant ST131-H30 isolates were present, indicating that dogs may serve as an occasional reservoir of this important human pathogen.
Virulence-associated genes.
Fewer than 60% of canine UTI E. coli isolates met our definition of ExPEC. The virulence gene with the lowest prevalence that we observed, afa-dra, is highly prevalent (>90%) in feline-UTI-source E. coli isolates (17), suggesting a possible difference in virulence requirements for E. coli binding to urothelium in different species. Because virulence-associated genes tend to be similar across isolates of the same ST, this may account for the dominant STs that vary across animal species. Compared to female dogs, males were more likely to have ExPEC isolates than non-ExPEC isolates, indicating that virulence-associated genes may be necessary to overcome the anatomical barriers to UTIs in males. The blaCTX-M-positive isolates tended to have lower virulence scores in our canine-source isolates. The exception was the ferric aerobactin receptor gene iutA, which was associated with blaCTX-M positive isolates, as has been previously described in human-source isolates and was also associated with blaCMY-2 isolates (55).
blaCTX-M.
Although there is widespread prevalence of blaCTX-M-positive E. coli in human clinical isolates, there were no blaCTX-M E. coli isolates obtained at two of the veterinary laboratories, and the overall antimicrobial resistance rates in canine isolates were lower than rates reported in community-acquired UTIs in people (54, 56). For example, the mean ampicillin resistance prevalence in human-uropathogenic E. coli isolates across multiple studies before 2010 was almost twice that of the ampicillin resistance prevalence that we observed in dogs (57). Most of the blaCTX-M-positive isolates carried the CTX-M-15 allele, as is the case in human ESBL-producing ExPEC isolates (56). We also found blaCTX-M-55 in two isolates of different STs from different locations, a blaCTX-M allele that has not been previously described in clinical veterinary E. coli isolates in the United States.
We also identified risk factors for blaCTX-M isolation, such as transitional cell carcinoma of the urinary bladder and antimicrobial treatment at the time of urine collection. Animals with bladder tumors lack some of the anatomical defenses against UTI, which may expose them to the administration of multiple rounds of antimicrobials for UTI therapy. These decreased defenses may also allow colonization and infection of the urinary bladder by isolates with fewer virulence-associated genes, which includes most blaCTX-M-carrying isolates that we isolated. These animals may also be more likely to be in a hospital environment with potential exposure to nosocomial pathogens, such as ESBL-producing E. coli isolates. This information could potentially be used by veterinarians to identify high-risk dogs in similar patient populations, such as referral practices and veterinary teaching hospitals, that may be infected with blaCTX-M E. coli of public health importance.
blaCMY-2.
Unlike blaCTX-M, blaCMY-2 prevalence did not vary dramatically across locations. Most of the blaCMY-2 isolates were from ST372, while blaCTX-M isolates were more phylogenetically diverse. This suggests a different geographic distribution of blaCMY-2 and blaCTX-M within canine populations. ST372 may be acquiring widely disseminated blaCMY-2-carrying plasmids within dogs, while the blaCTX-M-carrying isolates tended to be seen in minor canine STs that are known to harbor ESBLs in other animal species (17, 46, 58, 59). It is possible that there are geographic differences in canine exposure to blaCTX-M-positive isolates in other animals, which could contribute to the variability in blaCTX-M prevalence observed in this study.
Study limitations.
The limitations of this study include the reliance upon veterinarians to submit clinical samples for diagnostic workups as a source of isolates and the large number of isolates for which clinical data were completely or partially unavailable. The population of patients and bacterial isolates evaluated here may not represent uncomplicated UTIs in dogs, many of which are treated empirically and diagnosed without urine culture. We included both laboratories that serve general practice veterinarians, as well as those that serve large referral centers to attempt to better represent the diversity of canine urine isolates. Additionally, clinical information was limited to the information that was provided to the laboratory by the submitting veterinarian or, in some cases, the data that were available in an electronic medical record. Missing data were estimated based on parameters for the dogs for which clinical data were provided to the laboratories, and this may not truly represent the clinical status of the dogs that did not have clinical data available.
Summary and conclusions.
