Chronic infection with Helicobacter pylori causes peptic ulcers and stomach cancer in a subset of infected individuals. While standard eradication therapy includes multiple antibiotics, treatment failure due to resistance is an increasing clinical problem.
KEYWORDS: droplet digital PCR, noninvasive detection, Helicobacter pylori, heteroresistance, clarithromycin resistance
ABSTRACT
Chronic infection with Helicobacter pylori causes peptic ulcers and stomach cancer in a subset of infected individuals. While standard eradication therapy includes multiple antibiotics, treatment failure due to resistance is an increasing clinical problem. Accurate assessment of H. pylori antimicrobial resistance has been limited by slow growth and sampling of few isolates per subject. We established a method to simultaneously quantify H. pylori clarithromycin-resistant (mutant) and -susceptible (wild-type) 23S rRNA gene alleles in both stomach and stool samples using droplet digital PCR (ddPCR). In 49 subjects, we assessed the performance of these assays alongside clarithromycin MIC testing of up to 16 H. pylori isolates per subject and included both cancer (25 subjects) and noncancer (24 subjects) cases. Gastric ddPCR and H. pylori culture showed agreement with urea breath test (UBT) detection of infection in 94% and 88% of subjects, respectively, while stool ddPCR showed agreement with UBT in 92% of subjects. Based on MIC testing of 43 culture-positive cases, 20 subjects had only susceptible isolates, 14 had a mix of susceptible and resistant isolates, and 9 had only resistant isolates. ddPCR of gastric samples indicated that 21 subjects had only wild-type alleles, 13 had a mixed genotype, and 9 had only mutant alleles. Stool ddPCR detected mutant alleles in four subjects for which mutant alleles were not detected by stomach ddPCR, and no resistant isolates were cultured. Our results indicate that ddPCR detects H. pylori clarithromycin resistance-associated genotypes, especially in the context of heteroresistance.
INTRODUCTION
The Gram-negative pathogen Helicobacter pylori (H. pylori) infects half of the world's population. Long-term infection with H. pylori increases the risk of chronic gastritis, gastric and duodenal ulcers, gastric cancer, and gastric mucosa-associated lymphoid tissue lymphoma. Eradication of H. pylori is recommended in all infected individuals to prevent or postpone the development of these diseases. Treatment regimens contain various combinations of a proton pump inhibitor, two antibiotics, and bismuth. Treatment failure may result, in part, from increasing prevalence of antibiotic resistance, especially to clarithromycin (1).
Clarithromycin is an antibiotic that binds to the peptidyl transferase loop of domain V of 23S rRNA to inhibit protein synthesis in H. pylori (2). Point mutations in this domain cause resistance to clarithromycin, with A2143G and A2142(G/C) (3) accounting for the majority. Additional resistance-related point mutations, such as T2182C, C2611A, and T2717C, have been reported to be associated with low-level resistance (4, 5) but remain controversial (6, 7). Interstrain and intrastrain genetic diversity of H. pylori during persistent infection can cause heteroresistance in genotype and phenotype (8).
Noninvasive methods for detecting H. pylori include the urea breath test (UBT), antigen in stool, and antibody in blood, among which UBT is the gold standard. Invasive methods are based on endoscopic biopsy, including isolation and culture, as well as molecular methods to detect H. pylori existence and antibiotic resistance-associated genotypes, with a higher price and less frequent use. In the clinical laboratory, susceptibility testing is generally performed by sampling three to five colonies isolated in culture and performing testing. However, antibiotic-susceptible and -resistant H. pylori isolates can be simultaneously present in an individual before antibiotic treatment (9, 10), which might lead to false-negative results in antimicrobial susceptibility testing in the clinical laboratory by the traditional culture method.
To raise the sensitivity of resistance detection, molecular methods have been introduced, among which real-time PCR was regarded as the most efficient and sensitive method (11–14). In our previous study, we developed a new stool-based method for detection of H. pylori infection and the cagA gene using droplet digital PCR (ddPCR) (15). Here, we developed additional ddPCR assays to simultaneously quantify clarithromycin-susceptible and -resistant 23S RNA alleles. We also performed extensive phenotypic analysis of H. pylori strain populations within individuals to show that ddPCR gives accurate quantitative results for the proportion of clarithromycin-resistant strains.
MATERIALS AND METHODS
Study population and sample collection.
Gastric mucus samples were collected from 49 urea breath test (UBT)-positive patients seen at Henan Cancer Hospital, including 25 gastric cancer (3 female and 22 male) and 24 noncancer (12 female and 12 male) patients, ranging in age from 27 to 76 years old (24). Subjects who were H. pylori positive by UBT were offered participation in this research study, and all participants provided informed consent prior to participation. Participating subjects had gastric mucosal brushings collected and were asked to fill out a questionnaire that covered demographic characteristics, medical conditions, and medications and provide a stool sample. In total, samples were collected from 49 subjects, including 25 gastric cancer subjects diagnosed by pathology who were undergoing gastric resection surgery and 24 noncancer subjects with no history of gastric tumor or surgery who were undergoing upper gastrointestinal (GI) endoscopy either as indicated because of symptoms (n = 8) or as asymptomatic volunteers (n = 16). All noncancer cases showed histologic evidence of gastritis, and no ulcers were detected. All 49 subjects had not taken antibiotics in the past month. All procedures were approved by the Henan Cancer Hospital Medical Research Institution Review Board (document 2016oct005).
Gastric mucosal brushing samples were collected from two different anatomical sites in the stomach, the antrum and the corpus, using cytology brushes (Puritan Medical Products Co., LLC). From each anatomical site, two gastric mucosal brushings were collected. A sample for total DNA extraction was collected and stored in a cryovial containing 1 ml of flow medium (minimal essential medium plus 10% dimethyl sulfoxide [DMSO], 5% fetal calf serum, 5 mM HEPES) (26). A sample for culture was similarly collected and stored in 1 ml of brain heart infusion broth (Oxoid) containing 20% glycerol. Cryovials containing the gastric mucosal brushing samples were immediately placed and kept at −80°C. The gastric mucosal brushings were collected from the gastric cancer subjects during the gastric resection surgery and from noncancer subjects during upper GI endoscopy. Stool samples were collected 1 to 2 days before either surgery or endoscopy. Participants undergoing surgery collected the stool sample at the hospital, and participants undergoing endoscopy collected the stool sample at home and delivered it to the hospital on the same day. Stool samples were collected by the participants into a vial containing 5 ml of RNAlater nucleic acid preservative (Ambion) and were frozen at −20°C upon receipt. All five samples (four gastric brushes and one stool sample) were collected from each subject.
