LETTER
The deluge of data produced by the Xpert MTB/RIF test (Cepheid) can help improve global rifampin-resistant tuberculosis (RR-TB) control strategies through molecular epidemiological surveillance (1, 2). Recently, a new version of the test, Xpert Ultra (hereinafter called Ultra), was released (3). Determining the relationship between RR-conferring rpoB mutations, Ultra probes, and melting temperature shifts (ΔTm), i.e., the difference between mutant and wild-type melting temperatures, allows Ultra results to be utilized for rapid detection of RR-TB strains and related underlying rpoB mutations.
To determine the reliability of Ultra results for predicting specific mutations, we tested 13 rifampin-susceptible (RS)-TB strains and 104 RR-TB strains harboring 33 unique RR-conferring mutations from the Belgian Coordinated Collections of Microorganisms in the Institute of Tropical Medicine Antwerp according to a protocol previously described (2) (see the supplemental material). Of note, the Glu250Gly (n = 2) and Arg299Cys (n = 1) mutations were among the RS-TB strains. We then compared Ultra raw results with available rpoB sequences of the strains.
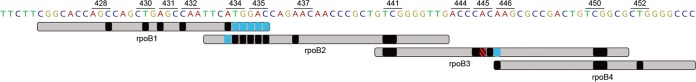
Overall, 29/30 (97%) mutations inside the rifampin resistance-determining region (RRDR) were correctly identified by Ultra. Of concern, mutation His445Arg gave a “RIF Resistance Indeterminate” result among 3/4 strains tested, while it was reported as RR in the initial validation study (3). The silent mutation Thr444Thr was not reported as RR (Fig. 1). The RR-conferring mutations on codons 170 and 491 situated outside the RRDR were not detected.
FIG 1.
Overview of Xpert Ultra test results. The observed probe reactions for each RRDR mutation were laid over the claimed probe coverage (light gray). Shown in black are probe reactions concordant with manufacturer claims, in blue are probe reactions missed by one probe but captured by another probe, and in red is a probe reaction representing a “RIF Resistance Indeterminate” result from 3 out of 4 strains tested. Results in the hatched pattern were superimposed for greater visibility.
The probe reactions observed were largely in agreement with previous results (3), although we noted that mutations Met434Val, Met434Thr, and those in codon 435 were captured only by probe rpoB2, Ser450Leu and Ser450Trp were captured by both probe rpoB3 and probe rpoB4a, His445Arg was captured only by probe rpoB3, and Lys446Gln was captured only by probe rpoB4.
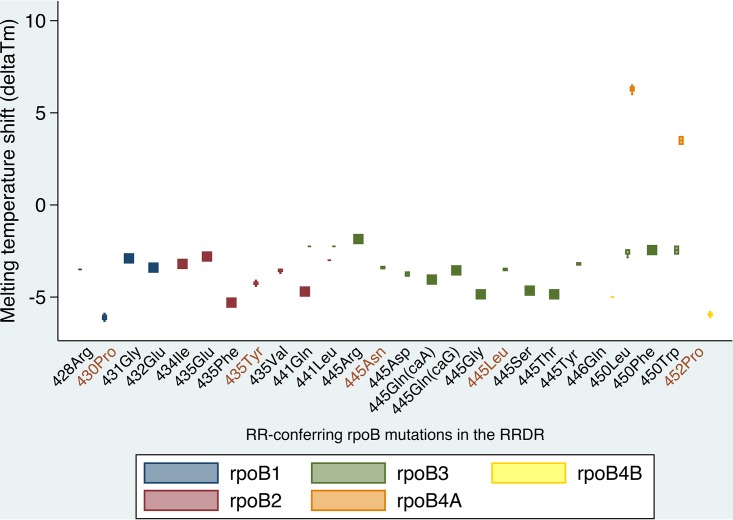
All mutations except those in codon 450 were associated with a negative ΔTm (Fig. 2). The combination of ΔTm values with the capturing probes enabled us to differentiate mutations in codons 430, 431, 434, 435, 441, 446, 450, and 452, including disputed mutations (4) (Table 1). Mutation Asp435Tyr was unambiguously distinguished from Asp435Val with the ∣ΔTm∣ of probe rpoB2, while mutations Ser441Gln and Ser441Leu were discriminated from the rest by the ∣ΔTm∣ values of probes rpoB2 and rpoB3. Mutations His445Asp and His445Tyr were distinguished from disputed mutations His445Leu and His445Asn through the ∣ΔTm∣ of probe rpoB3. Ser450Leu was distinguished from Ser450Trp by the ∣ΔTm∣ of probe rpoB4A. The indeterminate result associated with His445Arg may be caused by its ∣ΔTm∣ being equal to 1.8°C, unlike the ∣ΔTm∣ values for other mutations, which typically exceed 2°C. Our recent experience with Ultra on diagnostic sputum samples pertained only to the Ser450Leu and His445Asp mutations, for which the ΔTm corresponded exactly with the ΔTm that we observed for bacterial thermolysates. This should be validated more extensively, which is beyond the scope of our present study.