The clonal population structure of uropathogenic E. coli isolates that has been described in people is recapitulated in dogs, but the predominant canine strain is ST372. Additionally, human-associated ExPEC STs that produce broad-spectrum β-lactamases were isolated from clinical infections in dogs, albeit infrequently. While a direct assessment of zoonotic and reverse-zoonotic transfer of uropathogens requires further study, we present evidence of host sharing of clinical UTI STs that cause both human and veterinary disease; people are clinically affected by ST372 and human-associated STs made up many of the blaCTX-M-carrying isolates in canine UTI. However, this sharing appears to be minimal compared to the higher prevalence of extraintestinal infections caused by a predominant strain for each host species. The cross-species sharing of STs, including ST131-H30, and resistance genes of public health concern in clinical isolates highlight the importance of evaluating companion-animal pathogens, along with risk factors for their acquisition, to more completely elucidate the epidemiology of ExPEC.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award T32AI007025 from the National Institute of Allergy and Infectious Diseases.
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. T.E.L. received additional support from an ARCS fellowship from Washington State University.
We thank Lisa Jones for her technical expertise and Rance Sellon for his input on study design and clinical impact.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00788-18.
REFERENCES
- 1.Kahlmeter G. 2003. Prevalence and antimicrobial susceptibility of pathogens in uncomplicated cystitis in Europe. The ECO.SENS study. Int J Antimicrob Agents 22(Suppl 2):S49–S52. [DOI] [PubMed] [Google Scholar]
- 2.Ronald A. 2003. The etiology of urinary tract infection: traditional and emerging pathogens. Dis Mon 49:71–82. doi: 10.1067/mda.2003.8. [DOI] [PubMed] [Google Scholar]
- 3.Rogers BA, Aminzadeh Z, Hayashi Y, Paterson DL. 2011. Country-to-country transfer of patients and the risk of multi-resistant bacterial infection. Clin Infect Dis 53:49–56. doi: 10.1093/cid/cir273. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R, Robicsek A, Kuskowski MA, Porter S, Johnston BD, Sokurenko E, Tchesnokova V, Price LB, Johnson JR. 2013. Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-β-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob Agents Chemother 57:6385–6388. doi: 10.1128/AAC.01604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathers AJ, Peirano G, Pitout JDD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine M-H, Bertrand X, Madec J-Y. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrester SD, Troy GC, Dalton MN, Huffman JW, Holtzman G. 1999. Retrospective evaluation of urinary tract infection in 42 dogs with hyperadrenocorticism or diabetes mellitus or both. J Vet Intern Med 13:557–560. doi: 10.1111/j.1939-1676.1999.tb02209.x. [DOI] [PubMed] [Google Scholar]
- 8.Norris CR, Williams BJ, Ling GV, Franti CE, Johnson, Ruby AL. 2000. Recurrent and persistent urinary tract infections in dogs: 383 cases (1969–1995). J Am Anim Hosp Assoc 36:484–492. doi: 10.5326/15473317-36-6-484. [DOI] [PubMed] [Google Scholar]
- 9.Ling GV, Norris CR, Franti CE, Eisele PH, Johnson DL, Ruby AL, Jang SS. 2001. Interrelations of organism prevalence, specimen collection method, and host age, sex, and breed among 8,354 canine urinary tract infections (1969–1995). J Vet Intern Med 15:341–347. doi: 10.1111/j.1939-1676.2001.tb02327.x. [DOI] [PubMed] [Google Scholar]
- 10.Cohn LA, Gary AT, Fales WH, Madsen RW. 2003. Trends in fluoroquinolone resistance of bacteria isolated from canine urinary tracts. J Vet Diagn Invest 15:338–343. doi: 10.1177/104063870301500406. [DOI] [PubMed] [Google Scholar]
- 11.Platell JL, Cobbold RN, Johnson JR, Trott DJ. 2010. Clonal group distribution of fluoroquinolone-resistant Escherichia coli among humans and companion animals in Australia. J Antimicrob Chemother 65:1936–1938. doi: 10.1093/jac/dkq236. [DOI] [PubMed] [Google Scholar]
- 12.Harada K, Nakai Y, Kataoka Y. 2012. Mechanisms of resistance to cephalosporin and emergence of O25b-ST131 clone harboring CTX-M-27 β-lactamase in extraintestinal pathogenic Escherichia coli from dogs and cats in Japan. Microbiol Immunol 56:480–485. doi: 10.1111/j.1348-0421.2012.00463.x. [DOI] [PubMed] [Google Scholar]
- 13.Dahmen S, Haenni M, Châtre P, Madec J-Y. 2013. Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J Antimicrob Chemother 68:2797–2801. doi: 10.1093/jac/dkt291. [DOI] [PubMed] [Google Scholar]