DNA extraction.
For the gastric mucosal brushings, sample medium and brush were transferred from the cryovial to a microcentrifuge tube and centrifuged, followed by removal of the supernatant and brush. DNA was then extracted from the pellet using an UltraClean Tissue and Cells DNA isolation kit (MoBio) according to the manufacturer's instructions.
Stool samples were first transferred to a microcentrifuge tube and centrifuged to remove the RNAlater. DNA was then extracted using a QIAamp Stool DNA minikit (Qiagen) according to the manufacturer's instructions, with the lysis step performed at 95°C. Stool DNA concentration was measured using a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Scientific), and the concentration was adjusted to 100 ng/μl.
Primer and probe design.
The primer and probe sequences for the H. pylori 23S clarithromycin resistance ddPCR assay are listed in Table 1. Probes were designed to distinguish between the H. pylori wild-type (clarithromycin-susceptible) 23S allele and the three most common clarithromycin resistance-conferring mutations in the H. pylori 23S gene (A2143G, A2142G, and A2142C).
TABLE 1.
Primers and probes used for the droplet digital PCR assay to detect H. pylori 23S clarithromycin resistance alleles
| Primer or probe name | Sequence and chemical modification(s)a |
|---|---|
| Hp23SF | 5′-TCCCGTTAGCAGTGCTAA-3′ |
| Hp23SR | 5′-AGATGGGAGCTGTCTCAAC-3′ |
| Hp23S_WT_HEX | 5′-HEX-AAGACGGAAAGACCCCGTG-BHQ1-3′ |
| Hp23S_A2143G_FAM | 5′-FAM-AAGACGGAGAGACCCCGT-BHQ1-3′ |
| Hp23S_A2142G_FAM | 5′-FAM-AAGACGGGAAGACCCCGT-BHQ1-3′ |
| Hp23S_A2142C_FAM | 5′-FAM-AAGACGGCAAGACCCCGT-BHQ1-3′ |
HEX, hexachlorofluorescein; FAM, 6-carboxyfluorescein; BHQ1, Black Hole quencher 1.
Droplet digital PCR.
H. pylori-specific droplet digital PCR (ddPCR) assays were performed using a QX200 ddPCR System (Bio-Rad) as described previously (15) for the stool DNA and the gastric mucosal brushing DNA. For the clarithromycin resistance assay, thermal cycling conditions were 95°C for 10 min, 45 cycles of 94°C for 30 s and 58°C for 1 min, and 1 cycle of 98°C for 10 min. Data were analyzed using QuantaSoft software, version 1.6.6 (Bio-Rad). The thresholds for positive droplets were set at 2,000 for wild-type alleles (hexachlorofluorescein [HEX] channel) and at 4,000 for resistance mutations (6-carboxyfluorescein [FAM] channel). To decrease nonspecific detection of wild-type and resistance alleles, the concentration of wild-type and resistance alleles was set to zero if less than five positive droplets were detected in the respective channel.
The concentration of the gastric mucosal brushing DNA sample was adjusted so that the copy number of the ddPCR assay target was within the dynamic range of the assay. DNA samples were run as either undiluted or diluted 1:10, 1:100, 1:1,000, or 1:10,000. Stool DNA concentration was adjusted to 100 ng/μl as needed so that no more than 1 μg of stool DNA was added to each ddPCR reaction.
Isolation and verification of H. pylori from gastric brush samples.
Gastric brush samples were serially diluted 5-fold with brain heart infusion (Oxoid) broth, and 100 μl of undiluted or diluted (1:5, 1:25, and 1:125) sample was inoculated on Karmali agar base (CM 0935; Oxoid) containing 15% defibrinated sheep blood and H. pylori selective supplement SR0147E (Oxoid). Plates were incubated at 37°C under microaerobic conditions (5% O2, 85% N2, 10% CO2) for 48 h to 2 weeks until up to 16 single colonies or all the H. pylori colonies were collected. Individual isolates were collected as soon as they reached a sufficient size for harvesting, and every effort was made to include the diversity of colony morphologies and/or growth rates present in our sampling. A stringent confirmation process was applied in the following sequence to confirm all included isolates as H. pylori: Gram stain microscopy; urease, catalase, and oxidase tests; matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Microflex LT system; Bruker) analysis supplemented with the Biotyper database (subdatabase of H. pylori updated) and MicroID software (subdatabase of H. pylori).
Clarithromycin susceptibility test.
The MIC of clarithromycin was determined by Etest (bioMérieux) strips. After 48 h of growth on plates, isolates were suspended in sterile saline and adjusted to a McFarland turbidity standard of 2, as recommended by the Clinical and Laboratory Standards Institute (CLSI). The bacterial suspension was spread onto Karmali blood agar plates with a sterile cotton swab. Plates with clarithromycin Etest strips were incubated under microaerobic conditions for 48 h. The MIC was the lowest concentration that inhibited visible growth. As CLSI recommended, a clarithromycin resistance breakpoint of ≥0.75 mg/liter and a sensitive breakpoint of ≤0.25 mg/liter were used, while MIC values of 0.38 and 0.5 ml/liter were considered intermediate. If isolates grew well and showed a clear inhibition ring on the initial test plates, we used a single measurement to determine the MIC. Some isolates (16 isolates from 7 subjects, 1 to 3 isolates per subject) gave sparse growth on the initial Etest plate. In this case, we repeated the Etest and used only plates with uniform growth and clear inhibition rings to determine the MIC. For such isolates, we reported values that were consistent for at least two replicates. Four isolates from four different subjects gave clarithromycin MICs of 1 mg/liter in the initial test. We performed two additional Etest assays for these four isolates and observed MIC values of 1 mg/ml for all replicates for three of these isolates while for one isolate the MIC was 0.75 mg/ml for one replicate and 1 mg/ml for two replicates. H. pylori strain ATCC 43504 was used as the quality control strain. MIC values of H. pylori ATCC 43504 were 0.047 to 0.064 mg/liter, within the range of 0.016 to 0.12 as recommended by the CLSI. An Etest performed on CLSI-recommended Mueller-Hinton agar (MHA) with sheep blood and Karmali agar with sheep blood for H. pylori ATCC 43504 showed the same MIC value of 0.064 mg/liter. All testing was conducted at the National Institute for Communicable Disease Control and Prevention of the Chinese Center for Disease Control and Prevention.