FIG 2.
Melting temperature shifts (ΔTms) observed upon detection of a rifampin resistance (RR)-conferring rpoB mutation in the RR-determining region (RRDR) by Xpert Ultra. The y axis reflects the melting temperature difference (ΔTm) between mutant and wild-type probe-amplicon hybrids, while the x axis shows the mutations that we tested. The data points on the graph are ΔTm values grouped by their associated Ultra probes (differentiated by color), which correspond to a specific rpoB mutation. x axis labels in brown are disputed mutations.
TABLE 1.
Xpert Ultra raw resultsa
| Mutation(s)b | No. of strains tested | Nucleotide change(s) | Xpert Ultra probe(s) | Wild-typeTm range(s) (mean[s]) | Mutant Tm range(s) | ∣ΔTm∣ mean(s) or range(s) |
|---|---|---|---|---|---|---|
| Val170Phe | 3 | GTC→TTC | ND | ND | ND | ND |
| Glu250Gly# | 2 | GAG→GGG | ND | ND | ND | ND |
| Arg299Cys# | 1 | CGC→TGC | ND | ND | ND | ND |
| *Leu430Pro | 8 | CTG→CCG | rpoB1 | 69.1–69.5 (69.3) | 63.0–63.4 | 5.9–6.3 |
| Leu430Pro + *Met434Ile | 1 | CTG→CCG; ATG→ATA | rpoB1; rpoB2 | 69.1–69.5 (69.3); 72.8–73.2 (73) | 63.2; 69.8 | 6.1; 3.2 |
| Leu430Pro + Met434Val | 1 | CTG→CCG; ATG→GTG | rpoB1 | 69.1–69.5 (69.3) | 63.0 | 6.3 |
| Leu430Pro + His445Gln | 1 | CTG→CCG; CAC→CAG | rpoB1; rpoB3 | 69.1–69.5 (69.3); 75.5–76.0 (75.75) | 63.5; 72.2 | 5.8; 3.6 |
| Leu430Pro + His445Gln | 1 | CTG→CCG; CAC→CAA | rpoB1; rpoB3 | 69.1–69.5 (69.3); 75.5–76.0 (75.75) | 63.1; 71.7 | 6.2; 4.1 |
| Asp435Gly + Met434Thr | 1 | GAC→GGC; ATG→ACG | rpoB2 | 72.8–73.2 (73) | 69.7 | 3.3 |
| *Asp435Phe | 1 | GAC→TTC | rpoB2 | 72.8–73.2 (73) | 67.7 | 5.3 |
| *Asp435Tyr | 11 | GAC→TAC | rpoB2 | 72.8–73.2 (73) | 68.6–69.0 | 4.0–4.4 |
| Asp435Tyr + Asn437Asp | 1 | GAC→TAC; AAC→GAC | rpoB2 | 72.8–73.2 (73) | 66.6 | 6.4 |
| Asp435Tyr + Met434Ile | 1 | GAC→TAC; ATG→ATT | rpoB2 | 72.8–73.2 (73) | 68.5 | 4.5 |
| *Asp435Val | 5 | GAC→GTC | rpoB2 | 72.8–73.2 (73) | 69.3–69.5 | 3.5–3.7 |
| Asp435Val + Gln432Glu | 1 | GAC→GTC; CAA→GAA | rpoB2; rpoB1 | 72.8–73.2 (73); 69.1–69.5 (69.3) | 70.5; 65.9 | 2.5; 3.4 |
| *Ser441Gln | 1 | TCG→CAG | rpoB2; rpoB3 | 72.8–73.2 (73); 75.5–76.0 (75.75) | 68.3; 73.5 | 4.7; 2.3 |
| *Ser441Leu | 1 | TCG→TTG | rpoB2; rpoB3 | 72.8–73.2 (73); 75.5–76.0 (75.75) | 70.0; 73.5 | 3.0; 2.3 |
| His445Gly | 1 | CAC→GGC | rpoB3 | 75.5–76.0 (75.75) | 70.9 | 4.9 |
| His445Thr | 1 | CAC→ACC | rpoB3 | 75.5–76.0 (75.75) | 70.9 | 4.9 |