- 14.Reeves PR, Liu B, Zhou Z, Li D, Guo D, Ren Y, Clabots C, Lan R, Johnson JR, Wang L. 2011. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 6:e26907. doi: 10.1371/journal.pone.0026907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ukah UV, Glass M, Avery B, Daignault D, Mulvey MR, Reid-Smith RJ, Parmley EJ, Portt A, Boerlin P, Manges AR. 2018. Risk factors for acquisition of multidrug-resistant Escherichia coli and development of community-acquired urinary tract infections. Epidemiol Infect 146:46–57. doi: 10.1017/S0950268817002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner S, Gally DL, Argyle SA. 2014. Multidrug-resistant Escherichia coli from canine urinary tract infections tend to have commensal phylotypes, lower prevalence of virulence determinants and ampC-replicons. Vet Microbiol 169:171–178. doi: 10.1016/j.vetmic.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Thungrat K, Boothe DM. 2015. Multilocus sequence typing and virulence profiles in uropathogenic Escherichia coli isolated from cats in the United States. PLoS One 10:e0143335. doi: 10.1371/journal.pone.0143335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weese JS, Blondeau JM, Boothe D, Breitschwerdt EB, Guardabassi L, Hillier A, Lloyd DH, Papich MG, Rankin SC, Turnidge JD, Sykes JE. 2011. Antimicrobial use guidelines for treatment of urinary tract disease in dogs and cats: Antimicrobial Guidelines Working Group of the International Society for Companion Animal Infectious Diseases. Vet Med Int 2011:263768. doi: 10.4061/2011/263768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy CP, Reid-Smith RJ, Boerlin P, Weese JS, Prescott JF, Janecko N, McEwen SA. 2012. Out-patient antimicrobial drug use in dogs and cats for new disease events from community companion animal practices in Ontario. Can Vet J 53:291–298. [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 3rd ed CLSI supplement VET01S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 22.Bauer AW, Kirby WM, Sherris JC, Turck M. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 23.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Fakhr MK, Nolan LK. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097–2110. doi: 10.1099/mic.0.27499-0. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JR, O'Bryan TT. 2004. Detection of the Escherichia coli group 2 polysaccharide capsule synthesis gene kpsM by a rapid and specific PCR-based assay. J Clin Microbiol 42:1773–1776. doi: 10.1128/JCM.42.4.1773-1776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother 47:2161–2168. doi: 10.1128/AAC.47.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aarestrup MF, Hasman H, Veldman K, Mevius D. 2010. Evaluation of eight different cephalosporins for detection of cephalosporin resistance in Salmonella enterica and Escherichia coli. Microb Drug Resist 16:253–261. doi: 10.1089/mdr.2010.0036. [DOI] [PubMed] [Google Scholar]
- 29.Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother 47:3724–3732. doi: 10.1128/AAC.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitout JDD, Hossain A, Hanson ND. 2004. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J Clin Microbiol 42:5715–5721. doi: 10.1128/JCM.42.12.5715-5721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Chen J, Kang Y, Jiang N, An SC, Gao ZC. 2012. Prevalence and characterization of plasmid-mediated blaESBL with their genetic environment in Escherichia coli and Klebsiella pneumoniae in patients with pneumonia. Chin Med J (Engl) 125:894–900. [PubMed] [Google Scholar]
- 32.Wittum TE, Mollenkopf DF, Daniels JB, Parkinson AE, Mathews JL, Fry PR, Abley MJ, Gebreyes WA. 2010. CTX-M-type extended-spectrum β-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog Dis 7:1575–1579. doi: 10.1089/fpd.2010.0615. [DOI] [PubMed] [Google Scholar]
- 33.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol 78:1353–1360. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roer L, Tchesnokova V, Allesøe R, Muradova M, Chattopadhyay S, Ahrenfeldt J, Thomsen MCF, Lund O, Hansen F, Hammerum AM, Sokurenko E, Hasman H. 2017. Development of a web tool for Escherichia coli sub-typing based on fimH alleles. J Clin Microbiol 55:2538–2543. doi: 10.1128/JCM.00737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 36.Scherer R, Pallmann P. 2017. simboot: simultaneous inference for diversity indices. R package version 0.2-6. https://cran.r-project.org/web/packages/simboot/simboot.pdf.