23S rRNA gene amplification and sequence.
The 23S rRNA genes were amplified with forward (F) (5′-AGGCGATGAAGGACGTA-3′) and reverse (R) (5′-CTTAGATGCYTTCAGC-3′) primers. A reaction mixture of 50 μl contained 1× polymerase buffer, 200 μM each deoxyribonucleotide, 2.5 U of PrimeStar Taq DNA polymerase (TaKaRa Bio, Inc., Japan), and 0.5 μM each primer (Sangon Biotech Co., Shanghai, China). Step-down PCR was used for amplification (25) as follows: activation of polymerase at 94°C for 5 min; 8 cycles of denaturation at 94°C for 45 s, annealing at 62°C to 55°C for 45 s (dropping 1°C per cycle), and elongation at 72°C for 3 min; and 22 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 3 min, followed by a final elongation step of 7 min at 72°C. The V domain of the H. pylori 23S rRNA gene was amplified with primers F (5′-AGCGATGTGGTCTCAGCA-3′) and R (5′-CAAGGGTGGTATCTCAAGG-3′). A 25-μl reaction mixture contained 1× polymerase buffer, 200 μM each deoxyribonucleotide, 1 U of PrimeStar Taq DNA polymerase (TaKaRa Bio, Inc., Japan), and 0.5 μM each primer (Sangon Biotech Co., Shanghai, China). The PCR program was as follows: activation of polymerase at 94°C for 5 min and 35 cycles of denaturation at 94°C for 45 s, annealing at 56°C for 45 s, and elongation at 72°C for 45 s, followed by a final elongation step of 7 min at 72°C. PCR-amplified fragments were purified and sequenced by Beijing Tianyi Huiyuan Bioscience and Technology, Inc.
Statistical analysis.
Comparison of the clarithromycin resistance percentage in subjects with heteroresistance between the stomach ddPCR assay and culture and between stomach and stool ddPCR assays was analyzed using the Spearman correlation coefficient with Fisher's z-transformation. All statistical analyses were performed in SAS, version 9.4 (SAS Institute, Inc.).
RESULTS
Efficient isolation of H. pylori from gastric mucus cytology brush samples.
We used a cohort of 49 urea breath test (UBT)-positive subjects from Henan Cancer Hospital with gastritis (n = 24) or gastric cancer (n = 25) to investigate the prevalence of clarithromycin resistance. For each subject, four gastric brush samples (two from the antrum and two from the corpus) and one stool sample were collected. One sample from each stomach site was used for culture of H. pylori and collection of up to 16 single colony isolates per sample. Among the 49 UBT-positive subjects, we collected 1,084 isolates by culture, 560 from 39 (80%) gastric antrum samples, and 524 from 39 (80%) gastric corpus samples. Combining gastric corpus and antrum results, 43 (88%) of 49 subjects showed positive results in culture, among which 35 subjects were culture positive in both corpus and antrum samples, 6 subjects were culture negative in both corpus and antrum samples, 4 subjects were corpus positive and antrum negative, and 4 subjects were antrum positive and corpus negative. From 25 subjects, we collected 32 isolates, including 16 from antrum and 16 from corpus samples. In 9 subjects, we collected fewer than 16 isolates from either the antrum (3 subjects) or the corpus (6 subjects), and in 6 subjects we collected no isolates from the antrum (2 subjects) or the corpus (4 subjects) in spite of collecting 16 isolates from the other site. In 3 subjects, we collected fewer than 16 isolates in total, including 1 subject with a single isolate from both the corpus and antrum samples and 2 subjects with 1 and 12 isolates in antrum samples and no isolates from the corpus samples.
We successfully cultured H. pylori from 21/25 (84%) gastric cancer subjects (range, 16 to 32 isolates; median, 32 isolates) and 22/24 (92%) noncancer subjects (range, 1 to 32 isolates; median, 19 isolates).
Diversity of clarithromycin resistance in gastric isolates.
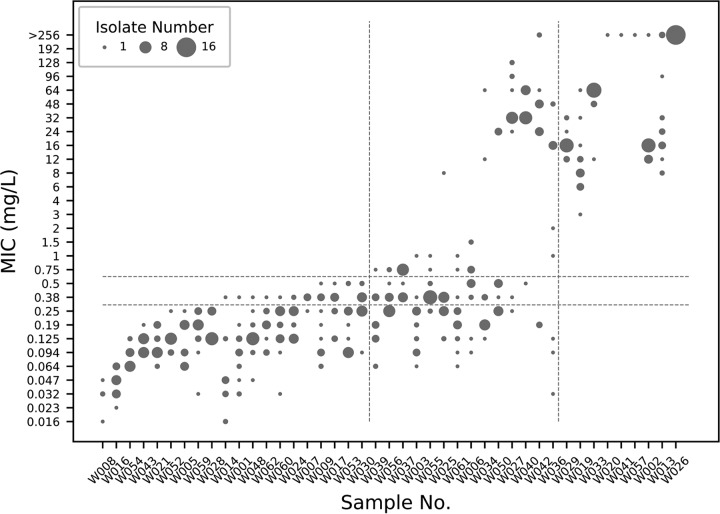
To evaluate clarithromycin resistance, we performed a clarithromycin Etest on 16 single-colony isolates or as many isolates as were recovered from the 43 culture-positive subjects (range, 1 to 16 isolates; median, 16 isolates). For subjects who were culture positive in the antrum, we performed the Etest only on antrum isolates and tested corpus isolates for the remaining subjects. Based on the resistance breakpoint of 1 mg/liter, these subjects were divided into three groups: one group in which all isolates were susceptible (MIC of <0.25 mg/liter) or showed intermediate resistance (MIC of 0.38 or 0.5 mg/liter) to clarithromycin, one group in which all isolates were resistant to clarithromycin (MIC of ≥0.75 mg/liter), and one group which consisted of a mixture of susceptible and resistant isolates. Among 43 subjects, 20 had only susceptible isolates, 14 subjects had a mix of susceptible and resistant isolates, and 9 subjects had only resistant isolates. As shown in Fig. 1, MIC values among all subject isolates and even among the 16 isolates sampled from each subject showed considerable variation (range, 0.023 to >256 mg/liter; median, 0.25 mg/liter). Among the 14 subjects with a mix of susceptible and resistant isolates, some (W025, W027, and W042) had isolates with a wide range of MICs above the resistance breakpoint while some had isolates with very few MICs near the resistance breakpoint and many susceptible isolates (W003, W006, and W055). While 23 of 34 subjects who had susceptible isolates also had isolates with intermediate susceptibility, in none of the subjects with only resistant isolates were the MIC values <3 mg/liter.