| His445Ser | 1 | CAC→AGC | rpoB3 | 75.5–76.0 (75.75) | 71.1 | 4.7 |
| His445Ser + *Lys446Gln + Thr444Thr | 1 | CAC→TCC; AAG→CAG; ACC→ACG | rpoB4B | 67.0–67.6 (67.3) | 62.3 | 5.0 |
| His445Asp | 3 | CAC→GAC | rpoB3 | 75.5–76.0 (75.75) | 71.9–72.1 | 3.7–3.9 |
| His445Leu | 2 | CAC→CTC | rpoB3 | 75.5–76.0 (75.75) | 72.2–72.3 | 3.5–3.6 |
| His445Asn | 2 | CAC→AAC | rpoB3 | 75.5–76.0 (75.75) | 72.3–72.4 | 3.4–3.5 |
| His445Asn + *Asp435Glu | 1 | CAC→AAC; GAC→GAA | rpoB3; rpoB2 | 75.5–76.0 (75.75); 72.8–73.2 (73) | 72.4; 70.2 | 3.4; 2.8 |
| His445Tyr | 4 | CAC→TAC | rpoB3 | 75.5–76.0 (75.75) | 72.5–72.6 | 3.2–3.3 |
| *His445Arg | 4 | CAC→CGC | rpoB3 | 75.5–76.0 (75.75) | 73.9 | 1.9 |
| His445Arg + Ser428Arg | 1 | CAC→CGC; AGC→AGG | rpoB1 | 69.1–69.5 (69.3) | 65.8 | 3.5 |
| Ser450Phe | 1 | TCG→TTC | rpoB3 | 75.5–76.0 (75.75) | 71.8 | 4.0 |
| *Ser450Leu | 14 | TCG→TTG | rpoB3; rpoB4A | 75.5–76.0 (75.75); 67.0–67.6 (67.3) | 72.9–73.3; 73.3–73.8 | 2.5–2.9; 6.0–6.5 |
| Ser450Leu + Thr482Asn | 2 | TCG→TTG; ACC→AAC | rpoB2; rpoB3; rpoB4A | 72.8–73.2 (73); 75.5–76.0 (75.75); 67.0–67.6 (67.3) | 69.2–69.5; 73.1–73.3; 73.6–73.7 | 3.5–3.8; 2.5–2.7; 6.3–6.4 |
| Ser450Leu + Ile491Val | 2 | TCG→TTG; ATC→GTC | rpoB2; rpoB3; rpoB4A | 72.8–73.2 (73); 75.5–76.0 (75.75); 67.0–67.6 (67.3) | 70.0; 73.2–73.3; 73.6–73.7 | 3.0; 2.5–2.6; 6.3–6.4 |
| *Ser450Trp | 3 | TCG→TGG | rpoB3; rpoB4A | 75.5–76.0 (75.75); 67.0–67.6 (67.3) | 73.1–73.5; 70.6–71.0 | 2.3–2.7; 3.3–3.7 |
| Ser450Trp + *Ser431Gly | 1 | TCG→TGG; AGC→GGC | rpoB3; rpoB4A; rpoB1 | 75.5–76.0 (75.75); 67.0–67.6 (67.3); 69.1–69.5 (69.3) | 73.2; 70.7; 66.4 | 2.6; 3.4; 2.9 |
| *Leu452Pro | 12 | CTG→CCG | rpoB4B | 67.0–67.6 (67.3) | 61.2–61.6 | 5.7–6.1 |
| Ile491Phe | 10 | ATC→TTC | ND | ND | ND | ND |
Capturing probes, wild-type melt peak temperature (Tm) ranges and means, mutant Tm ranges, and absolute values of melting temperature shift (ΔTm) ranges associated with specific rpoB mutations in the strains tested and the corresponding nucleotide changes. ND, strains that harbored corresponding mutations outside the RRDR yielded a “RIF Resistance Not Detected” result. *, rifampin resistance-determining region (RRDR) mutation unambiguously identified by unique combinations of Ultra probes and ΔTms, including disputed ones (in italics). #, rifampin susceptible according to phenotypic testing.