- 37.Oksanen J, Blanchet JG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2017. vegan: community ecology package. R package version 2.4-4. https://cran.r-project.org/web/packages/vegan/index.html.
- 38.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Vaz C. 2017. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 39.Freshman JL, Reif JS, Allen TA, Jones RL. 1989. Risk factors associated with urinary tract infection in female dogs. Prev Vet Med 7:59–67. doi: 10.1016/0167-5877(89)90037-8. [DOI] [Google Scholar]
- 40.Budreckis DM, Byrne BA, Pollard RE, Rebhun RB, Rodriguez CO Jr, Skorupski KA. 2015. Bacterial urinary tract infections associated with transitional cell carcinoma in dogs. J Vet Intern Med 29:828–833. doi: 10.1111/jvim.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou G. 2004. A modified Poisson regression Approach to prospective studies with binary data. Am J Epidemiol 159:702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 42.Honaker J, King G, Blackwell M. 2011. Amelia II: a program for missing data. J Stat Softw 45:1–47. doi: 10.18637/jss.v045.i07. [DOI] [Google Scholar]
- 43.King G, Honaker J, Joseph A, Scheve K. 2001. Analyzing incomplete political science data: an alternative algorithm for multiple imputation. Am Polit Sci Rev 95:49–69. [Google Scholar]
- 44.Zeileis A, Hothorn T. 2002. Diagnostic checking in regression relationships. R News 2:7–10. [Google Scholar]
- 45.Zeileis A. 2004. Econometric computing with HC and HAC covariance matrix estimators. J Stat Softw 11:1–17. doi: 10.18637/jss.v011.i10. [DOI] [Google Scholar]
- 46.Hertz FB, Nielsen JB, Schønning K, Littauer P, Knudsen JD, Løbner-Olesen A, Frimodt-Møller N. 2016. Population structure of drug-susceptible, -resistant and ESBL-producing Escherichia coli from community-acquired Urinary Tract Infections. BMC Microbiol 16:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 46:1076–1080. doi: 10.1128/JCM.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horner C, Fawley W, Morris K, Parnell P, Denton M, Wilcox M. 2014. Escherichia coli bacteraemia: 2 years of prospective regional surveillance (2010–12). J Antimicrob Chemother 69:91–100. doi: 10.1093/jac/dkt333. [DOI] [PubMed] [Google Scholar]
- 49.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 50.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 51.Manges AR, Tabor H, Tellis P, Vincent C, Tellier P-P. 2008. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg Infect Dis 14:1575–1583. doi: 10.3201/eid1410.080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brisse S, Diancourt L, Laouénan C, Vigan M, Caro V, Arlet G, Drieux L, Leflon-Guibout V, Mentré F, Jarlier V, Nicolas-Chanoine M-H, Coli β Study Group . 2012. Phylogenetic distribution of CTX-M- and non-extended-spectrum-β-lactamase-producing Escherichia coli isolates: group B2 isolates, except clone ST131, rarely produce CTX-M enzymes. J Clin Microbiol 50:2974–2981. doi: 10.1128/JCM.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35:316–321. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Lee S, Yu JK, Park K, Oh E-J, Kim S-Y, Park Y-J. 2010. Phylogenetic groups and virulence factors in pathogenic and commensal strains of Escherichia coli and their association with blaCTX-M. Ann Clin Lab Sci 40:361–367. [PubMed] [Google Scholar]
- 56.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 57.Foxman B. 2010. The epidemiology of urinary tract infection. Nat Rev Urol 7:653–660. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 58.Davis MA, Sischo WM, Jones LP, Moore DA, Ahmed S, Short DM, Besser TE. 2015. Recent emergence of Escherichia coli with cephalosporin resistance conferred by blaCTX-M on Washington state dairy farms. Appl Environ Microbiol 81:4403–4410. doi: 10.1128/AEM.00463-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peirano G, van der Bij AK, Gregson DB, Pitout JDD. 2012. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J Clin Microbiol 50:294–299. doi: 10.1128/JCM.06025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.