FIG 1.
MIC distribution of H. pylori isolates in culture-positive gastric samples. Each column represents the MIC distribution of a gastric antrum or corpus sample (W008, W014, W019, and W029 are from corpus samples, and the rest are from antrum samples). The diameter of each dot represents the number of isolates that show a corresponding MIC value (y axis) in each sample (x axis). Horizontal dotted lines separate three zones of clarithromycin resistance: resistant (≥0.75 mg/liter), intermediate (0.38 and 0.5 mg/liter), and sensitive (≤0.25 mg/liter). Vertical dotted lines separate subjects with only susceptible and intermediate isolates (left), with a mixture of susceptible, intermediate, and resistant isolates (middle), and with only resistant isolates (right).
Considering disease states, we detected resistance in 10/21 cancer cases (48%, including 2 subjects with only resistant isolates) and 13/22 noncancer cases (59%, including 7 subjects with only resistant isolates).
Detection of H. pylori clarithromycin resistance-related 23S rRNA alleles.
We developed a ddPCR assay to assess clarithromycin resistance using probes bearing different fluorophores to simultaneously measure the wild type and the three most common 23S rRNA gene mutations that confer clarithromycin resistance (A2142G, A2142C, and A2143G). We tested the H. pylori clarithromycin resistance assay by spiking H. pylori genomic DNA of a clarithromycin-susceptible wild-type strain and three isogenic clarithromycin-resistant strains into H. pylori-negative stool DNA from a UBT-negative volunteer. The genomic DNA of each strain was spiked individually and also at different ratios to test the sensitivity of the assay to detect subpopulations of clarithromycin-resistant H. pylori. The clarithromycin resistance ddPCR assay correctly distinguished between wild-type and clarithromycin resistance alleles and detected subpopulations of clarithromycin-resistant mutants at least down to 1% of the population (Fig. 2).
FIG 2.
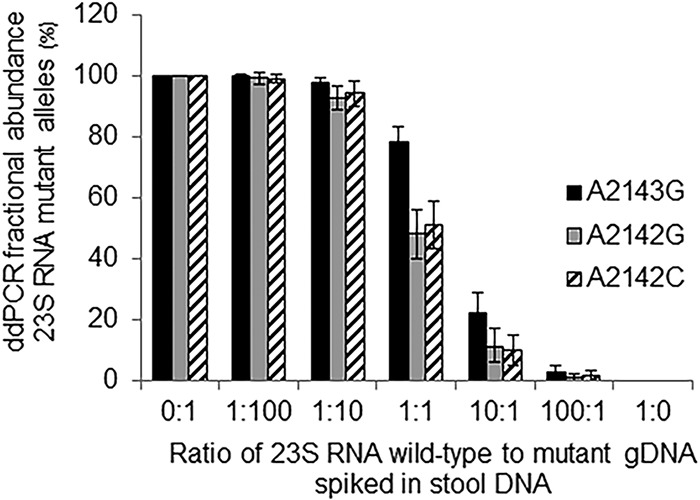
Fractional abundance analysis of clarithromycin resistance alleles measured by ddPCR. Three kinds of clarithromycin resistance strain DNAs (A2143G, A2142G, and A2142C) were mixed with the wild-type strain at ratio of 0:1, 1:100, 1:10, 1:1, 10:1, 100:1, and 1:0 before being spiked into stool DNA. Two ddPCRs were run for each sample, and the results were combined. Bars indicate Poisson 95% confidence intervals.
We next investigated the ability to measure resistance by ddPCR in the gastric mucosal brush sample total DNA. We detected 23S rRNA genes (wild type, mutant, or both) in all culture-positive samples and two additional samples that were culture negative. Of the 46 gastric brushing samples for which the H. pylori 23S gene was detected, 22 (48%) had only wild-type (clarithromycin susceptible) alleles detected and 24 (52%) had clarithromycin resistance alleles detected. Of the 24 with clarithromycin resistance alleles, 11 had only clarithromycin resistance alleles, and 13 had a mixed population of clarithromycin resistance and wild-type alleles (Table 2). For the 13 subjects with a mixed population in the gastric brushing samples, the percentage of the H. pylori population with clarithromycin resistance alleles ranged from 2% to 78%, with a median of 36%. All patients had concordant results between the antrum and the corpus brushing samples except for one patient for whom the 23S gene was detected in the corpus sample but not in the antrum sample.
TABLE 2.
Clarithromycin resistance alleles detected in gastric brushes and stool samples by ddPCR
| Clarithromycin resistance genotype | No. (%) of positive subjects by group |
|||
|---|---|---|---|---|
| Gastric brush |
Stool |
|||
| Gastric cancer (n = 25) | Noncancer (n = 24) | Gastric cancer (n = 25) | Noncancer (n = 24) | |
| Only susceptible alleles | 13 (52) | 9 (38) | 12 (48) | 5 (21) |
| Only resistance alleles | 2 (8) | 9 (38) | 1 (4) | 10 (42) |
| Mixed | 7 (28) | 6 (25) | 9 (36) | 8 (33) |
| 23S not detected | 3 (12) | 0 (0) | 3 (12) | 1 (4) |
Comparing ddPCR detection of resistance alleles to culture Etest results showed high concordance for the 42 subjects from which culture data were available (Table 3). Among the subjects with mixed resistance detected by both culture and ddPCR, the percentage of resistance detected was strongly correlated (Spearman correlation coefficient, 0.69; P = 0.004) (Fig. 3A). For two subjects, 1 of 16 isolates showed resistance at the 1 mg/liter breakpoint by Etest, but resistance alleles were not detected by ddPCR. Additionally, there were four subjects for whom resistance was detected at a frequency of 10 to 63% by ddPCR, but no resistant clones were detected by Etest. Of these four discordant cases, three had 16 individual isolates tested, while one had only 5 isolates available for susceptibility testing.