Our findings confirm the ability of Ultra to unambiguously identify a wide range of RRDR mutations. With the unprecedented rollout of Xpert MTB/RIF and associated connectivity solutions, such as DataToCare (Savics, Belgium) and GXAlert (SystemOne, USA) (2), Ultra results may allow us to rule out transmission between RR-TB patients in a specific setting (Fig. S1), distinguish relapse from reinfection (5) (Fig. S2), and resolve discordance between an RR Ultra result and a low-level RS phenotypic result due to a disputed mutation. For such applications, it is key that ΔTm values are included in the exported results.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by Erasmus Mundus Joint Doctorate Fellowship grant 2016-1346 to K.C.S.N.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00686-18.
REFERENCES
- 1.Andre E, Isaacs C, Affolabi D, Alagna R, Brockmann D, de Jong BC, Cambau E, Churchyard G, Cohen T, Delmee M, Delvenne JC, Farhat M, Habib A, Holme P, Keshavjee S, Khan A, Lightfoot P, Moore D, Moreno Y, Mundade Y, Pai M, Patel S, Nyaruhirira AU, Rocha LE, Takle J, Trebucq A, Creswell J, Boehme C. 2016. Connectivity of diagnostic technologies: improving surveillance and accelerating tuberculosis elimination. Int J Tuberc Lung Dis 20:999–1003. doi: 10.5588/ijtld.16.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng KC, Meehan CJ, Torrea G, Goeminne L, Diels M, Rigouts L, de Jong BC, Andre E. 2018. Potential application of digitally linked tuberculosis diagnostics for real-time surveillance of drug-resistant tuberculosis transmission: validation and analysis of test results. JMIR Med Inform 6:e12. doi: 10.2196/medinform.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravorty S, Simmons AM, Rowneki M, Parmar H, Cao Y, Ryan J, Banada PP, Deshpande S, Shenai S, Gall A, Glass J, Krieswirth B, Schumacher SG, Nabeta P, Tukvadze N, Rodrigues C, Skrahina A, Tagliani E, Cirillo DM, Davidow A, Denkinger CM, Persing D, Kwiatkowski R, Jones M, Alland D. 2017. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. mBio 8:e00812-17. doi: 10.1128/mBio.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Deun A, Aung KJ, Bola V, Lebeke R, Hossain MA, de Rijk WB, Rigouts L, Gumusboga A, Torrea G, de Jong BC. 2013. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol 51:2633–2640. doi: 10.1128/JCM.00553-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosser A, Marx FM, Pareek M. 2018. Recurrent tuberculosis in the pre-elimination era. Int J Tuberc Lung Dis 22:139–150. doi: 10.5588/ijtld.17.0590. [DOI] [PubMed] [Google Scholar]
- 6.Coll F, Phelan J, Hill-Cawthorne GA, Nair MB, Mallard K, Ali S, Abdallah AM, Alghamdi S, Alsomali M, Ahmed AO, Portelli S, Oppong Y, Alves A, Bessa TB, Campino S, Caws M, Chatterjee A, Crampin AC, Dheda K, Furnham N, Glynn JR, Grandjean L, Minh Ha D, Hasan R, Hasan Z, Hibberd ML, Joloba M, Jones-Lopez EC, Matsumoto T, Miranda A, Moore DJ, Mocillo N, Panaiotov S, Parkhill J, Penha C, Perdigao J, Portugal I, Rchiad Z, Robledo J, Sheen P, Shesha NT, Sirgel FA, Sola C, Oliveira Sousa E, Streicher EM, Helden PV, Viveiros M, Warren RM, McNerney R, Pain A, Clark TG. 2018. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat Genet 50:307–316. doi: 10.1038/s41588-017-0029-0. [DOI] [PubMed] [Google Scholar]
- 7.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. 2009. Tuberculosis drug resistance mutation database. PLoS Med 6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.