TABLE 3.
Comparison of gastric ddPCR, stool ddPCR, and culture Etest in detecting clarithromycin resistance
| Resistance category | Etest (no. of isolates) | No. of isolates by genotype |
|||||
|---|---|---|---|---|---|---|---|
| Gastric ddPCR |
Stool ddPCR |
||||||
| Wild type | Mixed | Mutant | Wild type | Mixed | Mutant | ||
| Susceptible + intermediate | 20 | 16 | 4 | 0 | 12 | 6 | 2 |
| Mixed | 14 | 5 | 9 | 0 | 3 | 9 | 1 |
| Resistant | 9 | 0 | 0 | 9 | 0 | 1 | 8 |
FIG 3.
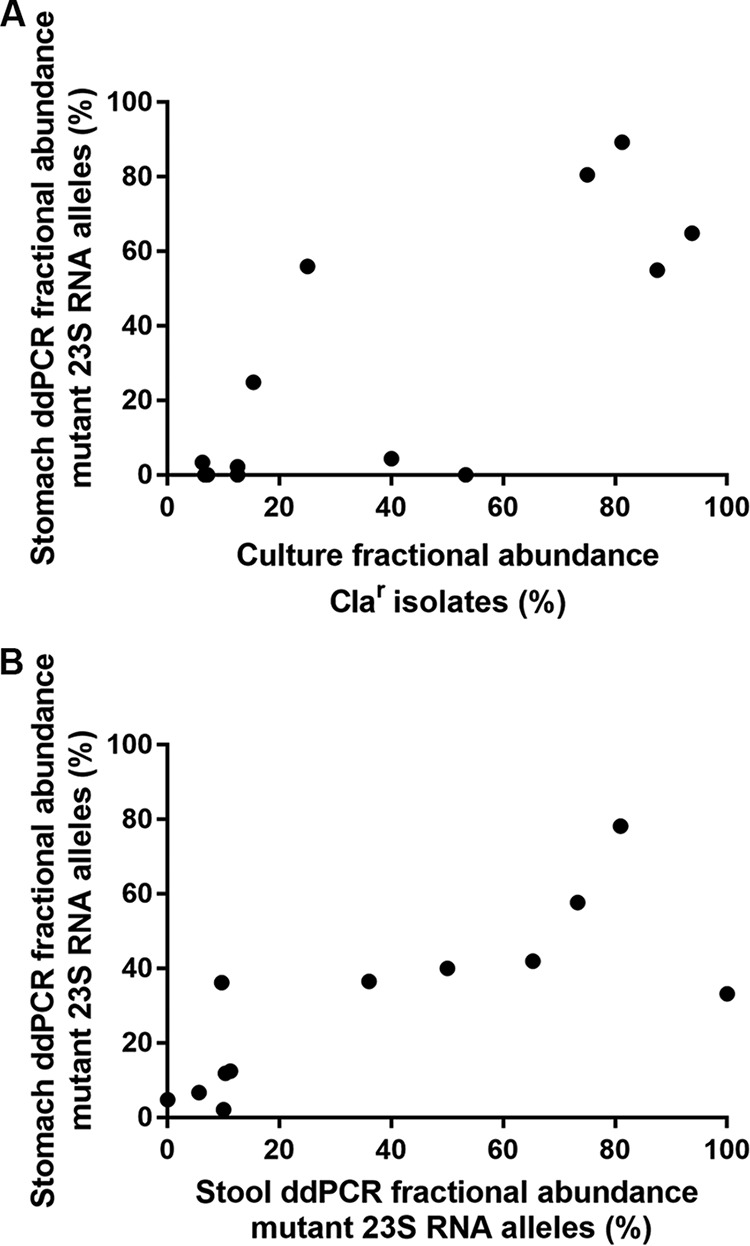
Comparison of phenotypic (Etest) and genotypic (ddPCR) clarithromycin resistance detected in the stomach and stool among heteroresistant subjects. Each point represents a single subject. The fractional abundance in culture of clarithromycin-resistant (Clar) isolates is defined as the number of isolates with clarithromycin MICs of ≥1 mg/liter divided by the total number of isolates and multiplied by 100. For stomach ddPCR, the percentage of 23S mutant alleles (number of mutant alleles divided by the sum of wild-type and mutant alleles and multiplied by 100) is reported for the same stomach region used in the MIC testing for that subject (A) or reported as the average of the antrum and corpus measurements (B). Spearman correlation coefficients are 0.69 for panel A (P = 0.004) 0.74 for panel B (P = 0.003).
Composition of H. pylori 23S alleles in stool is reflective of the stomach.
Stool samples from the 25 gastric cancer patients and 24 noncancer patients were also collected and analyzed for the presence of H. pylori clarithromycin resistance alleles using the stool-based ddPCR assay. The H. pylori 23S gene was detected in 45 of the 49 stool samples. Of these 45 stool samples, 17 (38%) had only wild-type (clarithromycin susceptible) alleles detected and 28 (62%) had clarithromycin resistance alleles detected. Of the 28 with clarithromycin resistance alleles, 11 had only clarithromycin resistance alleles, and 17 had a mixed population of clarithromycin resistance and wild-type alleles (Table 2). For the 17 subjects with a mixed population in the stool sample, the percentage of the H. pylori population with clarithromycin resistance alleles ranged from 1% to 98%, with a median of 35%.
The stool-based clarithromycin resistance assay detected resistance alleles in 21 (88%) of the 24 subjects, with clarithromycin resistance mutations detected in gastric brushings. Of the 22 subjects with only the wild-type susceptible allele in the gastric brushings, 15 (68%) had only the wild-type susceptible allele detected in the stool. Of the 49 subjects, 35 (71%) had concordant results for the clarithromycin resistance category (susceptible allele only, clarithromycin resistance allele only, mixed, and no detection of 23S) between the stool samples and the gastric brushing samples (Table 2). The 14 patients with discordant results between the stool sample and the gastric brushing samples had either a very low load of H. pylori in the stool or a mixed population detected in the stool with a low proportion of clarithromycin resistance or wild-type alleles.
Of 12 subjects with a mixed population in the gastric brushing samples, the percentage of the H. pylori population with resistance alleles was strongly correlated between the gastric brushing samples and the stool samples (Spearman correlation coefficient, 0.74; P = 0.003) (Fig. 3B). One subject with a mixed population in the gastric brushing samples did not have the 23S gene detected in the stool sample.
23S rRNA sequence analysis.
In a comparison of the results of the gastric ddPCR assay, two samples showed a mixture of clarithromycin-susceptible and -resistant clones by Etest, but no clarithromycin resistance alleles were detected by ddPCR, suggesting a false-negative result from ddPCR. To assess additional mutations in the 23S rRNA gene in these isolates, we amplified and sequenced the 23S rRNA gene of resistant isolates from each of these subjects (W003 and W055). Compared to the 23S rRNA reference gene of H. pylori ATCC 26695, the sequence showed no mutation at position 2142 or 2143 queried in the ddPCR assay, but we identified polymorphisms of A759T, G1513A, T1687C, A1821G, G1826A, T1830C, and T2182C in one subject. The second subject had these same polymorphisms and A1822T. Of these polymorphisms, T2182C has been implicated in low-level resistance previously (6, 20). Interestingly, for both of these isolates the MIC was 1 mg/liter, which is at the resistance breakpoint used in this study. We also amplified and sequenced domain V of the 23S rRNA gene in 32 isolates from 15 subjects for which the clarithromycin MIC values were 0.75 (5 isolates), 0.5 (7 isolates), and 0.38 (20 isolates) mg/liter, along with H. pylori strain ATCC 43504 and a sensitive isolate (MIC of 0.25 mg/liter clarithromycin). We observed only wild-type sequences at positions 2143 and 2142 in all of these samples. In four resistant isolates from four subjects for which the clarithromycin MIC values were 8 (1 isolate), 64 (1 isolate), and >256 (2 isolates) mg/liter, we observed the A2143G mutation exclusively.
DISCUSSION
While most studies of clarithromycin resistance within the H. pylori strain populations of individual subjects test 1 to 4 isolates per subjects, here up to 16 isolates were cultured and tested per patient sample. Given that there have been many reports of heteroresistance and that our ddPCR assays can quantify both wild-type and resistance-associated mutations at low frequency (<10%), this sampling strategy allowed us to capture rare resistant or susceptible clones. In order to test the MIC of such a large number of isolates (>500), we employed the Etest instead of the CLSI-recommended agar dilution method. We used Karmali blood agar instead of MHA, which we observe to give better growth for slow-growing clinical isolates. While agreement of Etest and agar dilution methods for clarithromycin resistance detection has been confirmed by multiple studies (17, 21, 22) and while simultaneous testing of control strains on Karmali blood agar and MHA yielded identical MIC measurements, our use of the Etest with nonstandard medium limits comparisons with prior studies and our ability to definitively establish the resistance profiles for the isolates characterized in this study. Additionally, except for cases of low growth and four isolates with MICs of 1 mg/liter, we used a single Etest measurement to determine the MIC, which limits the precision of the data reported. In addition to the Etest, we performed targeted sequencing of 36 isolates from 16 subjects with a range of susceptible and resistant MICs. In 32 isolates with clarithromycin MICs of ≤75 mg/liter, we did not observe any mutations at position 2142 or 2143, suggesting that we did not underestimate resistance with our method. For four isolates with clarithromycin MICs of ≥8 mg/liter, we observed only the A2143G mutation. While CLSI guidelines propose a clarithromycin MIC of 1 mg/liter as the breakpoint for resistance, we found that subjects where 100% of the bacterial population carried resistance alleles by ddPCR (n = 9), all of the isolates tested had MICs of ≥3 mg/liter under our culture conditions. Additionally, sequencing of the 23S RNA genes from two isolates with clarithromycin MICs of 1 mg/liter showed only wild-type sequences at positions 2142 and 2143. Thus, by using 1 mg/liter as the breakpoint for resistance under our culture conditions and Etest assays, we may be overestimating resistance.
Both Etest MIC testing and ddPCR analyses revealed a high rate of coinfection with susceptible and resistant isolates (33% and 30%, respectively). This high prevalence of mixed drug resistance suggests that the current clinical laboratory practice of culture-based drug resistance testing of a single clone or a few clones per subject could produce misleading results (18) and lead to failure in eradication treatment of H. pylori. Therefore, a rapid and accurate method to understand the profile of clarithromycin resistance in the stomach is critically needed. Among the 14 culture-positive subjects in our study with heteroresistance, the fraction of resistant isolates measured by Etest ranged from 6% to 94%, and nine subjects had a fraction of resistance of less than 50%. Testing a single isolate, as is often done in clinical practice, likely would not have revealed resistant clones in these nine subjects, but ddPCR detected resistance alleles in five. Thus, for this study population, ddPCR appears to provide more sensitive detection of resistance than might be expected from routine clinical resistance testing practice.
Comparison of the performance of ddPCR and culture in detecting H. pylori infection showed that agreement rates with UBT were 94% and 88%, respectively, indicating that the ddPCR method is equivalent, if not more sensitive, in stomach samples. Detection in stool was only slightly less sensitive with ddPCR than that in stomach samples (92%). The failure of culture in the discordant samples could be due to the very low growth rate of H. pylori compared to that of other microorganisms present in the stomach, especially in gastric cancer cases that often have higher stomach pH. There is a prevailing assumption that the colonization level of H. pylori is lower or difficult to detect in gastric cancer patients (16, 23). In this study, the ddPCR-positive rates for gastric cancer samples and noncancer samples were similar or slightly better than the culture detection rates in both cancer and noncancer groups. Although this study sample size is relatively small, it indicates that the ddPCR is a sensitive method for detection H. pylori in cancer cases.
Our ddPCR assays can also differentiate an H. pylori 23S mutation from wild type to predict clarithromycin resistance in a single assay. Using the current clinical breakpoint of a MIC value at ≥1 mg/liter, ddPCR was concordant with Etest for 79% of subjects (Table 3). Out of the nine samples that showed discrepancy between the two assays, four of the samples were classified as only susceptible or intermediate by Etest but mixed by ddPCR. Even though 16 isolates from the gastric brush sample were analyzed, this is likely still not a full representation of the stomach population. This concept is further supported by our finding that the stool-based assays detected mixed resistance in several samples that were only susceptible by both culture and stomach sample ddPCR. Thus, while stool-based H. pylori detection is slightly less sensitive, it may be advantageous for capturing resistance and has the benefit of not requiring endoscopy. Furthermore, there were two subjects for whom a single resistant clone at the MIC of 1 mg/liter was detected by Etest but only wild-type alleles were detected by ddPCR. Our ddPCR assay detects any of the three major mutations (A2142C, A2142G, and A2143G) in the 23S rRNA gene which are reported to cover >90% of clarithromycin resistance (19). Sequence analysis of the two weakly resistant isolates showed the presence of a T2182C mutation, which was reported to be related with low resistance levels (6, 20). While a more expansive coverage of mutations could be explored to improve ddPCR assay precision, the relevance of this mutation and of low-level resistance to therapy failure is not yet clear.
In addition to detecting heteroresistance, our phenotypic analysis revealed a wide range of clarithromycin MICs even among subjects with only susceptible or only resistant isolates. Independent retesting of individual isolates resulted in consistent clarithromycin MICs (within 2-fold) and did not correlate with growth rate (data not shown). Preliminary sequencing of a few housekeeping loci suggests identical genotypes among isolates with different MICs (data not shown). Further analysis will be needed to determine the genetic and/or epigenetic mechanisms that underlie the phenotypic variation in clarithromycin MICs among strains obtained from the same individual.
A major limitation of our study is the small number of subjects analyzed and the lack of healthy subjects to assess the specificity of our assays. Furthermore, our study design did not allow assessment of treatment outcomes. While it is likely that subjects with only resistant bacteria would be at higher risk of eradication therapy failure, most therapies include multiple antibiotics. At present it is not clear whether detection of any or a certain threshold percentage of resistant clones predicts therapy failure. Future larger studies that additionally evaluate the relationship between ddPCR-based resistance allele detection pretreatment and success of therapy will thus be needed.
The frequent concordance between stool and stomach sample results holds promise for development of a noninvasive test for detection of resistance. With further studies on additional hospital and population-based cohorts, this technology might develop into new tests to suggest alternative or additional antibiotics to clarithromycin for inclusion in therapy when resistance alleles are detected. Conversely, treatment could be simplified in cases where only susceptible alleles are detected, possibly reducing side effects and increasing compliance. Similar to any other PCR diagnostic assays, this ddPCR method could be extended to additional antibiotics whose resistance genotype is clear, for example, gyrA mutations that confer quinolone resistance. New patient-tailored therapy approaches could lead to more effective H. pylori eradication and possibly slow the spread of antibiotic resistance.
ACKNOWLEDGMENTS
This work was supported in part by a fund from the China Mega-Project for Infectious Disease (2011ZX10004-001), a grant from the National Technology R&D Program in the 12th Five-Year Plan of China (2012BAI06B02), a grant from the State Key Laboratory of Infectious Disease Prevention and Control (SKLID) (2014SKLID102), grants from the U.S. National Institutes of Health (R01 AI054423 to N.R.S. and K01 DK090103 to S.T.), and the Genomics Shared Resource of the NIH/NCI Cancer Center Support Grant (P30 CA015704).
The contents are solely the responsibility of the authors and do not necessarily represent the official views of these funding agencies.
REFERENCES
- 1.Nishizawa T, Maekawa T, Watanabe N, Harada N, Hosoda Y, Yoshinaga M, Yoshio T, Ohta H, Inoue S, Toyokawa T, Yamashita H, Saito H, Kuwai T, Katayama S, Masuda E, Miyabayashi H, Kimura T, Nishizawa Y, Takahashi M, Suzuki H. 2015. Clarithromycin versus metronidazole as first-line Helicobacter pylori eradication: a multicenter, prospective, randomized controlled study in Japan. J Clin Gastroenterol 49:468–471. doi: 10.1097/MCG.0000000000000165. [DOI] [PubMed] [Google Scholar]
- 2.Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, Tanaka SK, Graham DY, Go MF. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother 40:477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera-Correa JJ, Urruzuno P, Barrio J, Martinez MJ, Agudo S, Somodevilla A, Llorca L, Alarcón T. 2017. Detection of Helicobacter pylori and the genotypes of resistance to clarithromycin and the heterogeneous genotype to this antibiotic in biopsies obtained from symptomatic children. Diagn Microbiol Infect Dis 87:150–153. doi: 10.1016/j.diagmicrobio.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Fontana C, Favaro M, Minelli S, Criscuolo AA, Pietroiusti A, Galante A, Favalli C. 2002. New site of modification of 23S rRNA associated with clarithromycin resistance of Helicobacter pylori clinical isolates. Antimicrob Agents Chemother 46:3765–3769. doi: 10.1128/AAC.46.12.3765-3769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JM, Kim JS, Kim N, Kim YJ, Kim IY, Chee YJ, Lee CH, Jung HC. 2008. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J Microbiol Biotechnol 18:1584–1589. [PubMed] [Google Scholar]
- 6.Khan R, Nahar S, Sultana J, Ahmad MM, Rahman M. 2004. T2182C mutation in 23S rRNA is associated with clarithromycin resistance in Helicobacter pylori isolates obtained in Bangladesh. Antimicrob Agents Chemother 48:3567–3569. doi: 10.1128/AAC.48.9.3567-3569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burucoa C, Landron C, Garnier M, Fauchère JL. 2005. T2182C mutation is not associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother 49:868 (Letter.) doi: 10.1128/AAC.49.2.868-870.2005 (Reply, 49: 868–870. doi: 10.1128/AAC.49.2.868-870.2005.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farzi N, Malekian T, Alebouyeh M, Vaziri F, Zali MR. 2015. Genotype diversity and quasispecies development of Helicobacter pylori in a single host. Jpn J Infect Dis 68:351. doi: 10.7883/yoken.JJID.2015.E002. [DOI] [PubMed] [Google Scholar]
- 9.van der Ende A, van Doorn LJ, Rooijakkers S, Feller M, Tytgat GN, Dankert J. 2001. Clarithromycin-susceptible and-resistant Helicobacter pylori isolates with identical randomly amplified polymorphic DNA-PCR genotypes cultured from single gastric biopsy specimens prior to antibiotic therapy. J Clin Microbiol 39:2648–2651. doi: 10.1128/JCM.39.7.2648-2651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao CY, Lee AY, Huang AH, Song PY, Yang YJ, Sheu SM, Chang WL, Sheu BS, Wu JJ. 2014. Heteroresistance of Helicobacter pylori from the same patient prior to antibiotic treatment. Infect Genet Evol 23:196–202. doi: 10.1016/j.meegid.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Monno R, Giorgio F, Carmine P, Soleo L, Cinquepalmi V, Ierardi E. 2012. Helicobacter pylori clarithromycin resistance detected by Etest and TaqMan real-time polymerase chain reaction: a comparative study. APMIS 120:712–717. doi: 10.1111/j.1600-0463.2012.02896.x. [DOI] [PubMed] [Google Scholar]
- 12.Vécsei A, Innerhofer A, Binder C, Gizci H, Hammer K, Bruckdorfer A, Riedl S, Gadner H, Hirschl AM, Makristathis A. 2010. Stool polymerase chain reaction for Helicobacter pylori detection and clarithromycin susceptibility testing in children. Clin Gastroenterol Hepatol 8:309–312. doi: 10.1016/j.cgh.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Shukla SK, Prasad KN, Tripathi A, Ghoshal UC, Krishnani N, Nuzhat H. 2011. Quantitation of Helicobacter pylori ureC gene and its comparison with different diagnostic techniques and gastric histopathology. J Microbiol Methods 86:231–237. doi: 10.1016/j.mimet.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Siopi M, Spyros P, Joseph M. 2017. Comparative evaluation of Sensititre YeastOne and CLSI M38-A2 reference method for antifungal susceptibility testing of Aspergillus spp. against echinocandins. J Clin Microbiol 55:1714–1719. doi: 10.1128/JCM.00044-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talarico S, Safaeian M, Gonzalez P, Hildesheim A, Herrero R, Porras C, Cortes B, Larson A, Fang FC, Salama NR. 2016. Quantitative detection and genotyping of Helicobacter pylori from stool using droplet digital PCR reveals variation in bacterial loads that correlates with cagA virulence gene carriage. Helicobacter 21:325–333. doi: 10.1111/hel.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wroblewski LE, Peek RM Jr, Wilson KT. 2010. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev 23:713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osato MS, Reddy R, Reddy SG, Penland RL, Graham DY. 2001. Comparison of the Etest and the NCCLS-approved agar dilution method to detect metronidazole and clarithromycin resistant Helicobacter pylori. Int J Antimicrob Agents 17:39–44. doi: 10.1016/S0924-8579(00)00320-4. [DOI] [PubMed] [Google Scholar]
- 18.Selgrad M, Tammer I, Langner C, Bornschein J, Meißle J, Kandulski A, Varbanova M, Wex T, Schlüter D, Malfertheiner P. 2014. Different antibiotic susceptibility between antrum and corpus of the stomach, a possible reason for treatment failure of Helicobacter pylori infection. 2014. World J Gastroenterol 20:16245–16251. doi: 10.3748/wjg.v20.i43.16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu G, Xu X, He L, Ding Z, Gu Y, Zhang J, Zhou L. 2011. Primary antibiotic resistance of Helicobacter pylori isolated from Beijing children. Helicobacter 16:356–362. doi: 10.1111/j.1523-5378.2011.00856.x. [DOI] [PubMed] [Google Scholar]
- 20.Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2008. Novel mutation in 23S rRNA that confers low-level resistance to clarithromycin in Helicobacter pylori. Antimicrob Agents Chemother 52:3465–3466. doi: 10.1128/AAC.00445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glupczynski Y, Broutet N, Cantagrel A, Andersen LP, Alarcon T, López-Brea M, Mégraud F. 2002. Comparison of the Etest and agar dilution method for antimicrobial susceptibility testing of Helicobacter pylori. Eur J Clin Microbiol Infect Dis 21:549–552. doi: 10.1007/s10096-002-0757-6. [DOI] [PubMed] [Google Scholar]
- 22.Lang L, García F. 2004. Comparison of Etest and disk diffusion assay to evaluate resistance of Helicobacter pylori isolates to amoxicillin, clarithromycin, metronidazole and tetracycline in Costa Rica. Int J Antimicrob Agents 24:572–577. doi: 10.1016/j.ijantimicag.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Karnes WE Jr, Samloff IM, Siurala M, Kekki M, Sipponen P, Kim SW, Walsh JH. 1991. Positive serum antibody and negative tissue staining for Helicobacter pylori in subjects with atrophic body gastritis. Gastroenterology 101:167–174. doi: 10.1016/0016-5085(91)90474-Y. [DOI] [PubMed] [Google Scholar]
- 24.Talarico ST, Leverich C, Wei B, Ma J, Cao X, Guo Y, Han G, Yao L, Self S, Zhao Y, Salama NR. 2018. Increased H. pylori stool shedding and EPIYA-D cagA alleles are associated with gastric cancer in an East Asian hospital. bioRxiv doi: 10.1101/284448. [DOI] [PMC free article] [PubMed]
- 25.Dewhirst FE, Shen Z, Scimeca MS, Stokes LN, Boumenna T, Chen T, Paster BJ, Fox JG. 2005. Discordant 16S and 23S rRNA gene phylogenies for the genus Helicobacter implications for phylogenetic inference and systematics. J Bacteriol 187:6106–6118. doi: 10.1128/JB.187.17.6106-6118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gall A, Fero J, McCoy C, Claywell BC, Sanchez CA, Blount PL, Li X, Vaughan TL, Matsen FA, Reid BJ, Salama NR. 2015. Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett's esophagus cohort. PLoS One 10:(6)e0129055. doi: 10.1371/journal.pone.0129055. [DOI] [PMC free article] [PubMed] [Google Scholar]